Abstract
The activity of GATA factors is regulated, in part, at the level of protein-protein interactions. LIM domain proteins, first defined by the zinc finger motifs found in the Lin11, Isl-1, and Mec-3 proteins, act as coactivators of GATA function in both hematopoietic and cardiovascular tissues. We have identified a novel GATA-LIM interaction between GATA6 and LMCD1/dyxin. The LIM domains and cysteine-rich domains in LMCD1/dyxin and the carboxy-terminal zinc finger of GATA6 mediate this interaction. Expression of LMCD1/dyxin is remarkably similar to that of GATA6, with high-level expression observed in distal airway epithelium of the lung, vascular smooth muscle, and myocardium. In contrast to other GATA-LIM protein interactions, LMCD1/dyxin represses GATA6 activation of both lung and cardiac tissue-specific promoters. Electrophoretic mobility shift and chromatin immunoprecipitation assays show that LMCD1/dyxin represses GATA6 function by inhibiting GATA6 DNA binding. These data reveal an interaction between GATA6 and LMCD1/dyxin and demonstrate a novel mechanism through which LIM proteins can assert their role as transcriptional cofactors of GATA proteins.
GATA factors regulate gene expression in a variety of cell types, including cardiovascular, pulmonary, and hematopoietic tissues (for reviews, see references 21 and 43). There are six mammalian GATA factors, each of which contains two zinc fingers that are required for sequence-specific DNA binding. Mammalian GATA factors can be subdivided into two groups: GATA1/2/3, which are expressed primarily in hematopoietic tissues, and GATA4/5/6, which are expressed primarily in cardiovascular, lung, and gut tissues (for reviews, see references 21, 27, and 47). Loss of function of each of these genes results in specific defects in developmental processes, ranging from erythrocyte development (GATA1) to heart formation (GATA4) (13, 22, 29, 41).
As with other transcriptional regulators, GATA factors are regulated by protein-protein interactions with cofactors. Members of the Nkx family of homeodomain transcription factors, including Nkx2.1 and Nkx2.5, interact with members of the GATA4/5/6 subfamily and synergistically regulate lung and cardiac gene expression (9, 30). Members of a novel family of zinc finger transcriptional cofactors, FOG-1/2, interact with GATA family members and regulate distinct aspects of both hematopoietic and cardiovascular development (35, 36).
In addition to these interactions, GATA factors also interact with members of the LIM domain family of zinc finger proteins. Interactions between GATA1 and LMO2 regulate erythroid gene expression and development (28, 39). Interactions between GATA4 and GATA6 and the cysteine-rich protein (CRP) family of LIM domain proteins regulate cardiac and vascular smooth-muscle gene expression (7). In all previously reported GATA-LIM interactions, LIM proteins act as coactivators of GATA-mediated gene transcription. The importance of these interactions is underscored by the finding that LMO2- and GATA1-deficient mice exhibit similar blocks in erythropoiesis (29, 45).
GATA6 is expressed in myocardium, lung epithelium, and vascular smooth muscle and has been shown to regulate promoters specific for all of these tissues (6, 23, 37, 38). To identify additional cofactors that regulate GATA6 function in lung and cardiac development, we performed a yeast two-hybrid screen for GATA6-interacting proteins. These studies revealed an interaction between GATA6 and the LIM protein LMCD1/dyxin. LMCD1/dyxin expression overlaps extensively with that of GATA6 in distal lung epithelium, cardiac myocytes, and vascular smooth muscle. Interaction between GATA6 and LMCD1/dyxin inhibits GATA6 DNA binding, resulting in repression of GATA6 transcriptional activation of downstream target genes. The GATA6-LMCD1 interaction defines a novel mechanism to restrict GATA6 function during lung and heart development.
MATERIALS AND METHODS
Yeast two-hybrid screen.
The full-length mouse GATA6 cDNA was cloned into the pSOS vector of the CytoTrap yeast two-hybrid system (Stratagene) and used to screen a human adult lung cDNA library cloned into the pMYR plasmid as described in the manufacturer's protocols (3). A total of 1.4 × 106 primary clones were screened for growth on galactose-containing medium at 37°C in the cdc25H strain of Saccharomyces cerevisiae. Positive clones were retransformed into the cdc25H yeast strain with the pSOS plasmid or the pSOS/GATA6 plasmid to determine the necessity of GATA6 for activation of the Ras pathway. From this screen, three different LMCD1/dyxin clones were isolated. The full-length mouse LMCD1/dyxin cDNA was generated by reverse transcriptase PCR from mouse lung cDNA based on the published sequence (5).
Protein-protein interaction assays.
Full-length and domain-specific regions of mouse LMCD1/dyxin were generated in pGEX4T1 using the following oligonucleotides: full-length sense, 5′-CAG GAA TTC ATG GCA AAA GTG GCT AA-3′; full-length antisense, 5′-CTC GAG TCA GGA GCG TTT TGA CT-3′; cysteine-rich domain sense, 5′-CGC GAA TTC GCA AAA GTG GCT AAG GAC CTC AAC CC-3′; cysteine-rich domain antisense, 5′-GCC TCG AGT CAT AGG CAG TGC TCC TCC TGG-3′; Prickle-Espinas-Testin (PET) domain sense, 5′-CGC GAA TTC AGC TCT GAC CTG GAC GAT GAT CGG-3′; PET domain antisense, 5′-GCC TCG AGT CAC TCT GGC TTT TCC TGT GTC TTG-3′; LIM domain sense, 5′-CGC GAA TTC GGC ACA GAG ACC ACT-3′; LIM domain antisense, 5′-CTC GAG TCA GGA GCG TTT TGA CT-3′. G lutathione S -transferase (GST) fusion proteins were generated by inducing protein expression with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 hours and purifying bacterial lysates over glutathione-agarose. For pull-down assays, in vitro-translated GATA6 proteins were incubated with equal amounts of each GST-LMCD1 fusion protein (1 μg) for 2 hours at 4°C in the following buffer: 20 mM Tris (pH 7.5), 300 mM NaCl, 1 mM dithiothreitol, 0.4% NP-40, 0.5 mg/ml bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride. Protein complexes were washed three times with the above-described buffer and then one time with the same buffer containing 150 mM NaCl but lacking bovine serum albumin. Protein complexes were resolved on either 12% or 4 to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gradient gels, which were dried and exposed to film.
Coimmunoprecipitation assays were performed using whole-cell extracts from 1 × 108 MLE-15 cells, which are known to express GATA6 (42). Cells were extracted as previously described (42), and proteins were precipitated with a commercially available antibody against GATA6 (Santa Cruz; C-20). Precipitated proteins were resolved on SDS-PAGE gels, blotted onto polyvinylidene difluoride membranes, and probed with the antibodies indicated in the figure legends.
