Abstract
L1Tc, a non-long terminal repeat retrotransposon from Trypanosoma cruzi, is a 4.9-kb actively transcribed element which contains a single open reading frame coding for the machinery necessary for its autonomous retrotransposition. In this paper, we analyze the protein encoded by the L1Tc 3′ region, termed C2-L1Tc, which contains two zinc finger motifs similar to those present in the TFIIIA transcription factor family. C2-L1Tc binds nucleic acids with different affinities, such that RNA > tRNA > single-stranded DNA > double-stranded DNA, without any evidence for sequence specificity. C2-L1Tc also exhibits nucleic acid chaperone activity on different DNA templates that may participate in the mechanism of retrotransposition of the element. C2-L1Tc promotes annealing of complementary oligonucleotides, prevents melting of perfect DNA duplexes, and facilitates the strand exchange between DNAs to form the most stable duplex DNA in competitive displacement assays. Mapping of regions of C2-L1Tc using specific peptides showed that nucleic acid chaperone activity required a short basic sequence accompanied by a zinc finger motif or by another basic region such as RRR. Thus, a short basic polypeptide containing the two C2H2 motifs promotes formation of the most stable duplex DNA at a concentration only three times higher than that required for C2-L1Tc.
Trypanosoma cruzi is the etiological agent of Chagas' disease, a parasitism which affects millions of people in Central and South America (43). Apart from the impact on human health, T. cruzi has been extensively studied due to the interesting molecular characteristics that the Trypanosomatidae family has, such as polycistronic transcription, trans splicing, RNA editing, nonchromatin condensation into chromosomes during cell division (27), and a high percentage of repeated sequences. Approximately 15% of the T. cruzi genome is made of mobile retroelements. Among them, long terminal repeat (LTR) retrotransposons (38), SINE (34), and LINE retrotransposons (21, 30) have been widely described. SINEs are the most abundant elements and are probably mobilized by LINEs (4, 30).
There are about 1,000 copies of the non-LTR retrotransposon L1Tc per T. cruzi haploid genome (4), which has been suggested to confer a significant genomic polymorphism and a high degree of plasticity to the parasite (11, 39). L1Tc is actively transcribed in the three stages of the parasite life cycle (21) and codes for the enzymatic machinery involved in its retrotransposition including AP endonuclease, 3′ phosphatase, 3′ phosphodiesterase (28, 31), reverse transcriptase (RT) (13), and RNase H (29) activities. The C-terminal domain of the L1Tc element contains two cysteine motifs of the C2H2 type, which are structurally homologous to the zinc fingers of transcription factors of multicellular organisms (21). This C2H2 motif has been also observed in some non-LTR retroelements included in the R2 clade, the trypanosome CRE/SLACS clade, the nematode NeSL clade, and the Giardia lamblia GENIE family (5, 12, 20).
Mobilization of non-LTR retrotransposons occurs by a mechanism termed target-primed reverse transcription (TPRT), originally described for the insect R2Bm non-LTR element (19, 24). In TPRT, the endonuclease encoded by the element cleaves the genomic DNA minus strand, generating a 3′ hydroxyl end that is used as a primer by the element-encoded reverse transcriptase for first-strand cDNA synthesis using the element RNA as template. Some non-LTR retrotransposons, including L1Tc, encode an RNase H activity (29) which may be responsible for removing the RNA from the resulting RNA/cDNA hybrid. Following second-strand cleavage, the host factors involved in DNA synthesis or the RT encoded by the element (13) synthesize the second-strand cDNA. Thus, the integrated LINEs contain structural hallmarks such as frequent 5′ truncation, 3′ poly(A) tails, and variable-length target site duplications. Retroviruses and LTR retrotransposons utilize a distinct retrotransposition mechanism where double-stranded DNA (dsDNA) generation in the cytoplasm is mediated by RT and gag-nucleic acid chaperone (NAC) activities (40), and then this dsDNA is subsequently integrated into the chromosomal DNA.
