Abstract
Tumor necrosis factor alpha (TNF-α) is an important mediator of inflammation, apoptosis, and the development of secondary lymphoid structures. Multiple polymorphic microsatellites have been identified in and around the gene, and there are also multiple single-base pair biallelic polymorphisms in the introns and promoter. The TNF-α −308 promoter polymorphism is a G-to-A transition which has been statistically associated with various autoimmune disorders. Some studies have found that it may directly mediate the increased transcription of TNF-α in some circumstances. This study characterizes proteins interacting at the polymorphic promoter site. Affinity purification of binding proteins and confirmatory chromatin immunoprecipitation assays were used to identify the proteins. Electrophoretic mobility shift analyses and surface plasmon resonance were used to define binding characteristics. Proteins interacting at this site include GCF2/LRRFIP1 and Ets-1. GCF2/LRRFIP1 appears to act as a repressor and occupies the −308 site in cells that do not make TNF-α. Cells competent to produce TNF-α have Ets-1 bound to the −308 promoter site. Active transcription is accompanied by NF-κB and c-Jun binding to the proximal promoter. Thus, dynamic changes on the TNF-α promoter, particularly at the −308 site, accompany the transition from repressed to active transcription. GCF2/LRRFIP1 is the first TNF-α repressor identified.
Tumor necrosis factor (TNF-α) is a proinflammatory cytokine which plays a role in humoral immunity, chemokine expression, the regulation of adhesion molecule expression, and apoptosis. The expression of TNF-α is regulated at many levels, including transcription, message turnover, protein production, and protein release (15, 21, 25, 36). The transcription of TNF-α is complex, with tissue-specific expression and stimulus-inducible expression, which in turn can be tissue specific. Some of the TNF-α promoter motifs which have been implicated in transcription include binding sites for NF-κB, NFAT, ATF-2, Ets-1, C/EBPβ, cis-acting replication element, AP-1, and AP-2 (13, 14, 24, 26-28, 35, 37, 39, 40, 48, 49, 58, 59). Several promoter polymorphisms have been identified, and a polymorphism at −308 has been implicated in the regulation of TNF-α transcription (24, 29, 52, 56). (This work uses the traditional numbering system for the promoter polymorphisms [55].)
The TNF-α promoter polymorphism at −308 involves a biallelic single-base pair transition from G to A. The polymorphic sequence, designated as TNF-α −308A, has gene frequencies of approximately 0.12 to 0.17 in Caucasians and 0.08 in African Americans (9, 10, 42, 45, 4). Studies have found associations of TNF-α −308A with systemic lupus erythematosus, sarcoidosis, alopecia areata, rheumatoid arthritis, and dermatitis herpetiformis (9, 17, 18, 34, 42, 43, 45, 54). Other studies have found no association of TNF-α −308A with rheumatoid arthritis, ankylosing spondylitis, and Felty's syndrome (5, 51, 53). Certain infectious diseases, such as cerebral malaria and mucocutaneous leishmania, also appear to occur with increased frequency in patients with TNF-α −308A (8, 33). These studies suggest that inheritance of the TNF-α −308A allele is biologically relevant, and other studies have shown that inheritance of the TNF-α −308A allele is associated with increased production of TNF-α by peripheral blood mononuclear cells, suggesting a possible mechanism for the association with autoimmune disease (2, 3, 41). Another study, using haplotype-specific chromatin immunoprecipitation (CHIP) analysis, demonstrated that the −308A polymorphism is associated with increased lymphotoxin α transcription in B cell lines (23).
Nine studies have attempted to directly demonstrate transcriptional effects of TNF-α −308A in vitro using transient transfections. Three studies failed to demonstrate any difference in transcription driven by the wild type compared to that driven by the polymorphic TNF-α promoter in transient transfection analyses (6, 44, 50). Two studies utilized T cell and B cell lines, and the other used a murine monocyte line. An alternative strategy using allele-specific PCR of cDNA showed that the −308G and −308A alleles from heterozygous individuals produced equivalent levels of transcripts (6, 7). Six other studies, using similar strategies, demonstrated that transcription driven by the polymorphic promoter was 2- to 10-fold more efficient than that driven by the wild-type promoter (1, 4, 29, 30, 56, 57). These studies utilized T cell and B cell lines as well as baby hamster kidney cells and a human myelomonocytic cell line, U937. Our own data demonstrated that the TNF-α −308A promoter was capable of driving transcription at a level markedly higher than that of the wild-type TNF-α −308G promoter in 3T3 cells stimulated with interleukin 1 alpha and UVB light (52). Possible explanations for the differing findings include the use of different constructs, different cells, different kinetics, or different stimuli.
In spite of the discrepancies, these provocative studies suggested that TNF-α −308A might directly confer an increased transcription of TNF-α in macrophages. We wished to examine potential mechanisms underlying this phenomenon. This promoter region was found to bind GCF2/LRRFIP1 and Ets-1, with different cells favoring one protein or the other depending on the cells' transcriptional competence. GCF2/LRRFIP1 is a poorly understood protein with little homology to recognized transcription factor families. It has been previously identified as a protein interacting with the human ortholog of Flightless-I of Drosophila melanogaster (16). Our studies suggest that it can also function in the repression of TNF-α.
MATERIALS AND METHODS
TNF-α production.
Three different experiments using six normal control donors who are heterozygous for the polymorphic variant and four experiments with six donors homozygous for the wild-type allele were performed. A total of 5 × 105 mononuclear cells were used per well, and triplicate wells were either mock treated or stimulated. Macrophages were purified by adherence, differentiated in culture by adherence, and stimulated with lipopolysaccharide (LPS; 1 μg/ml) or granulocyte-macrophage colony-stimulating factor (GM-CSF; 5 ng/ml). This strategy typically leads to 90%-pure macrophages, as defined by flow cytometry. The nonadherent cells, which are predominantly T cells, were removed and stimulated with phorbol myristate acetate (PMA; 5 ng/ml) and phytohemagglutinin (15 μg/ml). The T cell purity was typically 90%, as defined by flow cytometry. Cell supernatants were harvested at 24 h and analyzed for TNF-α production by specific enzyme-linked immunosorbent assay (Endogen, Woburn, MA). The mean number of macrophages per culture was 2.5 × 104. The mean number of lymphocytes per culture was 3 × 105. The interindividual variation in cell number was <15%. P values were calculated using the Mann-Whitney method.
Electrophoretic mobility shift analyses.