Nuclear extracts were prepared from 293 cells transfected with a GATA6 expression plasmid (pCMVGATA6) as previously described (42). Far-Western blotting was performed essentially as described previously (11). Briefly, nuclear extracts were resolved on SDS-PAGE gels, blotted onto polyvinylidene difluoride membranes, and probed with GST or GST-LMCD1 fusion proteins (diluted to 0.5 μg/ml) in 50 mM Tris (pH 7.5), 150 mM NaCl, and 0.1% Tween 20 for 1 hour at room temperature. The blots were washed three times with incubation buffer and probed with anti-GST-LMCD1 antibody, which recognizes both GST and LMCD1 proteins, at a 1:500 dilution.
Histology and immunocytochemistry.
The LMCD1/dyxin in situ probe was generated from the full-length cDNA by PCR using an oligonucleotide containing a T7 site in the antisense orientation. Fixing, processing, and hybridization of tissue sections with a 35S-labeled riboprobe were performed as described previously (20). The anti-LMCD1 rabbit polyclonal antibody was generated using the full-length GST-LMCD1 fusion protein. Antiserum was used at a 1:100 dilution for immunohistochemistry on paraffin sections of mouse embryos. Immunohistochemistry was performed as described previously (19). Further details on the histology can be found at the University of Pennsylvania Molecular Cardiology Center website (http://www.uphs.upenn.edu/mcrc/). Immunocytochemistry was performed on 293 cells transfected with a FLAG-tagged LMCD1/dyxin expression plasmid. Cells were treated with 20 μM leptomycin B for 4 hours prior to fixation with 3.7% formaldehyde. Anti-FLAG mouse monoclonal antibody (M2; Sigma) was used at a 1:1,000 dilution to detect LMCD1/dyxin protein in cells.
Cotransfection, EMSAs, and chromatin immunoprecipitation assays.
NIH 3T3 cells were transfected with plasmids encoding full-length GATA6 (pCMVGATA6), plasmids encoding LMCD1/dyxin, and the indicated reporter plasmids. After 48 h, cells were harvested and luciferase assays were performed with a commercially available kit (Promega). Electrophoretic mobility shift assays (EMSAs) were performed as described previously, using the GATA6 DNA binding site oligonucleotide from the mouse Wnt7b promoter (42) and either in vitro-translated GATA6 protein (5 μl) or nuclear extracts (10 μg) from cells transfected with a GATA6 expression plasmid.
Chromatin immunoprecipitation (ChIP) assays were performed using a commercially available kit (Upstate Biologicals) and oligonucleotides spanning the GATA DNA binding sites located in the mouse surfactant protein C (SP-C) and cardiac troponin C (cTNC) promoters (16, 23) or the forkhead DNA binding site located in the mouse CC10 promoter (14, 32). The anti-GATA6 polyclonal antibody (C-20; Santa Cruz Biotechnology), anti-GATA4 polyclonal antibody (C-20; Santa Cruz Biotechnology), anti-GATA1 monoclonal antibody (N6; Santa Cruz Biotechnology), and the previously described Foxp4 polyclonal antibody were used for immunoprecipitation (19). Chromatin was isolated from 293 cells transfected with GATA6, GATA1, GATA4, LMCD1/dyxin, or Foxp4 expression plasmids as indicated in the figure legends, as well as the previously described pGL2/SP-C and pGH/cTNC reporter plasmids (23, 32). Fugene 6 (Roche Biochemicals) was used for all cell culture transfections. Quantitative reverse transcriptase PCR (Q-PCR) with the oligonucleotides described above was performed with an Applied Biosystems SYBR green PCR master mix and an MJ Research DNA Engine Opticon 2 real-time detection system according to the manufacturers' instructions.
Inhibition of LMCD1 expression by siRNA.
Oligonucleotides directed against LMCD1/dyxin were purchased from Dharmacon as a pool (SMARTpool catalog no. M-053725-00). MLE-15 cells were transfected using DharmaFECT with the amounts of small interfering RNA (siRNA) oligonucleotides indicated in the figure legends. Cells were harvested after 48 h, and total RNA or protein was extracted. Equal amounts of total cell protein were probed on Western blots for the presence of LMCD1 as described above. Q-PCR was performed as described above for the presence of mouse surfactant protein A (SP-A) using the following nucleotides: sense, 5′-CTG CAA ACA ATG GGA GTC CTC AGC-3′; antisense, 5′-CT GCA GGC AGC CCT TAT CAT TCC-3′.
RESULTS
Identification of LMCD1/dyxin as a GATA6-interacting protein.
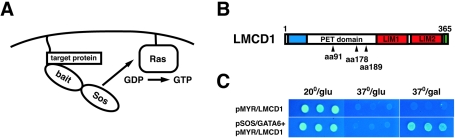
The CytoTrap yeast two-hybrid system was used to screen a human adult lung cDNA library using the full-length mouse GATA6 open reading frame to identify new GATA6-interacting proteins (Fig. 1A) (3). Several clones were obtained that corresponded to the human orthologue of mouse LMCD1/dyxin (Fig. 1B) (5). The longest of these clones, encoding amino acids (aa) 91 to 365, was retransformed into the cdc25H yeast strain, along with either the pSOS bait plasmid itself or the bait plasmid containing the GATA6 cDNA. These results show that the activation of the Ras signaling transduction pathway in the Cytotrap two-hybrid system relied upon coexpression of both GATA6 and LMCD1/dyxin (Fig. 1C).
FIG. 1.
Identification of LMCD1/dyxin as a GATA6 binding protein. (A) The Cytotrap yeast two-hybrid screening strategy. The bait protein, in this case GATA6, is expressed in frame with human Sos. Target proteins in the library are expressed with a myristoylation sequence, causing them to locate on the inner side of the plasma membrane. Interaction between bait and target proteins colocalizes Sos with Ras, which activates the Ras signal transduction pathway, allowing growth of yeast strain cdc25H on galactose at 37°C. (B) Screening of a human adult lung cDNA library with GATA6 as the bait protein resulted in the identification of three clones homologous to human LMCD1/dyxin. LMCD1/dyxin has a conserved cysteine-rich domain at the amino-terminal end (blue), two LIM domains (red), and a putative nuclear localization motif (green). The amino-terminal end of the proteins encoded by the partial cDNAs isolated is noted by black arrowheads. (C) The LMCD1/dyxin cDNA clone encoding aa 91 to 365 was retransformed into the cdc25H yeast strain with blank pSOS plasmid or with pSOS/GATA6. Activation of the Ras signal transduction pathway, which results in the ability of the cdc25H yeast strain to grow at 37°C on galactose, occurred only with coexpression of GATA6.
Domains within LMCD1/dyxin and GATA6 mediating protein-protein interaction.