NAC activities encoded by Drosophila melanogaster I factor and mouse L1 non-LTR retroelements have been reported (9, 23). I-factor NAC activity relies on ORF1 in a region containing CCHC, similar to that described in the gag proteins from retroviruses and some LTR retroelements (9). Homologous CCHC regions have been described in several clades of the non-LTR group of retrotransposons (20). In contrast, the mouse L1 NAC activity is associated with a region without known homology to the previously mentioned gag proteins (23). The wide distribution of this domain argues that NAC activity is a general requirement for retrotransposition of non-LTR elements, as it has been described previously for the mobilization of retroviruses and LTR retroelements. As diversity is one of the major characteristics of LINEs due to host-LINE coevolution, divergences of the location and sequence of the domain responsible for NAC activity should be expected (9, 23).
In the present paper, we show that the protein encoded by the C-terminal end of the LINE L1Tc, C2-L1Tc, has in vitro NAC activity. This C-terminal domain contains two cysteine motifs of the C2H2 type (21), similar to that present in the TFIIIA family of transcription factors. C2-L1Tc promotes annealing of complementary oligonucleotides, prevents melting of perfect DNA duplexes, and does not affect melting of mismatched DNA duplexes. Furthermore, C2-L1Tc, as well as some synthetic peptides covering the C2H2 region and the basic residues upstream of them, facilitates, in competitive displacement assays, the strand exchange between DNAs to form the most stable duplex DNA. The differential ability of the C2-L1Tc protein to bind different types of nucleic acids is probably related to its function in the mobilization and integration mechanism of L1Tc.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides used in annealing, strand exchange, and melting assays were purified by electrophoresis through denaturing polyacrylamide gels before use. One oligonucleotide of each pair was 5′ end labeled using [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Roche), and unincorporated isotope was removed by gel filtration chromatography (Sephadex G25). Oligonucleotide names and sequences are as follows: primer 14 (5′-AAAAACGAATGGAGACCCTGGCTCATCCAG-3′), 14c (5′-CTGGATGAGCCAGGGTCTCCATTCGTTTTT-3′), 52 (5′-AAAAAGTATCTTTGGCGCAAATCTTCT-3′), 52c (5′-AGAAGATTTGCGCCAAAGATACTTTTT-3′), 62 (5′-GAACGAAATGCAGACATCAGGTCGTTATTT-3′), 62c (5′-AAATAACGACCTGATGTCTGCATTTCGTTC-3′), 29 (5′-AAAAAGTACACAGTCTAACATCAACTCGC-3′), 29c (5′-GCGAGTTGATGTTAGACTGTGTACTTTTT-3′), mm29c (5′-GCGAGTTGACGTCAGACCGTGCACTTTTT-3′), and 25c (5′-GCGAGTTGATGTTAGACTGTGTACT-3′). “c” indicates the inverse complement oligonucleotide. The nucleotides in the mm29c primer that form mismatches when hybridized to 29 oligonucleotides are underlined.
DNA templates and RNA synthesis.
The DNA encoding the first 77 nucleotides of the L1Tc element were PCR amplified using the pBAC62 clone (GenBank accession no. AF208537) (30) as a template and R775′ (5′-GCATAGATATCCCTGGCTCAG-3′) and R773′ (5′-GCATTAAGCTTCAGCAGGCGC-3′) primers which contain EcoRV and HindIII restriction sites (underlined), respectively, and cloned into the pGEM-T vector (Promega), generating the pGR77 vector. One hundred forty-four-nucleotide (nt) RNA was generated using HindIII-digested pGR77 plasmid as a template and control RNA from pGEM-T using EcoRV-digested pGEM-T vector. One hundred thirty-nt RNA was produced using the HindIII-digested TcKMP11n clone (GenBank accession no. AJ000077) (37). In vitro transcription was carried out using 1 μg of linearized DNA and T7 RNA polymerase as described previously by Barroso-del Jesus et al. (1). A total of 3 μCi of [α-32P]UTP (3,000 Ci · mmol−1) was added to the reaction mixture for radiolabeling the in vitro-synthesized transcripts. Specific activity was determined using a Bioscan QC.2000 counter. RNAs were eluted from denaturing polyacrylamide gel, precipitated, and resuspended in diethyl pyrocarbonate-treated water.