Binding reactions were analyzed by electrophoresis on an 8% acrylamide gel at 4°C in 0.25× Tris-borate-EDTA. Each binding reaction consisted of 20 ng of radiolabeled double-stranded oligonucleotide, 10% glycerol, 1 μg poly(dI:dC), and 10 to 100 ng of nuclear extract in a final volume of 10 μl. Nuclear extracts were prepared by a modification of the method of Dignam et al., namely, substituting KCl for NaCl (11). Various cell lines were used as sources of nuclear extracts: HepG2 is a hepatoma line, K562 is an erythroleukemia line, CCL-156 is a B cell line, Jurkat is a T cell line, D54MG is a glioma line, and THP-1 and U937 are monocytic cell lines. Distamycin (2.5 μM) and 15 μM chromomycin (final concentrations) were added to the probes 10 min prior to incubation with the nuclear extracts. The oligonucleotides used for the electrophoretic mobility shift assays (EMSAs) include the following (lowercase letters in sequences indicate variations from TNFAWT): TNFAWT, (−320) 5′-TTGAGGGGCATGGGGACGGGGTTCA-3′ (−296); TNFAPOL, (−320) 5′-TTGAGGGGCATGaGGACGGGGTTCA-3′ (−296); 5′SHIFT, (−327) 5′-ataggttTTGAGGGGCATGGGGAC-3′ (−304); 3′SHIFT, (−313) 5′-GCATGGGGACGGGGTTCAgcctcca-3′ (−289); M1, (−320) 5′-TTGAGGttCATGGGGACGGGGTTCA-3′ (−296) (−314/−313 mutated); M2, (−320) 5′-TTGAGGGGCgcGGGGACGGGGTTCA-3′ (−296) (−311/−310 mutated); M3, (−320) 5′-TTGAGGGGCATttGGACGGGGTTCA-3′ (−296) (−309/−308 mutated); M4, (−320) 5′-TTGAGGGGCATGGttACGGGGTTCA-3′ (−296) (−307/−306 mutated); and M5, (−320) 5′-TTGAGGGGCATGGGGtgGGGGTTCA-3′ (−296) (−305/−304 mutated).
Protein analyses.
Biotinylated oligonucleotides corresponding to the polymorphic promoter motif were bound to streptavidin-coated magnetic beads by using a ratio of 40 μg of double-stranded oligonucleotides per 5 mg of beads (CPG, Lincoln Park, NJ). Optimization experiments in which the salt concentrations of the binding and elution buffer were varied demonstrated that the following solutions were optimal for the recovery of the C3 complex. U937 nuclear extract was incubated with the oligonucleotide-coated beads in binding buffer (12% glycerol, 12 mM HEPES, 4 mM Tris, 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol) and 25 μg of salmon sperm DNA for 15 min at room temperature. The beads were washed three times with additional binding buffer, and the proteins bound to the DNA were eluted in 12% glycerol, 20 mM Tris, 1.6 M KCl, 5 mM MgCl, 1 mM EDTA, and 1 mM dithiothreitol. The eluted material was diluted 10-fold and repurified over the beads. Gel analyses utilized a 4 to 12% Bis-Tris morpholineethanesulfonic acid (MES)-reducing protein gel (Novex, San Diego, CA) with size standards.
The Western blotting against GCF2 was performed using antisera which were a generous gift from Alfred Johnson at a 1:5,000 dilution in BLOTTO and donkey anti-rabbit at 1:5,000 in BLOTTO. As a control, anti-actin at 1:20,000 in BLOTTO and goat anti-mouse at 1:20,000 in BLOTTO were used. Western blots were developed in Super Signal West Pico (Pierce Biotechnology, Inc., Rockford, IL). With the exception of the anti-GCF2 antisera, the antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Biosensor analysis.
A BIA3000 instrument (Biacore, Piscataway, NJ) was used to detect binding to the TNF-α −308G and TNF-α −308A promoter sequences. Biotinylated probes, consisting of 25-bp double-stranded oligonucleotides bracketing the polymorphism and a 6-bp spacer, were captured to the CM5 sensor chip via streptavidin. The proteins, as analytes, were suspended in running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.005% Tween 20). The experiments were performed at 25°C with a flow rate of 30 μl/min. A reference surface with a control biotinylated oligonucleotide was generated as a control. The molarity of the heterodimer complex in U937 nuclear extract was estimated from equilibrium binding to serial dilutions of radiolabeled oligonucleotides. Kinetic constants were obtained by fitting the data to a simple one-to-one binding model using BIA EVALUATION 3.1 software. Three runs were performed with identical results.
CHIP assay.
The protocol from Upstate Biotechnology (Lake Placid, NY) was used with some modifications (32). Ets-1 (sc-111), c-Jun (sc-1694), and NFκBp 65 (sc-372) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). GCF2 antisera were a gift from Alfred Johnson. Anti-bovine serum albumin (BSA) was purchased from Sigma-Aldrich, St. Louis, MO.
We used real-time PCR to quantitate the amount of DNA from specific regions of the TNF-α promoter and used the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter as an internal control. Each DNA sample was subjected to four PCRs for the four sets of primers and probes to analyze the four regions of the TNF-α promoter (32). The regions detected by each amplimer are the following: TNF1 (+99/−42), TNF2 (+32/−119), TNF3 (−100/−250), TNF4 (−195/−345), and GAPDH (−152/−211). Known concentrations of genomic DNA were used as standards and positive controls with each TaqMan run. Output was normalized to the GAPDH signal in each well according to the manufacturer's instructions.
Retroviral system.
J774.1 cells (a murine monocytic cell line) were transduced with the pREV TET-ON virus, and stable transfectants were selected with G418. GCF2 and DN-ETS1 cDNAs were subcloned into pREVTRE (BD Biosciences, San Jose, CA), virus was prepared according to manufacturer's instructions, and transfections into J774.1 TETON cells were performed. Stable transfectants were grown in G418 and hygromycin. The integration of both constructs was confirmed by PCR. Time course studies and dose titration analyses established 16 h with 1 μg/ml of doxycycline as optimal for the induction of expression. At 16 h, 90% of the cells were viable.
Real-time PCR was performed on the transfected cells. cDNA was made from RNA using random primers, and AMV-RT and cDNA were quantitated using real-time PCR. The murine TNF-α and 18S ribosomal primers and probes were obtained from Applied Biosystems (Foster City, CA). The TNF-α message was normalized to the rRNA internal control.
RESULTS
TNF-α production by macrophages and lymphocytes from donors with and without the polymorphism.
Previous investigators have demonstrated that total peripheral blood mononuclear cell cultures stimulated with anti-CD3 and anti-CD28 from donors who carry the TNF-α −308A allele produce increased levels of TNF-α compared to cultures from wild-type individuals (3). Transient transfection studies have suggested that differences in expressions from the −308A and −308G promoters might be cell type specific or stimulus specific. We examined two cell types and three stimuli. LPS and GM-CSF stimulation of macrophages and phytohemagglutinin-plus-PMA stimulation of lymphocytes revealed significantly increased TNF-α protein production by cells from people heterozygous for the TNF-α −308A allele (Table 1). The homozygous TNF-α −308G donors had the following DRB1 alleles: 01, 04; 01, 04; 14, 13; 04, 09; 11, 16; and 11, 1001. The heterozygous TNF-α −308A donors had the following DRB1 alleles: 12, 07; 11, 08; 01, 08; 03, 11; 03, 16; and 03, 14. There was no difference between the levels of TNF-α production for the TNF-α −308A donors who also carried DRB1 *03 and for those who did not, suggesting that the effect is TNF-α specific rather than dependent on a gene in linkage disequilibrium.