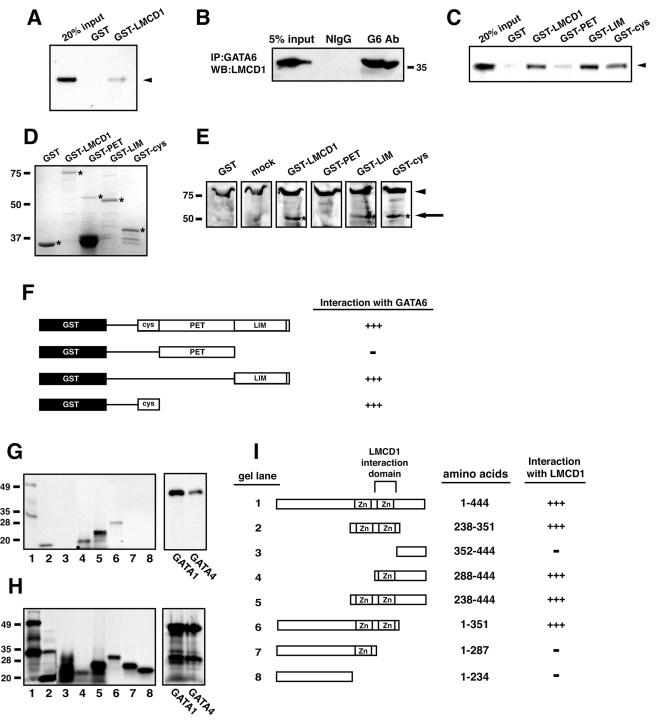
To verify that LMCD1/dyxin directly interacted with GATA6, a GST fusion protein was generated using the full-length mouse LMCD1/dyxin cDNA. In vitro-translated GATA6 interacted with GST-LMCD1 but not GST (Fig. 2A). Coimmunoprecipitation assays using GATA6- and LMCD1-specific antibodies demonstrate endogenous in vivo association of GATA6 and LMCD1 in the lung epithelial cell line MLE-15 (Fig. 2B). LMCD1/dyxin is a modular protein containing at least three domains: (i) a cysteine-rich domain (CYS domain) at the amino terminus that contains zinc finger-like motifs, (ii) a PET domain based on its similarity to the Drosophila prickle protein (10), and (iii) two LIM domains. To determine which domains in LMCD1/dyxin were required for interaction with GATA6, GST fusion proteins were generated with the CYS, PET, and LIM domains. GST pull-down assays revealed that GATA6 bound to the LIM as well as CYS domains (Fig. 2C and D). Binding to the PET domain was not observed at significant levels (Fig. 2C). Far-Western blot analyses were also performed using nuclear extracts from 293 cells transfected with an expression plasmid encoding the full-length GATA6 cDNA or cells mock transfected with empty plasmid. Extracts transferred to membranes were probed with the GST fusion proteins described above. These experiments demonstrated binding of GATA6 to full-length LMCD1/dyxin and to the LIM and CYS domains (Fig. 2E). These results confirm that the LIM and CYS domains, but not the PET domain, interact with GATA6 (Fig. 2F).
FIG. 2.
Protein-protein interaction between GATA6 and LMCD1/dyxin. (A) GST pull-down assay showing the interaction between GATA6 and full-length LMCD1/dyxin. (B) Coimmunoprecipitation of GATA6 and LMCD1 from MLE-15 cell extracts. Extracts were immunoprecipitated with a GATA6 antibody and then analyzed by Western blotting with the LMCD1/dyxin polyclonal antibody. “Input” represents 5% of the extract used in the immunoprecipitation. Position of the 35-kDa molecular mass marker is shown. Expression of GATA6 in MLE-15 cells has already been reported (6, 31). IP, immunoprecipitation; WB, Western blot; NIgG, normal IgG; G6 Ab, GATA6 antibody. (C) GST pull-down assay showing interaction between the LIM and CYS domains and GATA6. Arrowheads in panels A and C indicate radiolabeled GATA6 interaction with GST-LMCD1 fusion proteins. (D) Coomassie blue-stained gel of GST fusion proteins. Asterisks denote full-length fusion proteins. Molecular mass standards in kilodaltons are noted on the left. (E) Far-Western blot showing interaction between GATA6 and the LIM- and cysteine-rich domains of LMCD1/dyxin. The arrow and asterisks denote GATA6 interaction with GST fusion proteins. The arrowhead denotes a nonspecific band that reacts with anti-GST-LMCD1 antisera. Mock-transfected cell extracts were probed with full-length GST-LMCD1. Molecular mass standards in kilodaltons are noted on the left. (F) Summary of the domains within LMCD1/dyxin that interact with GATA6. −, no interaction; +++, interaction. (G) GST pull-down assay to assess the abilities of different regions of GATA6 that interact with LMCD1/dyxin, as well as full-length GATA1 and GATA4. Molecular mass standards in kilodaltons are noted on the left. (H) Input of radiolabeled GATA6, GATA1, and GATA4 proteins. Molecular mass standards in kilodaltons are noted on the left. (I) Summary of domains within GATA6 that interact with LMCD1/dyxin. These regions delineate the carboxy-terminal zinc finger as the domain in GATA6 required for interaction with LMCD1/dyxin. Gel lane numbers in panels G and H correspond to the GATA6 proteins indicated in panel I.
To determine the domains within GATA6 responsible for mediating interaction with LMCD1, GST pull-down assays were performed with full-length GST-LMCD1 and in vitro-translated proteins representing various domains in GATA6. All GATA6 proteins containing the carboxy-terminal zinc finger interacted with LMCD1, indicating that this domain is responsible for mediating GATA6-LMCD1/dyxin interactions (Fig. 2G through I). LMCD1 also bound to GATA1 and GATA4, consistent with the highly conserved carboxy-terminal zinc finger mediating this interaction (Fig. 2G and H).
Expression of the LMCD1/dyxin gene during lung and heart development.
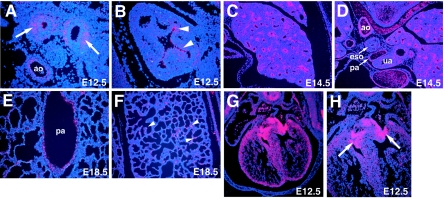
GATA6 is expressed in myocardium, lung epithelium, and vascular smooth-muscle cells during development (23, 24). To determine the mRNA expression pattern of LMCD1/dyxin during mouse gestation, in situ hybridization was performed using a specific riboprobe to LMCD1/dyxin. At embryonic day 12.5 (E12.5), LMCD1/dyxin expression is observed in distal epithelium of the developing lung and in the underlying mesenchyme (Fig. 3A and B). This expression pattern continues through gestation, and by E18.5, expression of LMCD1/dyxin is observed in a small subset of alveolar epithelial cells in a pattern consistent with that of type 2 alveolar epithelial cells (Fig. 3C through F). Expression of LMCD1/dyxin is observed in vascular smooth-muscle tissues, including the thoracic aorta and large pulmonary blood vessels (Fig. 3D and E). LMCD1/dyxin mRNA expression is also observed in the ventricular myocardium as well as at high levels in the developing endocardial cushions (Fig. 3G and H). Expression of LMCD1/dyxin in the heart has been previously reported (4).