Peptide synthesis.
Peptides were synthesized by the simultaneous multiple-solid-phase synthetic method (36). The peptides were assembled using the standard t-Boc solid-phase peptide synthesis strategy on a p-methylbezhydrilamide resin (15, 33). Purity was checked by high-performance liquid chromatography. Peptide sequences are shown in Fig. 9A and Table 1. Peptide 4814 (CGYSLFQKEKMVLYSLFQKEKMVLYSLFQKEKMVLGC) was used as a control peptide. Peptides were dissolved in sterile 1× phosphate-buffered saline containing 30 μM zinc chloride at a 500 μM final concentration.
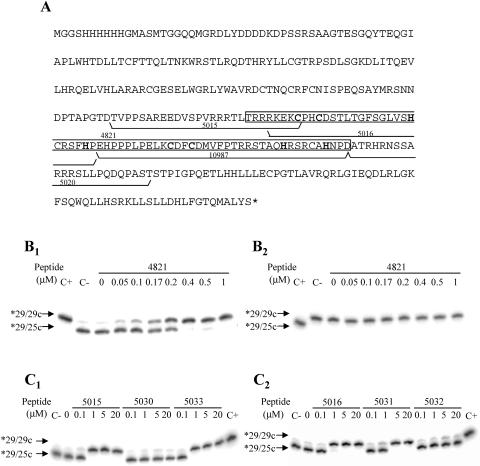
FIG. 9.
NAC activity of C2-L1Tc-derived peptides. (A) Deduced amino acid sequence of the recombinant protein encoded by the pCAS-C2-L1Tc vector. The His tag and enterokinase cleavage site derived from pCAS vector are included in the protein sequence. The two C2H2 motifs are in boldface type. The sequences of the peptides used in the assays are underlined and numbered. The sequence of peptide 4821 is enclosed in a white box. (B) Strand exchange assays using 29/25c (B1) or 29/29c (B2) preformed DNA duplexes at the indicated concentrations of peptide 4821. (C) Strand exchange assays using 29/25c preformed DNA duplex and different concentrations of 5015, 5030, and 5033 (C1) and 5016, 5031, and 5032 (C2) peptides (sequences are in Table 1). In all cases, the radiolabeled primer was the 29 oligonucleotide (*29). Preformed *29/29c and *29/25c duplexes were loaded in all gels as size markers of the preannealed duplex before the reaction (C−), and the expected size of the duplex formed after strand exchange is shown (C+).
TABLE 1.
Sequences of the peptides derived from 5015 and 5016 peptides which contain mutations, additions, and deletions
| Peptide | Sequencea |
|---|---|
| 5015 | TVPPSAREEDVSPVRRRTLTRRRKEKC |
| 5030 | TVPPSAREEDVSPV---TLTRRRKEKC |
| 5016 | RKEKCPHCDSTLTGFSGLVSHCRSFHP |
| 5031 | RKEKSPHSDSTLTGFSGLVSHCRSFHP |
| 5032 | LTCPHCDSTLTGFSGLVSHCRSFHP |
| 5033 | RRRKEKCPHSDCTLTGFSGLVSHCRSFHP |
Regions that are essential for NAC activity are in boldface type. The C2H2 zinc finger motif is underlined.
Cloning and protein purification of the recombinant C2-L1Tc.