TABLE 1.
TNF-α production by macrophages and lymphocytesa
| Macrophage or lymphocyte | Control (pg/ml) | LPS (pg/ml) | PHA and PMA (pg/ml) | GM-CSF (pg/ml) |
|---|---|---|---|---|
| Macrophages | ||||
| −308GG | 5.0 (2.2) | 629 (254) | 7.2 (2.8) | |
| −308GA | 6.1 (3.2) | 1,106 (363) | 10.2 (4.9) | |
| (P = 0.10) | (P < 0.0001) | (P = 0.001) | ||
| Lymphocytes | ||||
| −308GG | 7.4 (6.1) | 583 (334) | ||
| −308GA | 9.6 (7.8) | 958 (481) | ||
| (P = 0.15) | (P = 0.001) |
Values in parentheses are standard deviations.
The TNF-α −308 site binds several protein complexes.
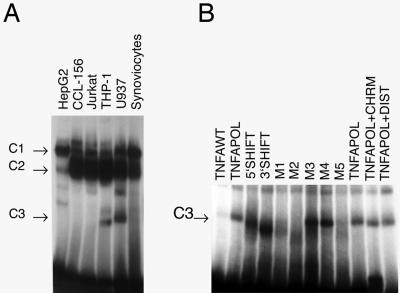
To examine the potential of this promoter polymorphism to mediate transcriptional effects, double-stranded oligonucleotides corresponding to either the wild-type promoter sequence (TNF-α −308G) or the polymorphic promoter sequence (TNF-α −308A) were used to identify potential DNA-binding proteins. Up to five complexes were identified, and additional minor bands were frequently observed. Two complexes were seen in nearly all nuclear extracts examined (C1 and C2), but the fastest-migrating complex (C3) was identified primarily in macrophage lineage cell extracts (U937 and THP-1) (Fig. 1A). This activity was absent from most other lineages tested.
FIG. 1.
(A) Tissue-specific expression of the DNA-binding complex. The wild-type sequence was used as a probe in this EMSA. Nuclear extracts from a variety of cell lines and primary tissues were examined. The C1 and C2 complexes were formed with DNA-binding proteins found in a variety of cell lines examined. The C3 binding activity appears to be largely restricted to macrophage lineage cells (THP-1 and U937). This is representative of three EMSAs. (B) Mutation analysis of the binding motif. This figure demonstrates the effect of sequence alterations on the formation of the C3 complex. The change in mobility induced by the 3′-shift and 5′-shift oligonucleotides is of uncertain cause but could be due to the bending of DNA. M1, M2, and M5 mutations abrogate binding, suggesting that these bases are essential for recognition by the DNA-binding proteins. Chromomycin (CHRM) and distamycin (DIST) do not affect complex formation. The C1 and C2 complexes are not markedly affected by the mutations. WT, wild-type; POL, polymorphic.
Characterization of the DNA-binding complex.
To assess potential contact points of this DNA-binding complex with the target sequence, a series of mutations within the target oligonucleotide was generated. In addition, we determined whether either of the minor groove inhibitors, distamycin or chromomycin, inhibited binding. Figure 1B demonstrates that mutation of the bases at −310 to −313 eliminated binding activity, as did mutation at −304 to −305. This suggests that the relevant core sequence is delimited by −304 and −313 and that binding occurs in the major groove.
The DNA-binding complex has a higher affinity for the polymorphic sequence than for the wild-type sequence.
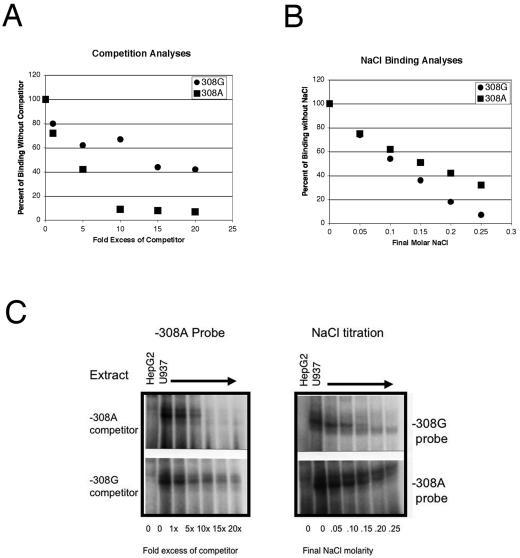
Extracts from a human macrophage cell line (U937) exhibited increased C3 binding in the reactions with the TNF-α −308A oligonucleotide compared to those with the wild-type oligonucleotide (TNF-α −308G). To define the relative affinities of the two oligonucleotides for the DNA-binding protein present in macrophages, cross competition experiments were performed using EMSAs. The TNF-α −308A polymorphism has higher affinity for the DNA-binding protein than the wild-type oligonucleotide does, and this polymorphism is capable of competing 50% of the DNA-binding protein activity at lower concentrations. Several independent experiments were performed with all four possible combinations of competitor and probe. In all cases, the polymorphic TNF-α −308A sequence demonstrated higher affinity for the DNA-binding protein than the TNF-α −308G sequence did (Fig. 2A and C). We additionally performed salt titration of the DNA-protein interaction to compare relative affinities. The DNA-binding protein bound the polymorphic TNF-α −308A sequence with higher affinity than it did the wild-type TNF-α −308G sequence (Fig. 2B and C). To more specifically characterize the interactions of proteins at the polymorphic promoter site, a surface plasmon resonance biosensor (BIA3000) was used to directly measure binding. U937 nuclear extract bound to both the TNF-α −308A sequence and the TNF-α −308G sequence. U937 nuclear extract demonstrated a faster on rate when binding to the TNF-α −308A target (Table 2). There was no overall difference in affinities, however. These runs were performed three times with identical results.
FIG. 2.