FIG. 3.
LMCD1/dyxin gene expression during cardiopulmonary development. In situ hybridization was performed with tissue sections from E12.5 (A, B, G, and H), E14.5 (C and D), and E18.5 (E and F) mouse embryos. Expression is observed in developing airway epithelium (A, arrows) with the highest expression at the distal tips of the airways (B, arrowheads). Expression is also observed in the underlying mesenchyme (A). Expression continues in the lung airways at E14.5 (C) but decreases by E18.5 so that it is observed in only a small subset of alveolar epithelium (F, arrowheads). Expression in the cardiovascular system is observed in the muscular component of the dorsal aorta (D) and in the pulmonary arteries (E). Expression in the heart (H) is observed in the myocardium as well as the developing endocardial cushions (arrows). ao, aorta; ua, upper airways; eso, esophagus; pa, pulmonary artery.
Expression of LMCD1/dyxin protein during lung and heart development.
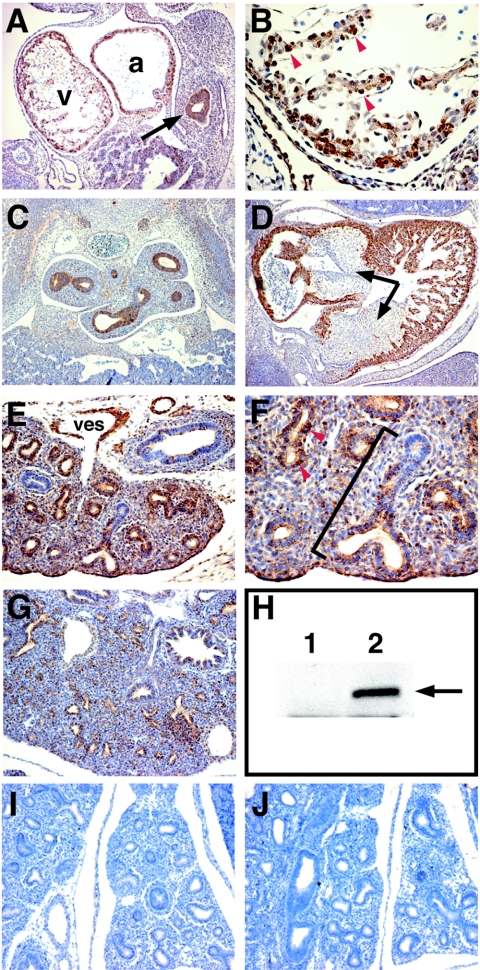
To analyze LMCD1/dyxin protein expression during cardiopulmonary development, a polyclonal antibody was generated to the GST-LMCD1 fusion protein. This antibody recognizes a 40-kDa protein in transfected but not untransfected 293 cells (Fig. 4H). Immunohistochemistry using the anti-LMCD1 antibody on staged sections of mouse embryos shows that LMCD1/dyxin protein is expressed in myocardium as early as E9.5, with some cells exhibiting apparent nuclear staining (Fig. 4A and B). Expression in the forming anterior foregut epithelium is also observed at E9.5 (Fig. 4A). By E12.5, LMCD1/dyxin expression is observed in the airways of the developing lung as well as in the myocardium (Fig. 4C and D). In contrast to LMCD1/dyxin mRNA expression, LMCD1/dyxin protein expression is not observed in the developing endocardial cushions of the heart (Fig. 4D).
FIG. 4.
Expression of LMCD1/dyxin protein during cardiopulmonary development. Immunohistochemistry was performed with tissue sections from E9.5 (A and B), E12.5 (C and D), E14.5 (E, F, I, and J), and E16.5 (G) mouse embryos. LMCD1/dyxin protein expression was observed in the developing heart at E9.5 (A and B) and E12.5 (D). LMCD1/dyxin expression is observed in the anterior foregut endoderm at E9.5 (A, arrow). In contrast to gene expression, LMCD1/dyxin protein expression is not observed in the developing endocardial cushions of the heart at E12.5 (D, arrows). Expression is observed throughout the branching airways of the lung at E12.5 (C). By E14.5, LMCD1/dyxin expression is found in the distal airway epithelium in the lung (E), with the highest levels observed in the distal regions (F, bracket). Expression is also observed in the developing pulmonary arteries (E). In both heart and lung sections, note the presence of nuclear LMCD1/dyxin expression in a subset of cells (B and F, arrowheads). The anti-GST-LMCD1 antibody recognizes LMCD1/dyxin protein from transfected (lane 2) but not untransfected (lane 1) 293 cells (H, arrow). Use of preimmune serum (I) and preincubation of LMCD1 antiserum with LMCD1 fusion protein (J) results in loss of staining in cardiopulmonary tissues. v, ventricle; a, atrium; ves, pulmonary vessel.
During development, airways in the lung develop along a distinct proximal-distal axis, which allows the differentiation of specific epithelial cell types required for postnatal respiration (for a review, see reference 40). At E14.5, when proximal-distal differentiation in the lung is proceeding rapidly, LMCD1/dyxin expression is observed at high levels in the distal but not proximal airways of the developing lung (Fig. 4E and F). As in the myocardium, LMCD1/dyxin expression is observed in the nucleus of a subset of airway epithelial cells at E14.5 (Fig. 4F). By E16.5, LMCD1/dyxin expression decreases in airway epithelium in the lung (Fig. 4G). Expression of LMCD1/dyxin protein is also observed in the vascular smooth muscle of pulmonary arteries (Fig. 4E). Use of preimmune antisera and preincubation of the anti-LMCD1 antisera with LMCD1 fusion protein resulted in loss of signal in all of these tissues (Fig. 4I and J). Together, these data demonstrate that LMCD1/dyxin expression overlaps extensively with that of GATA6 in the lung, heart, and vascular smooth muscle.
LMCD1/dyxin is retained in the nucleus by inhibition of nuclear export.