C2-5′ (5′-CCTAGAAGCTTTAATACAGTGCCATC-3′) and C2-3′ (5′-GGATAGGTACCGAAAGTGGACAATA-3′) oligonucleotides were used to PCR amplify the region of L1Tc at positions 3976 to 4851 (GenBank accession no. AF208537) using the pBAC62 clone (30) as a template. The KpnI and HindIII sites at oligonucleotide 5′ and 3′ ends were respectively added ad hoc in PCR (see underlined restriction sites in oligonucleotide sequences above). After KpnI and HindIII digestion, the 0.9-kb-long DNA was cloned into the pCAS B vector (active motif) previously digested with the same enzymes. The pCAS vector series contains a Psal promoter which is activated by salicylate through an xylS2 regulator, making a circuit for amplifying gene expression, and adds a six-histidine tract to the recombinant protein N terminus. The resulting clone, pCAS-C2-L1Tc, was fully sequenced and used to transform Escherichia coli strain TAP-F (active motif). Transformed bacteria were grown in SOB medium (20.0 g/liter tryptone, 5.0 g/liter yeast extract, 0.5 g/liter NaCl, 10 mM MgCl2, 2.5 mM KCl), and when cultures reached an optical density at 600 nm of 0.3, protein induction was carried out by 1 mM sodium salicylate addition and grown at 30°C for 5 h. Proteins solubilized by sonication in extraction buffer (20 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 2 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 0.1% Triton X-100, 20% glycerol, and 1 mg/ml lysozyme, pH 8) were centrifuged, and supernatant was incubated with Ni-nitrilotriacetic acid (NTA) resin (QIAGEN) for 3 h at 4°C. All subsequent steps were performed at 4°C. Resin was extensively washed with extraction buffer at pH 7 and 6, and protein was eluted on a 50 to 500 mM imidazole gradient performed in extraction buffer, pH 6. Eluted fractions were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining (42), and fractions containing purified C2-L1Tc were pooled and concentrated to 250 μl with a Centriprep 10 device (Millipore). C2-L1Tc concentrate was subsequently subjected to gel filtration using a Superdex 75 column in a fast protein liquid chromatography system (Pharmacia Biotech Inc.). Fractions containing purified C2-L1Tc were pooled and concentrated as mentioned above. The protein concentration was determined by the Bradford method (3). The same conditions of C2-L1Tc overexpression and purification process were carried out with pCAS-RT L1Tc-transformed TAP-F E. coli as a control.
UV cross-linking.
Two micrograms of C2-L1Tc was incubated with 50 ng of 144-nt RNA (106 cpm) in 20 μl of UV cross-linking buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 2 mM MgCl2, 2 mM dithiothreitol, 5 U RNasin [Ambion], and 5% glycerol). After 10 min at 30°C, the mixture was transferred to ice and irradiated for 5 min with UV light (252 nm, 3,000 μW/cm2) (UV cross-linker 1800; Stratagene). The complexes formed were digested with RNase A and RNase T1 in the presence of 1 mM phenylmethylsulfonyl fluoride, fractionated by 10% SDS-PAGE, and transferred onto a polyvinylidene difluoride membrane. The radiolabeled protein was visualized by phosphorimager analysis. Protein identity was confirmed by Western blot using a specific antibody directed against the histidine tag of the recombinant protein.
Electrophoretic mobility shift assays (EMSAs).
C2-L1Tc and 32P-labeled in vitro-transcribed 130-nt RNA (0.72 nM) and 144-nt RNA (0.65 nM) were incubated in 16 μl of UV cross-linking buffer containing 100 μg/ml bovine serum albumin for 30 min at 37°C. The reaction mixtures were transferred to ice, and 8 μl of dye solution (50% glycerol, 0.1% bromophenol blue, 0.1% xylenecyanol) was added. RNA-protein complexes were fractionated by electrophoresis though 5% native polyacrylamide gels (39:1 acrylamide/bisacrylamide) with 1% glycerol in 1% Tris-borate-EDTA. The gels were dried and visualized by phosphorimager analysis.
For competition experiments, 0.5 ng of 130-nt RNA (0.72 nM) and 144-nt RNA (0.65 nM) was mixed with a series of nucleic acid competitors before C2-L1Tc was added (0.31 μM and 0.67 μM, respectively). The mixture was incubated at 37°C with the exception of the case indicated in the Fig. 3 and 4 legends. A 77-mer primer corresponding to the first 77 nt of L1Tc (77-mer single-stranded DNA [ssDNA]) was purified from denaturing polyacrylamide gel before use. One hundred forty-four-nucleotide dsDNA was PCR amplified using the pGR77 vector as a template and T7 forward and R773′ primers. The 3.09-kb dsDNA corresponds to linearized pGR77 plasmid. One hundred forty-four-nucleotide ssDNA was produced by denaturing 144-nt dsDNA for 5 min at 95°C.