(A) The affinity of the TNF-α −308A sequence for the DNA-binding complex is higher than the affinity of the TNF-α −308G sequence. This is a representative example of several different independent experiments. The polymorphic −308A sequence was used as a radiolabeled probe in an EMSA in which cold competitors were added. HepG2 nuclear extract does not contain the DNA-binding complex and served as a negative control. The cognate competitor (−308A) was much more able to compete for the protein than the heterologous competitor (−308G), demonstrating that the −308A sequence has a higher affinity for the DNA-binding complex. A phosphorimager was used to quantitate the amount of bound probe in the C3 complex, and the results are expressed as percentages of binding in the absence of the competitor. Although this example uses −308A as both the probe and the competitor, the −308A was more able to compete off protein from the −308G probe as well. (B) Salt titration of the DNA-binding complex off of the wild-type and polymorphic DNA sequences. This is a representative example of several independent experiments. The DNA-protein interaction is progressively inhibited in the presence of increasing concentrations of NaCl. The molarity of the final NaCl concentration is indicated at the bottom of the figure. The protein interaction with the −308G probe was much more labile than the interaction between the protein and the −308A probe. A phosphorimager was used to quantitate the amount of bound probe in the C3 complex. The results are expressed as percentages of binding in the absence of additional NaCl. (C) Representative EMSAs. The −308A (left panel) was end labeled with 32P. Increasing amounts of cold −308G (lower panel) or cold −308A (upper panel) competitor were added as indicated. The bands decayed more rapidly with the −308A competitor. The right panel is representative of the salt titration experiments. The −308G probe has no protein bound at 0.20 M NaCl, while the −308A probe has significant protein bound at even higher NaCl concentrations.
TABLE 2.
Biosensor analysis of U937 nuclear extract binding to either the TNF-α −308G oligonucleotide or the TNFα −308A oligonucleotide
| Parametera | Value |
|---|---|
| On rate G (1/Ms) | 8.0 × 105 |
| On rate A (1/Ms) | 1.6 × 106 |
| Off rate G (1/s) | 5.1 × 10−4 |
| Off rate A (1/s) | 1.7 × 10−3 |
| KA G (1/M) | 3.1 × 106 |
| KA A (1/M) | 3.1 × 106 |
KA, association rate constant.
Purification and functional analysis of the DNA-binding proteins.
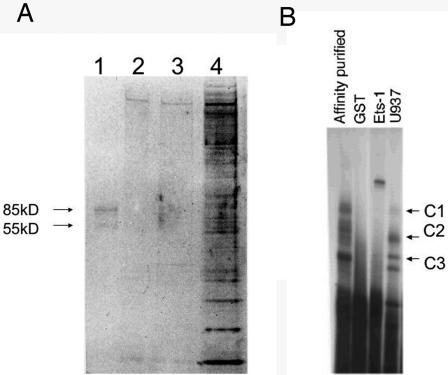
To further characterize this complex, we partially purified the DNA-binding proteins using a magnetic bead purification strategy. Two proteins of approximately 85 kDa and 55 kDa and of >95% purity were identified (Fig. 3A). These same bands were identified in material eluted from the C3 complex from high-density EMSAs (data not shown). Additionally, the purified material, when incubated with radiolabeled −308A probe, reconstituted the C3 complex seen in Fig. 1 (Fig. 3B). Mass spectroscopy revealed that these two proteins were Ets-1 (55 kDa) and GCF2/LRRFIP1 (85 kDa).
FIG. 3.
(A) Partial protein purification of the DNA-binding protein. Affinity purification of the DNA-binding protein was performed using oligonucleotides bound to magnetic beads. The purified product is shown in lane 1. Lanes 2 and 3 are high-salt washing steps between the two column purifications, and lane 4 is the protein material after a single column purification. Two proteins of 85 kDa and 55 kDa are seen. (B) Affinity-purified material reconstitutes the C3 binding complex. The affinity-purified material was diluted in binding buffer and used in an EMSA with radiolabeled TNF-308A probe (lane 1). The C3 complex is seen. Dialyzed GST does not bind, and Ets-1 cleaved from GST binds the probe strongly but does not reconstitute the C1, C2, or C3 complex. U937 is shown for comparison in lane 4. The U937 C1 complex is faint, as was true for Fig. 1.
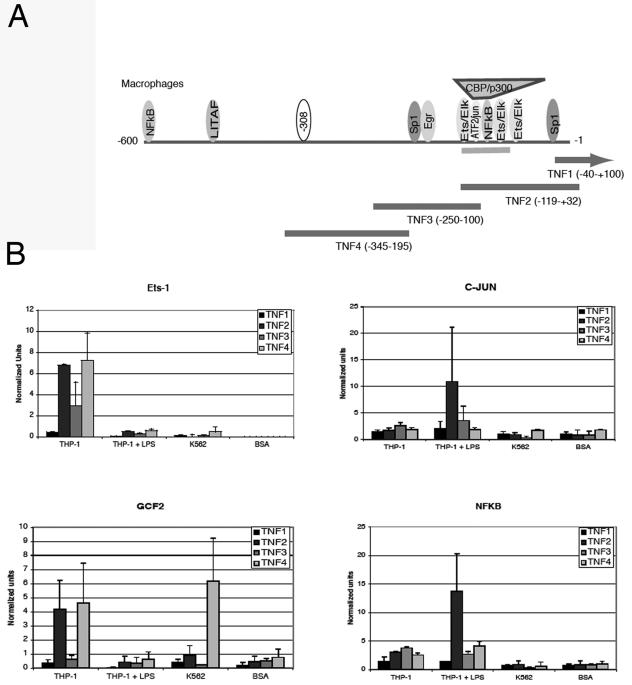
To confirm the proteins identified by affinity purification, we used CHIP assays to identify proteins bound to the promoter in vivo (Fig. 4A). The original EMSAs and protein purification were performed using nuclear extracts from a previously described subline of U937 (32). This subline produces minimal TNF-α when stimulated. For the CHIP assays, we compared two cell lines, K562 and THP-1. K562 cannot be induced to produce TNF-α using any common stimuli. THP-1, in contrast, is easily induced to express TNF-α, although only 30 to 80% of the cells produce TNF-α (data not shown). CHIP assays comparing GCF2/LRRFIP1, Ets-1, NF-κB, and c-Jun binding to the TNF-α promoter are shown (Fig. 4B). GCF2/LRRFIP1 is found at the highest levels on the promoter of K562. This suggests that it may be acting as a repressor. Ets-1, in contrast, is found at the highest levels in the quiescent THP-1 cell line. To confirm that stimulation is inducing the expected transcription factor binding, CHIP assays, which demonstrated binding to the proximal promoter, were performed for NF-κB and c-Jun.
FIG. 4.
(A) Chromatin immunoprecipitation primers and probes. The TNF4 amplimer encompasses the −308 site. The TNF2 amplimer encompasses the region known to bind Ets-1, c-Jun, and NF-κB. (B) Chromatin immunoprecipitation assays. These assays examine the in vivo binding of transcription factors by chromatin immunoprecipitation. Ets-1 (sc-111), GCF2/LRRFIP1, c-Jun (sc-1694), and NFκBp 65 (sc-372) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). THP-1 cells were either unstimulated or stimulated with 1 μg/ml of LPS for 30 min. K562 cells do not produce TNF-α. Antibodies to BSA were used as a negative control. Three separate experiments were averaged. Means and standard deviations are shown.