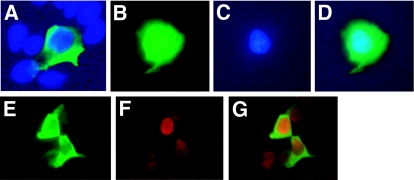
LIM proteins are often actively exported from the nucleus (12, 26). The physiological signals that regulate this process remain obscure. The above data suggest that LMCD1/dyxin is expressed in the nucleus in a subset of cardiac myocytes and lung epithelial cells. LMCD1/dyxin localizes primarily to the cytoplasm in 293 cells transfected with a plasmid encoding FLAG-tagged LMCD1/dyxin (Fig. 5A). To determine whether LMCD1/dyxin was actively exported from the nucleus, transfected 293 cells were treated with the nuclear export inhibitor leptomycin B. This resulted in an accumulation of LMCD1/dyxin in the nucleus (Fig. 5B through -D). To determine whether cytoplasmic expression of LMCD1 disrupted cellular localization of GATA6, both proteins were coexpressed in 293 cells, and immunocytochemistry was performed. In cells that express both GATA6 and LMCD1, GATA6 is still primarily localized to the nucleus (Fig. 5E through G). Thus, LMCD1/dyxin is actively exported from the nucleus and does not affect cellular localization of GATA6.
FIG. 5.
LMCD1/dyxin is retained in the nucleus of leptomycin B-treated cells. 293 cells were transfected with a myc-tagged LMCD1 expression construct and treated with 20 μM leptomycin B for 4 hours. Cells were stained with the anti-myc epitope antibody (9E10). Untreated cells showed primarily cytoplasmic staining of LMCD1/dyxin (A), whereas leptomycin B-treated cells showed an accumulation of protein within the nucleus. (B) Fluorescein isothiocyanate staining of LMCD1; (C) DAPI (4′,6′-diamidino-2-phenylindole) staining; (D) overlay of panels B and C. Coexpression of LMCD1 (E) and GATA6 (F) does not change cellular localization of GATA6. Panel G is an overlay of panels E and F.
LMCD1/dyxin represses GATA activation of target genes.
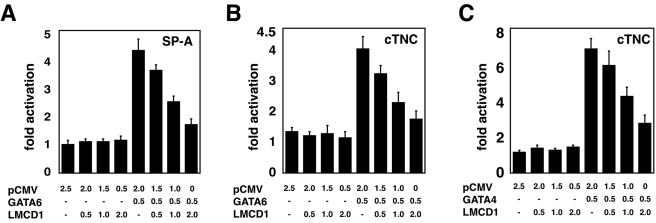
To determine the effect of LMCD1/dyxin on GATA6 function, trans-activation assays were performed using the previously characterized promoters for SP-A and cTNC (6, 23). GATA6 is known to activate both of these promoters in vitro (6, 23). As expected, expression of GATA6 increased SP-A and cTNC promoter activity (Fig. 6A and B). Expression of LMCD1/dyxin did not affect either SP-A or cTNC promoter activity in the absence of GATA6 (Fig. 6A and B). However, transfection of increasing amounts of LMCD1/dyxin expression plasmid caused dose-dependent repression of GATA6 trans activation of both the SP-A and cTNC promoters (Fig. 6A and B). Increasing levels of LMCD1 also repressed GATA4 activation of the cTNC promoter (Fig. 6C). These data suggest that LMCD1/dyxin acts as a repressor of GATA function in both lung and heart.
FIG. 6.
LMCD1/dyxin represses GATA6-mediated trans activation of lung- and cardiac tissue-specific promoters. NIH 3T3 cells were transfected with the indicated amounts (in micrograms) of GATA6 and LMCD1/dyxin expression plasmids or empty vector along with the mouse SP-A luciferase reporter plasmid 150 bp upstream of the transcriptional start site (A) or the −124-bp cTNC growth hormone reporter plasmid (B). An identical assay was performed with GATA4 and the cTNC promoter (C). Data represent the averages of results of three assays ± standard errors of the means.
LMCD1/dyxin does not contain inherent repression activity, and the full-length protein is required for repression.
There were at least two possible mechanisms by which LMCD1/dyxin could repress GATA6 activity. LMCD1/dyxin could contain inherent transcriptional repression activity and, by physically interacting with GATA6, repress gene transcription in much the same way as FOG-1/2 (34-36). Another possible mechanism is that LMCD1/dyxin could inhibit GATA6 DNA binding, resulting in decreased GATA6 activity.
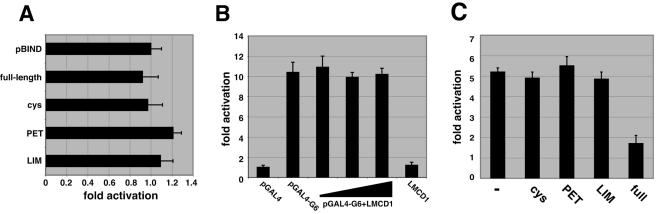
To determine whether LMCD1/dyxin had inherent transcriptional repression activity, full-length LMCD1/dyxin as well as different domains of this protein were fused to the GAL4 DNA binding domain and used in trans-activation assays with a GAL4-responsive simian virus 40 reporter plasmid. These data show that LMCD1/dyxin contains little, if any, inherent repression activity (Fig. 7A). To assess whether LMCD1 could effect GATA6 activation of target genes independent of GATA6 DNA binding, we used a GAL4-GATA6 fusion protein and a GAL4 luciferase reporter. The GAL4-GATA6 fusion protein vigorously activated the GAL4 reporter (Fig. 7B). However, increasing levels of LMCD1 did not affect this trans activation. These data also indicate that LMCD1 does not affect the activity of the GAL4 protein (Fig. 7B). These studies suggest that LMCD1 represses GATA function through other means.
FIG. 7.
LMCD1/dyxin does not have inherent transcriptional repression activity, and the full-length protein is required for repression. LMCD1/dyxin, or domains within, does not repress a GAL4-responsive reporter containing the simian virus 40 promoter (A). A GAL4-GATA6 fusion protein was used to activate the GAL4 response reporter, and increasing amounts of LMCD1 were coexpressed. LMCD1/dyxin does not affect the activity of GATA6 when fused to the GAL4 DNA binding domain (B). The indicated domains in LMCD1/dyxin were expressed, along with GATA6 and the SP-A luciferase reporter in 293 cells, to show that full-length LMCD1/dyxin can repress GATA6 activity on the SP-A promoter but that individual domains within LMCD1/dyxin cannot (C). Data represent the averages of results of three assays ± standard errors of the means.
Since LMCD1 contains multiple domains that can interact with GATA6, we assessed which of these domains were responsible for repressing GATA6 function. Using the SP-A promoter, expression plasmids encoding different domains of LMCD1 along with GATA6 were transfected into 293 cells. Surprisingly, while full-length LMCD1 was able to effectively repress GATA6 trans activation of the SP-A promoter, none of the individual domains tested were capable of repressing GATA6 activity (Fig. 7C). These data indicate that repression by LMCD1 required the full-length protein, which correlates with the ability of both the CYS and LIM regions to interact with GATA6.
LMCD1/dyxin represses GATA6 activity by inhibiting DNA binding.