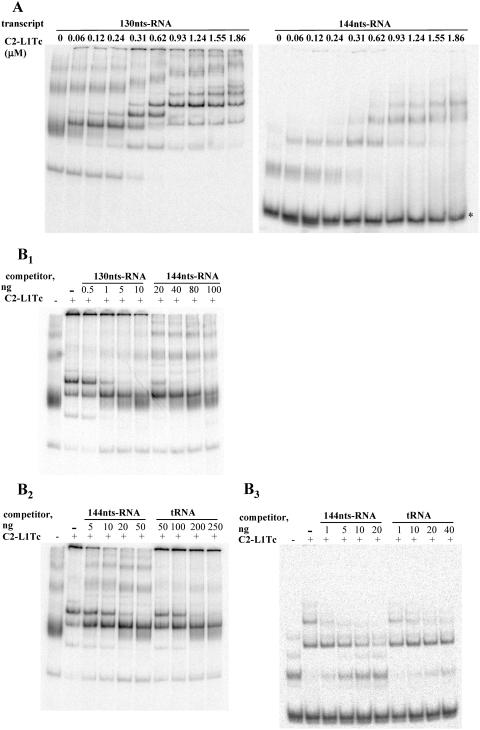
FIG. 3.
Binding of C2-L1Tc to RNA. (A) Binding of C2-L1Tc to RNAs of different sequences by EMSA. A total of 0.5 ng of 32P-labeled in vitro-transcribed 144-nt RNA (144nts-RNA) or 130-nt RNA (130nts-RNA) (0.65 nM and 0.72 nM, respectively) was preincubated with increasing amounts of C2-L1Tc (from 0.06 to 1.86 μM) at 37°C for 30 min. Reaction control was performed without protein. Dye solution was added, and samples were run on 5% native polyacrylamide gels. Results were visualized and quantified by phosphorimager analysis. Under the employed assay conditions, a fraction of the 144-nt RNA free form exhibits faster migration (*); C2-L1Tc apparently has a lower binding affinity for this RNA conformation. (B) Affinity of C2-L1Tc for different RNAs by EMSA. A total of 0.5 ng of 32P-labeled in vitro-transcribed 130-nt RNA (0.72 nM [B1, B2]) or 144-nt RNA (0.62 nM [B3]) was incubated with the indicated amount of unlabeled competitor, 130-nt RNA, 144-nt RNA, or tRNA, and 0.31 μM (B1, B2) or 0.67 μM (B3) of C2-L1Tc protein was added to the reaction mixture. After incubation at 37°C for 30 min, samples were resolved on 5% native polyacrylamide gels as described in the text.
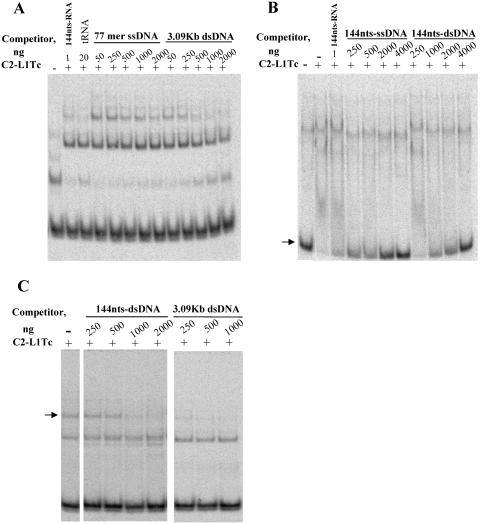
FIG. 4.