Ets-1 was found exclusively on resting THP-1 promoters. This was somewhat surprising. A binding site at the proximal promoter at −117, corresponding to the TNF2 amplimer, has been previously described as binding Ets-1 after LPS stimulation (47). Very little if any Ets-1 was found on the LPS-stimulated THP-1 promoter in these assays, although resting cells exhibited binding. The TNF4 amplimer brackets the −308 site, and these data therefore confirm that quiescent cells (such as the original resting U937 cells used for the affinity purification) can and do have Ets-1 occupying the distal promoter. GCF2/LRRFIP1 occupies the distal promoter in both a repressed cell type (K562) and the quiescent THP-1. A significant difference is that quiescent THP-1 also seems to demonstrate GCF2/LRRFIP1 binding to the proximal promoter corresponding to the TNF2 amplimer. This region is GC rich and does contain a potential GCF2/LRRFIP1 binding site.
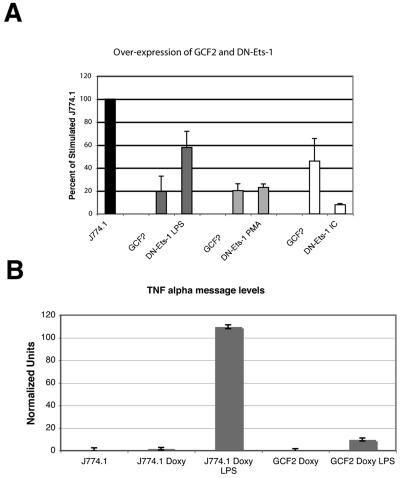
GCF2/LRRFIP1 is a poorly understood repressor. To better define the roles of GCF2/LRRFIP1 and Ets-1, an inducible retroviral system was used to transiently overexpress GCF2/LRRFIP1 and a dominant negative inhibitor of Ets-1 (31). Overexpression was associated with markedly reduced TNF-α protein produced in response to three different stimuli (Fig. 5A). To confirm that the effect of GCF2 overexpression is mediated through TNF-α message, real-time PCR was performed on the same cells. Vector-transfected cells served as the control, and GCF2-transfected cells were induced with doxycycline for 16 h as described above. GCF2 overexpression was associated with markedly diminished TNF-α transcript levels (Fig. 5B).
FIG. 5.
(A) Induction of GCF2/LRRFIP1 expression. An inducible retroviral system was used to introduce the cDNA for GCF2/LRRFIP1 into J774.1 cells under the control of doxycycline. A dominant negative inhibitor of Ets-1 (DN-Ets-1) was also introduced using the same system. Stable nonclonal cell lines were isolated, induced with doxycycline for 16 h, and stimulated with 10 ng/ml of LPS, 100 ng/ml of PMA, or 1 μg/ml of BSA-anti-BSA immune complexes (IC). Cell supernatants were collected 6 hours after stimulation. The vector-transfected cells were used in each case as the control (J774.1) and are set at 100% for each stimulus. The graph represents the averages of triplicate cultures of three separate experiments. P values were derived; ** indicates a P value of ≤0.001, and * indicates a P value of ≤0.01. (B) GCF2/LRRFIP1 overexpression is associated with diminished TNF-α message. The same transfected cells were stimulated with 1 μg/ml of LPS and harvested at 3 hours. TNF-α cDNA was quantitated and normalized to 18S rRNA. Two separate experiments consisting of quadruplicate samples were performed, and one example is shown. A t test demonstrated that the difference in TNF-α message levels between vector-transfected cells (J774.1 Doxy LPS) treated with LPS and GCF2-transfected cells (GCF2 Doxy LPS) has a P value of 0.019. The unstimulated cells produce negligible TNF-α message, and there is no difference between the vector-transfected cells (J774.1) and the vector-transfected cells treated with doxycycline (J774.1 Doxy).
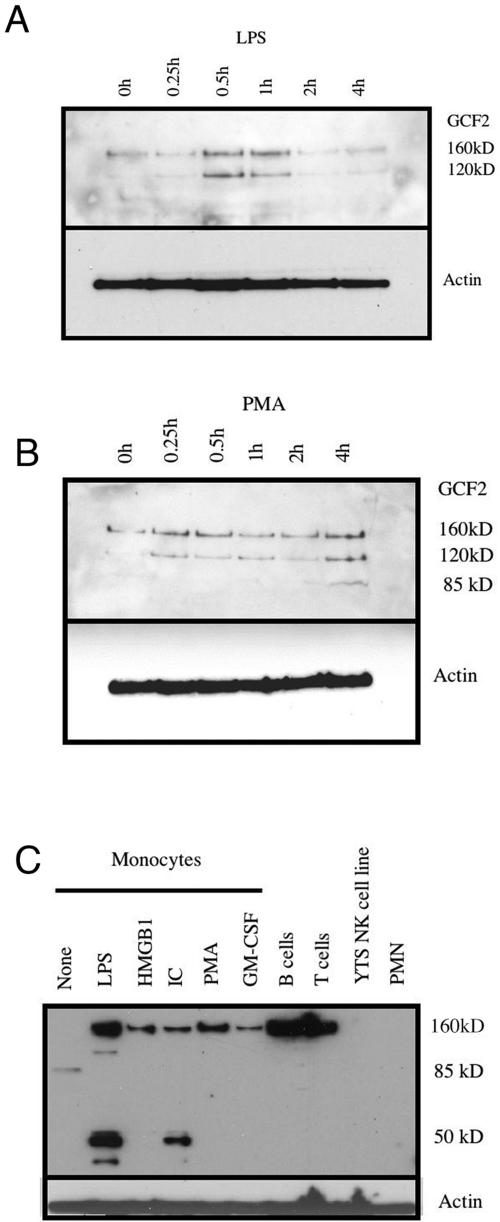
To determine whether GCF2/LRRFIP1 expression was regulated, Western blotting was performed (Fig. 6). GCF2 exists as several splice variants which have uncertain functional consequences. The original mass spectroscopy was performed on the 85-kDa band. Proteins of this size could be produced by splicing out either exon 5 or exon 11. Both of these splice variants have been identified in GenBank. The Western blots on THP-1 (Fig. 6A and B) reveal two bands of 120 kDa and 160 kDa. The 160-kDa form probably represents the full-length protein including all 11 exons. After stimulation with PMA or LPS, the levels rise, with kinetics that differ depending on the stimulus. LPS induces a rapid rise in both the 160-kDa and the 120-kDa bands, which return to baseline by 2 hours poststimulation (Fig. 6A). PMA induces a rapid rise in the 120-kDa band, which is sustained, and a slower rise in the 160-kDa band (Fig. 6B). In THP-1 cells under these conditions, the 85-kDa band is seen faintly after PMA stimulation. Figure 6C demonstrates that in primary monocytes the 85-kDa form predominates in the absence of stimulation. Stimulation induces the appearance of alternate forms. In these primary cells, the 160-kDa form dominates after stimulation. These data suggest that GCF2/LRRFIP1 exists as a number of different isoforms which may be differentially regulated. The distinct roles of these isoforms are not understood at present.
FIG. 6.