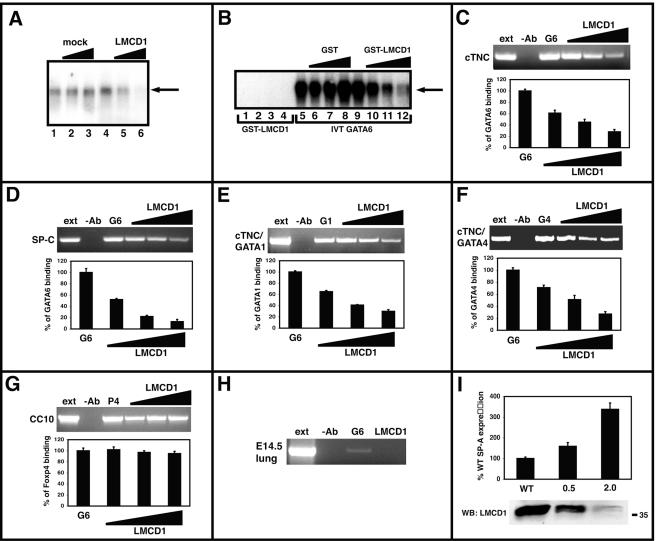
The results described above indicate that LMCD1 inhibits GATA function by a means other than conferring transcriptional corepression to GATA factors. Therefore, EMSAs were performed to determine whether LMCD1/dyxin inhibited GATA6 DNA binding. In vitro-translated GATA6 was initially used to bind to a known GATA6 DNA binding site found in the mouse Wnt7b promoter (42). Addition of increasing amounts of an in vitro translation reaction mixture using empty vector did not affect GATA6 DNA binding (Fig. 8A). However, increasing amounts of an in vitro translation reaction mixture using a full-length LMCD1/dyxin cDNA inhibited GATA6 DNA binding in a dose-dependent manner (Fig. 8A). EMSAs were also performed using nuclear extracts from 293 cells transfected with a GATA6 expression plasmid. Increasing amounts of GST-LMCD1/dyxin but not GST alone also inhibited GATA6 DNA binding in a dose-dependent manner in this assay (Fig. 8B). GST-LMCD1 did not exhibit detectable DNA binding (Fig. 8B and data not shown).
FIG. 8.
LMCD1/dyxin inhibits GATA6 DNA binding and represses GATA6 function in vivo. Gel shift assays were used to assess the ability of LMCD1/dyxin to affect GATA6 DNA binding. In vitro-translated GATA6 binds to the conserved GATA sites within the mouse Wnt7b promoter (A, lanes 1 and 4). Increasing expression of LMCD1/dyxin inhibits GATA6 DNA binding when LMCD1/dyxin is cotranslated in an in vitro transcription/translation reaction (A, lanes 5 and 6), while increasing amounts of an empty plasmid did not have any effect (A, lanes 2 and 3). Addition of increasing amounts of GST-LMCD1 (B, lanes 10 to 12), but not GST alone (B, lanes 6 to 8), inhibits GATA6 DNA binding. Note that GST-LMCD1 does not bind to DNA (B, lanes 1 to 4). ChIP assays show that increasing levels of LMCD1/dyxin inhibit GATA6 DNA binding to conserved sites located in the cTNC (C) and SP-C (D) promoters as well as that of GATA1 (E) and GATA4 (F) to the cTNC promoter. However, LMCD1/dyxin does not inhibit Foxp4 binding to the forkhead DNA binding site located in the mouse CC10 promoter (G). Q-PCR ChIP data are shown as a percentage of GATA or Foxp4 binding without LMCD1. GATA6 but not LMCD1/dyxin is found associated with the conserved GATA DNA binding site located in the mouse SP-C promoter (H). siRNA transfection of MLE-15 cells with the indicated amounts of oligonucleotides (in micrograms) resulted in inhibition of LMCD1/dyxin expression (indicated in Western blot in bottom panel) and an increase in SP-A expression (I). The position of the 35-kDa molecular mass marker is shown in panel I. IVT, in vitro translated; ext, extract; −Ab, without antibody; G6, GATA6; G1, GATA1; G4, GATA4; P4, Foxp4; WT, wild type; WB, Western blot.
To determine whether LMCD1/dyxin could inhibit GATA6 DNA binding on a target gene in a cellular context, ChIP assays were performed using the known GATA6 DNA binding sites in the SP-C and cTNC promoters as targets (16, 23). As expected, GATA6 binds to the well-characterized sites within these promoters (Fig. 8C and D). GATA6 DNA binding to both SP-C and cTNC promoters is inhibited in a dose-dependent manner by LMCD1/dyxin (Fig. 8C and D). Furthermore, LMCD1 inhibited DNA binding by GATA4 and GATA1 to the cTNC promoter (Fig. 8E and F), suggesting that LMCD1 can repress other GATA factors as well. However, LMCD1/dyxin did not inhibit DNA binding of another transcription factor, Foxp4, to its cognate DNA binding site located in the mouse CC10 promoter (Fig. 8G) (14, 32). Consistent with LMCD1 acting as a GATA6 DNA binding inhibitor, GATA6 but not LMCD1 was found associated with the endogenous mouse SP-C promoter in chromatin from E14.5 mouse lung tissue (Fig. 8H). To verify that inhibition of LMCD1/dyxin expression would translate into increased GATA6 target gene expression in vivo, MLE-15 cells were transfected with siRNA oligonucleotides directed against LMCD1/dyxin. These oligonucleotides inhibited LMCD1/dyxin protein expression and increased SP-A expression in a dose-dependent manner (Fig. 8I). Expression of β-actin was not affected by LMCD1/dyxin siRNA oligonucleotides (data not shown). Since SP-A is a known target of GATA6 (6), these data show that LMCD1/dyxin inhibits GATA6 DNA binding on promoters of target genes, resulting in repression of GATA6 activity in vivo, which reveals a novel mechanism to regulate GATA6 activity during lung and heart development.
DISCUSSION
GATA6 regulates both lung- and heart-specific gene expression (6, 23, 42, 46). Several cofactors have been shown to interact with GATA6 to either repress or activate GATA6-dependent transcription (7, 9, 28, 35, 36). We show that a novel interaction between GATA6 and LMCD1/dyxin represses GATA6 function by inhibiting GATA6 DNA binding. This repression is observed on both lung- and heart-specific target promoters. Given the known role of GATA-LIM protein interactions in other tissues, this interaction is likely to play an important role in the regulation of tissue-specific gene expression in lung, heart, and vascular smooth muscle.
Although several other cofactors have been shown to interact with GATA factors and repress their function, to our knowledge the LMCD1/dyxin interaction is the first example of a GATA cofactor that represses function through the inhibition of DNA binding. DNA binding inhibition by the formation of nonfunctional heterodimeric complexes has been demonstrated to play an important role in the regulation of several other transcription factors. For example, myocyte enhancer factor 2C DNA binding is inhibited by the binding of activated Notch ligand, resulting in the repression of skeletal muscle gene transcription (44). Id proteins also function by forming non-DNA binding heterodimers with E proteins, sequestering E proteins from forming functional heterodimers with other bHLH factors (33). Such sequestration likely plays key roles in regulating gene expression during development.