Western blot analysis of GCF2/LRRFIP1 expression. LPS at 1 μg/ml (A) or PMA at 100 ng/ml (B) was added to log-phase growth THP-1 cells, and cells were collected at the indicated times. (C) Primary monocytes purified by elutriation were either mock treated (None) or treated with 1 μg/ml of LPS, 400 ng/ml of HMGB1, 1 μg/ml of immune complexes (IC), 100 ng/ml of PMA, or 10 ng/ml of GM-CSF. Column-purified primary B cells and T cells were used freshly collected. YTS is a highly differentiated NK cell line. Neutrophils (PMN) were purified from human blood by dextran sedimentation and lysed immediately.
DISCUSSION
The goal of this study was to define proteins interacting with the −308 promoter site. A repressor and transcriptional activator both bind to that site. GCF2/LRRFIP1 is a protein originally identified as a repressor of the epidermal growth factor receptor gene (38). It also appears to act as a repressor (i) during imprinting and (ii) of platelet-derived growth factor (12, 22). Its binding pattern on the TNF-α promoter is consistent with repressor activity, because it was found bound to the promoter in cells not actively producing TNF-α. It was found bound to the expected region in K562 cells and quiescent THP-1 cells. In the competent but quiescent THP-1, it was also found bound to the proximal promoter region. This region is GC rich and could support GCF2/LRRFIP1 binding. In contrast, Ets-1 was found on the competent but quiescent promoter but not on the repressed promoter or on the active promoter. At this point, it is not certain whether the −308 site could support both GCF2/LRRFIP1 and Ets-1 binding. It seems more likely that the binding of these proteins is mutually exclusive, because the recognition motifs almost completely overlap. The THP-1 cells used for the CHIP analyses do not uniformly express TNF-α when stimulated. Intracellular flow cytometry (not shown) demonstrated that approximately 50% of the cells produced TNF-α at the time of these experiments. Therefore, the finding that both Ets-1 and GCF2 were bound to the distal region of the promoter could reflect the heterogeneity of the cell population. It is also likely that other proteins not yet identified interact at this site. The affinity-purified material, which represents GCF2/LRRFIP1 and Ets-1, reconstitutes only the C3 complex. Ets-1 alone does not produce C1, C2, or C3, suggesting it does not bind that sequence in isolation. Thus, it is likely that other proteins interact at this site.
Transient transfection studies using TNF-α promoter deletion mutants have generally concluded that the distal promoter upstream of −150 is dispensable (19, 20, 39, 40, 46). If this region is primarily involved in repression, transient transfection studies may not reflect that role if the repression requires higher-order chromatin structure. Differences in relative levels of Ets-1 and GCF2 might also affect transient transfection studies in a manner that would not occur in vivo if the promoter were constrained by chromatin structure.
The importance of this study is in the identification of additional regulatory mechanisms controlling TNF-α expression. Three previous studies evaluated the affinity of DNA-binding complexes for the −308 wild-type and polymorphic promoter sequences (29, 54, 57). Two of the studies defined a higher affinity for the polymorphic promoter sequence by using a variety of nuclear extracts in EMSA analyses (29, 57). Our analyses are consistent with altered protein interactions with the polymorphic promoter sequence. The faster on rate of the proteins binding to the TNF-α −308A sequence could drive an increased transcription of TNF-α. Although, based on the biosensor analysis, the overall affinities were not different, it is likely that the transcription factors become integrated into a higher-order complex after binding to the target sequence. Therefore, there may not be a simple off rate in vivo. The identification of proteins interacting with this site and the discovery of a repressor represent a significant advance in our understanding of the regulation of TNF-α. These findings could potentially explain the biological phenomena and transcriptional difference seen with the −308A and −308G promoters.
GCF2/LRRFIP1 is a poorly understood transcriptional repressor. TNF-α utilizes a variety of strategies to rapidly downregulate its production after stimulation. Chief among these are strategies regulating message turnover. The induction of GCF2/LRRFIP1 by LPS and PMA, two common inducers of TNF-α, raises the question of whether GCF2/LRRFIP1 acts as a negative-feedback mechanism for TNF-α acting at the level of transcription. Impressively, the overexpression of GCF2/LRRFIP1 demonstrated very potent downregulation of TNF-α protein and message. The real-time PCR data could not directly demonstrate whether GCF2/LRRFIP1 affected the transcription rate or message turnover. Given its binding to the promoter, it is likely that it exerts at least some of its effect as a transcriptional repressor. GCF2/LRRFIP1 also binds RNA and could have multiple effects on transcript levels. Inhibition of TNF-α has markedly improved the quality of life for patients with diverse inflammatory disorders, and the manipulation of GCF2/LRRFIP1 could represent a novel therapeutic target.
Acknowledgments
Mike Ostrowski provided the DN-Ets-1 construct (31), and Alfred Johnson provided the antisera and cDNA for GCF2. We also thank Alfred Johnson for helpful discussions of GCF2. Tushar Dharia confirmed the Ets-1 binding using the yeast one hybrid system. Cheu Manka performed transfections of the J774.1 cells, and Wentao Zhang provided assistance with the Biosensor instrument. We are grateful for their assistance.
This work was supported by R29 AR/AI43172, RO1 AI44127, RO1 AI051323, and the Wallace Chair of Pediatrics.
REFERENCES
- 1.Abraham, L. J., and K. M. Kroeger. 1999. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J. Leukoc. Biol. 66:562-566. [DOI] [PubMed] [Google Scholar]
- 2.Baseggio, L., J. Bienvenu, C. Charlot, J. Picollet, P. Felman, B. Coiffier, and G. Salles. 2001. Higher LPS-stimulated TNF-alpha mRNA levels in peripheral blood mononuclear cells from non-Hodgkin's lymphoma patients. Exp. Hematol. 29:330-338. [DOI] [PubMed] [Google Scholar]
- 3.Bouma, G., J. B. A. Crusius, M. Oudkerk Pool, J. J. Kolkman, B. M. E. Von Blomberg, P. J. Kostense, M. J. Giphart, G. M. T. Schreuder, S. G. M. Meuwissen, and A. S. Pena. 1996. Secretion of tumour necrosis factor α and lymphotoxin α in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand. J. Immunol. 43:456-463. [DOI] [PubMed] [Google Scholar]
- 4.Braun, N., U. Michel, B. P. Ernst, R. Metzner, A. Bitsch, F. Weber, and P. Rieckmann. 1996. Gene polymorphism at position -308 of the tumor-necrosis-factor-α (TNF-α) in multiple sclerosis and its influence on the regulation of TNF-a production. Neurosci. Lett. 215:75-78. [PubMed] [Google Scholar]
- 5.Brinkman, B. M., M. J. Giphart, A. Verhoef, E. L. Kaijzel, A. M. Naipal, M. R. Daha, F. C. Breedveld, and C. L. Verweij. 1994. Tumor necrosis factor alpha-308 gene variants in relation to major histocompatibility complex alleles and Felty's syndrome. Hum. Immunol. 41:259-266. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman, B. M., T. W. Huizinga, F. C. Breedveld, and C. L. Verweij. 1996. Allele-specific quantification of TNFA transcripts in rheumatoid arthritis. Hum. Genet. 97:813-818. [DOI] [PubMed] [Google Scholar]
- 7.Brinkman, B. M. N., D. Zuijdgeest, E. L. Kaijzel, F. C. Breedveld, and C. L. Verweij. 1996. Relevance of the tumor necrosis factor alpha (TNFα) -308 promoter polymorphism in TNFa gene regulation. J. Inflamm. 46:32-41. [PubMed] [Google Scholar]
- 8.Cabrera, M., M. A. Shaw, C. Sharples, H. Williams, M. Castes, J. Convit, and J. M. Blackwell. 1995. Polymorphism in tumor necrosis factor genes associated with mucocutaneous leishmaniasis. J. Exp. Med. 182:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danis, V. A., M. Millington, V. Hyland, R. Lawford, Q. Huang, and D. Grennan. 1995. Increased frequency of the uncommon allele of a tumour necrosis factor alpha gene polymorphism in rheumatoid arthritis and systemic lupus erythematosus. Dis. Markers 12:127-133. [DOI] [PubMed] [Google Scholar]
- 10.Danis, V. A., M. Millington, V. J. Hyland, and D. Grennan. 1995. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin. Exp. Immunol. 99:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam, J. D., R. M. Lebovitz, and R. C. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eden, S., M. Constancia, T. Hashimshony, W. Dean, B. Goldstein, A. C. Johnson, I. Keshet, W. Reik, and H. Cedar. 2001. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 20:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falvo, J. V., B. M. Brinkman, A. V. Tsytsykova, E. Y. Tsai, T. P. Yao, A. L. Kung, and A. E. Goldfeld. 2000. A stimulus-specific role for CREB-binding protein (CBP) in T cell receptor-activated tumor necrosis factor alpha gene expression. Proc. Natl. Acad. Sci. USA 97:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falvo, J. V., A. M. Uglialoro, B. M. Brinkman, M. Merika, B. S. Parekh, E. Y. Tsai, H. C. King, A. D. Morielli, E. G. Peralta, T. Maniatis, D. Thanos, and A. E. Goldfeld. 2000. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 20:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flier, J. S., and L. H. Underhill. 1996. The tumor necrosis factor ligand and receptor families. N. Engl. J. Med. 334:1717-1725. [DOI] [PubMed] [Google Scholar]
- 16.Fong, K. S., and H. G. de Couet. 1999. Novel proteins interacting with the leucine-rich repeat domain of human Flightless-I identified by the yeast two-hybrid system. Genomics 58:146-157. [DOI] [PubMed] [Google Scholar]
- 17.Fong, K. Y., H. S. Howe, S. K. Tin, M. L. Boey, and P. H. Feng. 1996. Polymorphism of the regulatory region of tumor necrosis factor alpha gene in patients with systemic lupus erythematosus. Ann. Acad. Med. Singapore 25:90-93. [PubMed] [Google Scholar]
- 18.Galbraith, G. M., and J. P. Pandey. 1995. Tumor necrosis factor alpha (TNF-alpha) gene polymorphism in alopecia areata. Hum. Genet. 96:433-436. [DOI] [PubMed] [Google Scholar]
- 19.Goldfeld, A. E., C. Doyle, and T. Maniatis. 1990. Human tumor necrosis factor α gene regulation by virus and lipopolysaccharide. Proc. Natl. Acad. Sci. USA 87:9769-9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldfeld, A. E., J. L. Strominger, and C. Doyle. 1991. Human tumor necrosis factor α gene regulation in phorbol ester stimulated T and B cell lines. J. Exp. Med. 174:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huizenga, T. W. J., B. M. N. Brinkman, and C. L. Verweij. 1996. Regulation of tumor necrosis factor alpha production: basic aspects and pharmacological modulation. J. Rheumatol. 23:416-419. [PubMed] [Google Scholar]
- 22.Khachigian, L. M., F. S. Santiago, L. A. Rafty, O. L. Chan, G. J. Delbridge, A. Bobik, T. Collins, and A. C. Johnson. 1999. GC factor 2 represses platelet-derived growth factor A-chain gene transcription and is itself induced by arterial injury. Circ. Res. 84:1258-1267. [DOI] [PubMed] [Google Scholar]
- 23.Knight, J. C., B. J. Keating, K. A. Rockett, and D. P. Kwiatkowski. 2003. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 33:469-475. [DOI] [PubMed] [Google Scholar]
- 24.Knight, J. C., I. Udalova, A. V. S. Hill, B. M. Greenwood, N. Peshu, K. Marsh, and D. Kwiatkowski. 1999. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat. Genet. 22:145-150. [DOI] [PubMed] [Google Scholar]
- 25.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 26.Kramer, B., T. Machleidt, K. Wiegmann, and M. Kronke. 1995. Superantigen-induced transcriptional activation of the human TNF gene promoter in T cells. J. Inflamm. 45:183-192. [PubMed] [Google Scholar]
- 27.Kramer, B., K. Wiegmann, and M. Kronke. 1995. Regulation of the human TNF promoter by the transcription factor Ets. J. Biol. Chem. 270:6577-6583. [DOI] [PubMed] [Google Scholar]
- 28.Kroeger, K. M., and L. J. Abraham. 1996. Identification of an AP-2 element in the -323 to -285 region of the TNF-α gene. Biochem. Mol. Biol. Int. 40:43-51. [DOI] [PubMed] [Google Scholar]
- 29.Kroeger, K. M., K. S. Carville, and L. J. Abraham. 1997. The -308 tumor necrosis factor—a promoter polymorphism effects transcription. Mol. Immunol. 34:391-399. [DOI] [PubMed] [Google Scholar]
- 30.Kroeger, K. M., J. H. Steer, D. A. Joyce, and L. J. Abraham. 2000. Effects of stimulus and cell type on the expression of the -308 tumour necrosis factor promoter polymorphism. Cytokine 12:110-119. [DOI] [PubMed] [Google Scholar]