Inhibition of DNA binding, combined with regulation of cellular localization, may be a particularly relevant mechanism for regulation of gene transcription by cofactors such as LIM proteins because they shuttle between the nucleus and cytoplasm (1, 12, 26). The retention of LMCD1 in the cytoplasm would undo its ability to repress GATA6 DNA binding. Upon as-yet-uncharacterized signals, LMCD1/dyxin could translocate to the nucleus or its nuclear export could be inhibited, allowing it to repress GATA6 DNA binding and function. Such a simple mechanism would allow for the rapid repression of GATA6 function without affecting GATA6 gene or protein expression. Precise regulation of GATA6 function is important, since overexpression of GATA6 leads to multiple defects in lung development and homeostasis, including reduced alveolar septation and lung function (17).
Other LIM proteins such as CRP1 and -2 interact with GATA4 and GATA6 to synergistically activate cardiac and vascular smooth-muscle gene expression (7). The LIM protein LMO2 performs a similar function with GATA1 (28, 39). How these LIM proteins function to regulate GATA-dependent transcription is still unclear, but several reports show that they can bridge GATA factors with other transcriptional regulators (2, 7, 28, 39). CRP1 and -2 bridge GATA factors with the important muscle transcription factor serum response factor, synergistically activating smooth-muscle gene expression (7). LMO2 is critical in forming a multimeric transcriptional complex with GATA1 or -2, TAL1, E2A, and Ldb1 (2, 39). Loss of LMO2 results in an abrogation of hematopoietic development, likely due to loss of interactions with TAL1 and GATA2 (2, 45).
The expression patterns of GATA6 and LMCD1/dyxin overlap extensively. This observation suggests that LMCD1/dyxin may play a role in regulating GATA6 function in many of the tissues where GATA6 is expressed. However, the finding that nucleus-localized LMCD1/dyxin is observed in only a small subset of both cardiac myocytes and lung epithelial cells in vivo suggests that GATA6 repression by LMCD1/dyxin is tightly controlled during development. LIM protein shuttling between the nucleus and cytoplasm is not well understood (1, 12, 26). CRP2 nuclear localization changes as proepicardial cells differentiate (7). Our data showing that inhibition of nuclear export can result in nuclear accumulation of LMCD1/dyxin suggest that this process is one possible mechanism that controls LMCD1/dyxin cellular localization. However, the presence of a putative nuclear localization sequence at the carboxy terminus of LMCD1/dyxin suggests that its cellular localization could be regulated by multiple pathways.
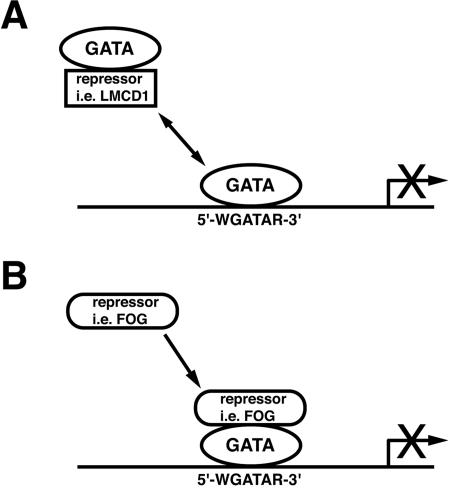
The expression of a corepressor in the same temporal and spatial pattern as an activator such as GATA6 may appear counterintuitive. However, as with other transcriptional regulators, GATA6 interacts with both coactivators and corepressors. Widely expressed factors such as p300 physically bind to GATA6, regulating vascular smooth-muscle gene transcription (38). Tissue-restricted transcription factors such as Nkx2.1 and Nkx3.2 further coactivate GATA6-dependent gene transcription (16, 25, 42). Thus, a precise balance between interaction with these coactivators and corepressors such as LMCD1/dyxin or FOG-1/2 is essential for the regulation of spatial and temporal activity of GATA6 as well as other GATA factors (Fig. 9).
FIG. 9.
DNA binding inhibition by LMCD1/dyxin defines a new mechanism to restrict GATA factor function. (A) LMCD1/dyxin represses GATA6 function through inhibition of DNA binding. Strict control over cellular localization of LMCD1/dyxin may define its regulatory function. (B) GATA factors also interact with a variety of corepressors, such as FOG-1/2. These interactions repress GATA function by recruiting repression function in trans, sometimes through interaction with additional corepressor proteins such as CtBP-1 (8, 15, 34). Both models effectively restrict GATA factor activity.
GATA factors are expressed in specific spatial and temporal patterns during development. Although the role of GATA6 in cardiac and vascular smooth-muscle development is still unclear, GATA6 has been demonstrated to play a critical role in lung epithelial differentiation, and sustained overexpression leads to defects in lung alveolar development (17, 18, 46). With its ability to inhibit GATA6 DNA binding and to repress GATA6 function, LMCD1/dyxin may provide an important mechanism to precisely regulate GATA6 function in cardiopulmonary tissues.
Acknowledgments
We thank John Lepore and Mark Kahn for helpful suggestions on these studies and Deborah Lang for assistance with the ChIP assays.
These studies were funded by an NIH grant to E.E.M. (HL064632). E.E.M. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Akazawa, H., S. Kudoh, N. Mochizuki, N. Takekoshi, H. Takano, T. Nagai, and I. Komuro. 2004. A novel LIM protein Cal promotes cardiac differentiation by association with CSX/NKX2-5. J. Cell Biol. 164:395-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekman, E., and D. Henrique. 2002. Embryonic expression of three mouse genes with homology to the Drosophila melanogaster prickle gene. Mech. Dev. 119(Suppl. 1):S77-S81. [DOI] [PubMed] [Google Scholar]
- 5.Bespalova, I. N., and M. Burmeister. 2000. Identification of a novel LIM domain gene, LMCD1, and chromosomal localization in human and mouse. Genomics 63:69-74. [DOI] [PubMed] [Google Scholar]
- 6.Bruno, M. D., T. R. Korfhagen, C. Liu, E. E. Morrisey, and J. A. Whitsett. 2000. GATA-6 activates transcription of surfactant protein A. J. Biol. Chem. 275:1043-1049. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 8.Deconinck, A. E., P. E. Mead, S. G. Tevosian, J. D. Crispino, S. G. Katz, L. I. Zon, and S. H. Orkin. 2000. FOG acts as a repressor of red blood cell development in Xenopus. Development 127:2031-2040. [DOI] [PubMed] [Google Scholar]
- 9.Durocher, D., F. Charron, R. Warren, R. J. Schwartz, and M. Nemer. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubb, D., C. Green, D. Huen, D. Coulson, G. Johnson, D. Tree, S. Collier, and J. Roote. 1999. The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev. 13:2315-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, R. A. 2004. Studying protein-protein interactions via blot overlay or Far Western blot. Methods Mol. Biol. 261:167-174. [DOI] [PubMed] [Google Scholar]
- 12.Kanungo, J., S. J. Pratt, H. Marie, and G. D. Longmore. 2000. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 11:3299-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 14.Li, S., J. Weidenfeld, and E. E. Morrisey. 2004. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell. Biol. 24:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, A. C., A. E. Roche, J. Wilk, and E. C. Svensson. 2004. The N termini of friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J. Biol. Chem. 279:55017-55023. [DOI] [PubMed] [Google Scholar]
- 16.Liu, C., S. W. Glasser, H. Wan, and J. A. Whitsett. 2002. GATA-6 and thyroid transcription factor-1 directly interact and regulate surfactant protein-C gene expression. J. Biol. Chem. 277:4519-4525. [DOI] [PubMed] [Google Scholar]
- 17.Liu, C., M. Ikegami, M. T. Stahlman, C. R. Dey, and J. A. Whitsett. 2003. Inhibition of alveolarization and altered pulmonary mechanics in mice expressing GATA-6. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L1246-L1254. [DOI] [PubMed] [Google Scholar]
- 18.Liu, C., E. E. Morrisey, and J. A. Whitsett. 2002. GATA-6 is required for maturation of the lung in late gestation. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L468-L475. [DOI] [PubMed] [Google Scholar]
- 19.Lu, M. M., S. Li, H. Yang, and E. E. Morrisey. 2002. Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech. Dev. 119(Suppl. 1):S197-S202. [DOI] [PubMed] [Google Scholar]
- 20.Lu, M. M., H. Yang, L. Zhang, W. Shu, D. G. Blair, and E. E. Morrisey. 2001. The bone morphogenic protein antagonist gremlin regulates proximal-distal patterning of the lung. Dev. Dyn. 222:667-680. [DOI] [PubMed] [Google Scholar]
- 21.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 22.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 23.Morrisey, E. E., H. S. Ip, M. M. Lu, and M. S. Parmacek. 1996. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 177:309-322. [DOI] [PubMed] [Google Scholar]
- 24.Morrisey, E. E., H. S. Ip, Z. Tang, M. M. Lu, and M. S. Parmacek. 1997. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev. Biol. 183:21-36. [DOI] [PubMed] [Google Scholar]
- 25.Nishida, W., M. Nakamura, S. Mori, M. Takahashi, Y. Ohkawa, S. Tadokoro, K. Yoshida, K. Hiwada, K. Hayashi, and K. Sobue. 2002. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J. Biol. Chem. 277:7308-7317. [DOI] [PubMed] [Google Scholar]
- 26.Nix, D. A., J. Fradelizi, S. Bockholt, B. Menichi, D. Louvard, E. Friederich, and M. C. Beckerle. 2001. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 276:34759-34767. [DOI] [PubMed] [Google Scholar]
- 27.Ohneda, K., and M. Yamamoto. 2002. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 108:237-245. [DOI] [PubMed] [Google Scholar]
- 28.Osada, H., G. G. Grutz, H. Axelson, A. Forster, and T. H. Rabbitts. 1997. LIM-only protein Lmo2 forms a protein complex with erythroid transcription factor GATA-1. Leukemia 11(Suppl. 3):307-312. [PubMed] [Google Scholar]
- 29.Pevny, L., M. C. Simon, E. Robertson, W. H. Klein, S. F. Tsai, V. D'Agati, S. H. Orkin, and F. Costantini. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349:257-260. [DOI] [PubMed] [Google Scholar]
- 30.Sepulveda, J. L., N. Belaguli, V. Nigam, C.-Y. Chen, M. Nemer, and R. J. Schwartz. 1998. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol. Cell. Biol. 18:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw-White, J. R., M. D. Bruno, and J. A. Whitsett. 1999. GATA-6 activates transcription of thyroid transcription factor-1. J. Biol. Chem. 274:2658-2664. [DOI] [PubMed] [Google Scholar]
- 32.Shu, W., H. Yang, L. Zhang, M. M. Lu, and E. E. Morrisey. 2001. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J. Biol. Chem. 276:27488-27497. [DOI] [PubMed] [Google Scholar]
- 33.Sun, X.-H., N. G. Copeland, N. A. Jenkins, and D. Baltimore. 1991. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 11:5603-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svensson, E. C., G. S. Huggins, F. B. Dardik, C. E. Polk, and J. M. Leiden. 2000. A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. J. Biol. Chem. 275:20762-20769. [DOI] [PubMed] [Google Scholar]
- 35.Svensson, E. C., R. L. Tufts, C. E. Polk, and J. M. Leiden. 1999. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA 96:956-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Rieff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada, H., K. Hasegawa, T. Morimoto, T. Kakita, T. Yanazume, M. Abe, and S. Sasayama. 2002. Calcineurin-GATA-6 pathway is involved in smooth muscle-specific transcription. J. Cell Biol. 156:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wada, H., K. Hasegawa, T. Morimoto, T. Kakita, T. Yanazume, and S. Sasayama. 2000. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J. Biol. Chem. 275:25330-25335. [DOI] [PubMed] [Google Scholar]
- 39.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburton, D., M. Schwarz, D. Tefft, G. Flores-Delgado, K. D. Anderson, and W. V. Cardoso. 2000. The molecular basis of lung morphogenesis. Mech. Dev. 92:55-81. [DOI] [PubMed] [Google Scholar]
- 41.Watt, A. J., M. A. Battle, J. Li, and S. A. Duncan. 2004. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 101:12573-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weidenfeld, J., W. Shu, L. Zhang, S. E. Millar, and E. E. Morrisey. 2002. The WNT7b promoter is regulated by TTF-1, GATA6, and Foxa2 in lung epithelium. J. Biol. Chem. 277:21061-21070. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, M. J., and S. H. Orkin. 1995. GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23:99-107. [PubMed] [Google Scholar]
- 44.Wilson-Rawls, J., J. D. Molkentin, B. L. Black, and E. N. Olson. 1999. Activated notch inhibits myogenic activity of the MADS-box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 19:2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada, Y., A. J. Warren, C. Dobson, A. Forster, R. Pannell, and T. H. Rabbitts. 1998. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 95:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, H., M. M. Lu, L. Zhang, J. A. Whitsett, and E. E. Morrisey. 2002. GATA6 regulates differentiation of distal lung epithelium. Development 129:2233-2246. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, M., and W. Ouyang. 2003. The function role of GATA-3 in Th1 and Th2 differentiation. Immunol. Res. 28:25-37. [DOI] [PubMed] [Google Scholar]