- 31.Langer, S. J., D. M. Bortner, M. F. Roussel, C. J. Sherr, and M. C. Ostrowski. 1992. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol. Cell. Biol. 12:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J. Y., N. A. Kim, A. Sanford, and K. E. Sullivan. 2003. Histone acetylation and chromatin conformation are regulated separately at the TNF alpha promoter in monocytes and macrophages. J. Leukoc. Biol. 73:862-871. [DOI] [PubMed] [Google Scholar]
- 33.McGuire, W., A. V. S. Hill, C. E. M. Allsopp, B. M. Greenswood, and D. Kwiatkowski. 1994. Variation in the TNF-α promoter region associated with susceptibility to cerebral malaria. Nature 371:508-511. [DOI] [PubMed] [Google Scholar]
- 34.Messer, G., G. Kick, A. Ranki, S. Koskimies, T. Reunala, and M. Meurer. 1994. Polymorphism of the tumor necrosis factor genes in patients with dermatitis herpetiformis. Dermatology 189(Suppl. 1):135-137. [DOI] [PubMed] [Google Scholar]
- 35.Paludan, S. R., S. Ellermann-Eriksen, V. Kruys, and S. C. Mogensen. 2001. Expression of TNF-alpha by herpes simplex virus-infected macrophages is regulated by a dual mechanism: transcriptional regulation by NF-kappa B and activating transcription factor 2/Jun and translational regulation through the AU-rich region of the 3′ untranslated region. J. Immunol. 167:2202-2208. [DOI] [PubMed] [Google Scholar]
- 36.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pope, R. M., A. Leutz, and S. A. Ness. 1994. C/EBPb regulation of tumor necrosis factor α gene. J. Clin. Investig. 94:1449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, A. L., H. Yamazaki, J. D. Kaufman, Y. Rubinstein, B. Murphy, and A. C. Johnson. 1998. Molecular cloning and characterization of a transcription regulator with homology to GC-binding factor. J. Biol. Chem. 273:21594-21602. [DOI] [PubMed] [Google Scholar]
- 39.Rhoades, K. L., S. Cai, S. H. Golub, and J. S. Economou. 1995. Granulocyte-macrophage colony-stimulating factor and interleukin-4 differentially regulate the human tumor necrosis factor-alpha promoter region. Cell. Immunol. 161:125-131. [DOI] [PubMed] [Google Scholar]
- 40.Rhoades, K. L., S. H. Golub, and J. S. Economou. 1992. The regulation of the human tumor necrosis factor α promoter region in macrophage, T cell, and B cell lines. J. Biol. Chem. 267:22102-22107. [PubMed] [Google Scholar]
- 41.Rudwaleit, M., S. Siegert, Z. Yin, J. Eick, A. Thiel, A. Radbruch, J. Sieper, and J. Braun. 2001. Low T cell production of TNFalpha and IFNgamma in ankylosing spondylitis: its relation to HLA-B27 and influence of the TNF-308 gene polymorphism. Ann. Rheum. Dis. 60:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudwaleit, M., M. Tikly, M. Khamashta, K. Gibson, J. Klinke, G. Hughes, and P. Wordsworth. 1996. Interethnic differences in the association of tumor necrosis factor promoter polymorphisms with systemic lupus erythematosus. J. Rheumatol. 23:1725-1728. [PubMed] [Google Scholar]
- 43.Seitzer, U., C. Swider, F. Stuber, K. Suchnicki, A. Lange, E. Richter, P. Zabel, J. Muller-Quernheim, H.-D. Flad, and J. Gerdes. 1997. Tumour necrosis factor alpha promoter gene polymorphism in sarcoidosis. Cytokine 9:787-790. [DOI] [PubMed] [Google Scholar]
- 44.Stuber, F., I. A. Udalova, M. Book, L. N. Drutskaya, D. V. Kuprash, R. L. Turetskaya, F. U. Schade, and S. A. Nedospasov. 1996. -308 tumor necrosis factor (TNF) polymorphism is not associated with survival in severe sepsis and is unrelated to lipopolysaccharide inducibility of the human TNF promoter. J. Inflamm. 46:42-50. [PubMed] [Google Scholar]
- 45.Sullivan, K. E., C. Wooten, B. Schmeckpeper, D. Goldman, and M. Petri. 1997. A promoter polymorphism of tumor necrosis alpha is associated with systemic lupus erythematosus in African Americans. Arthritis Rheum. 40:2207-2211. [DOI] [PubMed] [Google Scholar]
- 46.Trede, N. S., A. V. Tsytsykova, T. Chatila, A. E. Goldfeld, and R. S. Geha. 1995. Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B. J. Immunol. 155:902-908. [PubMed] [Google Scholar]
- 47.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfield. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai, E. Y., J. Yie, D. Thanos, and A. E. Goldfeld. 1996. Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/JUN. Mol. Cell. Biol. 16:5232-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uglialoro, A. M., D. Turbay, P. A. Pesaavento, J. C. Delgado, F. E. McKenzie, J. G. Gribben, D. Hartl, E. J. Yunis, and A. E. Goldfeld. 1998. Identification of three new single nucleotide polymorphisms in the human tumor necrosis factor-α gene promoter. Tissue Antigens 52:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verjans, G. M., B. M. Brinkman, C. E. Van Doornik, A. Kijlstra, and C. L. Verweij. 1994. Polymorphism of tumour necrosis factor-alpha (TNF-alpha) at position -308 in relation to ankylosing spondylitis. Clin. Exp. Immunol. 97:45-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werth, V. P., W. Zhang, K. Dortzbach, and K. Sullivan. 2000. Association of a promoter polymorphism of TNFα with subacute cutaneous lupus erythematosus and distinct photoregulation of transcription. J. Investig. Dermatol. 115:726-730. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, A. G., N. de Vries, L. B. van de Putte, and G. W. Duff. 1995. A tumour necrosis factor alpha polymorphism is not associated with rheumatoid arthritis. Ann. Rheum. Dis. 54:601-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, A. G., C. Gordon, F. S. di Giovine, N. de Vries, L. B. A. van de Putte, P. Emery, and G. W. Duff. 1994. A genetic association between systemic lupus erythematosus and tumor necrosis factor alpha. Eur. J. Immunol. 24:191-195. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, A. G., J. A. Symons, T. L. McDowell, F. S. diGiovine, and G. W. Duff. 1994. Effects of a tumor necrosis factor promoter base 167 transition on transcriptional activity. Br. J. Rheumatol. 33:89. [Google Scholar]
- 56.Wilson, A. G., J. A. Symons, T. L. McDowell, H. O. McDevitt, and G. W. Duff. 1997. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 94:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, W.-S., and K. L. McClain. 1997. DNA polymorphisms and mutations of the tumor necrosis factor-α (TNF-α) promoter in Langerhans cell histiocytosis (LCH). J. Interferon Cytokine Res. 17:631-635. [DOI] [PubMed] [Google Scholar]
- 58.Xu, Z., R. Dziarski, Q. Wang, K. Swartz, K. M. Sakamoto, and D. Gupta. 2001. Bacterial peptidoglycan-induced tnf-α transcription is mediated through the transcription factors Egr-1, Elk-1, and NF-κB. J. Immunol. 167:6975-6982. [DOI] [PubMed] [Google Scholar]
- 59.Yao, J., N. Mackman, T. S. Edgington, and S.-T. Fan. 1997. Lipopolysaccharide induction of the tumor necrosis factor—a promoter in human monocytic cells. J. Biol. Chem. 272:17795-17801. [DOI] [PubMed] [Google Scholar]