Binding of C2-L1Tc to DNA. EMSA was carried out after preincubation of 0.5 ng (0.65 nM) 32P-labeled 144-nt RNA (144nts-RNA) with different concentrations of unlabeled DNA competitor (as indicated): 77-mer ssDNA or 3.09-kb dsDNA (A), 144-nt ssDNA (144nts-ssDNA) or 144-nt dsDNA (144nts-dsDNA) (B), and 144-nt dsDNA or 3.09-kb dsDNA (C). Afterwards, 0.67 μM of C2-L1Tc (+) was added to each reaction. As a negative control, reactions were carried out without protein (−) or without competitor (−). One nanogram of cold 144-nt RNA (A, B) and 20 ng of tRNA (B) were also added as a reaction control. Reactions were performed at 37°C (A and C) or 4°C (B). The arrows indicate the free form that the 144-nt RNA adopts (B) and the complex of higher molecular weight which decreases in amount as the competitor concentration increases (C).
Annealing assays.
Annealing assays were performed as previously described by Martin and Bushman (23). Briefly, increasing protein concentrations were added to the annealing buffer (20 mM HEPES, pH 7.6, 1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100) containing 0.95 nM of γ-32P 5′-end-labeled oligonucleotide and 1 nM of its reverse complementary oligonucleotide. Reaction mixtures were incubated for 2 min at 37°C and stopped by the addition of a half-volume of stop mix (0.4 mg/ml tRNA, 0.2% SDS, 15% Ficoll, 0.2% bromophenol blue, and 0.2% xylene cyanol blue). dsDNA generation was monitored on native 15% polyacrylamide gels. The dried gel was analyzed with a phosphorimager system (Storm; Pharmacia).
DNA melting assays.
As indicated above, assays were performed as described previously (23). Briefly, preannealed duplex was mixed with the indicated amount of C2-L1Tc on ice. Tubes were incubated for 5 min at the indicated temperature, and the reaction was stopped as described above for the annealing assay. Electrophoretical analysis of the generated products was done as described above.
Strand exchange assays.
Strand exchange assays were performed as described previously by Martin and Bushman (23). When synthetic peptides other than 4821 were used, serial dilutions, from 20 μM to 0.1 μM, were assayed.
RESULTS
Cloning and purification of the C-terminal domain from LINE L1Tc.
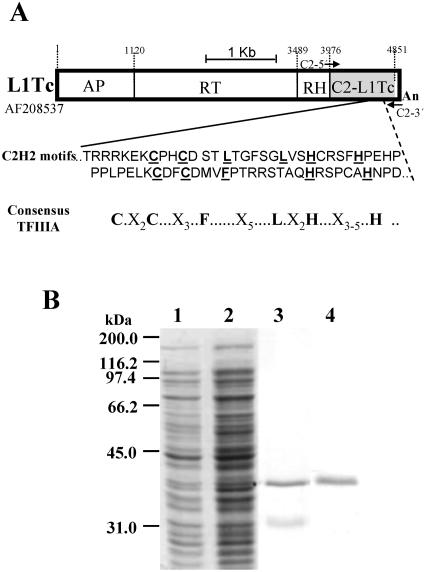
In an attempt to elucidate the function of the two C2H2 type cysteine motifs present in the C-terminal end of all reported L1Tc elements (21, 30), the region between nucleotides 3976 and 4851 of L1Tc (GenBank accession no. AF208537) (Fig. 1A) was cloned into the prokaryotic expression vector pCAS, generating the pCAS-C2-L1Tc plasmid. The deduced amino acid sequence derived from this sequence corresponds to a protein with a high content of positively charged amino acids and a theoretical isoelectric point of 8.70. Following salicylate addition to pCAS-C2-L1Tc-transformed TAP-F E. coli cells and 5 h of induction, a 37-kDa overproduced protein was observed by SDS-PAGE. The overexpressed C2-L1Tc protein was soluble in nondenaturing buffer. Thus, purification was performed by Ni-NTA chromatography under nondenaturing conditions. To eliminate an unknown nonabundant protein that copurified with C2-L1Tc (Fig. 1B, lane 3), the eluted protein fraction was subjected to gel filtration using a Superdex 75 column. As can be observed in Fig. 1B, lane 4, C2-L1Tc was recovered with a high efficiency and more than 95% purity.
FIG. 1.
Schematic representation of L1Tc and C2-L1Tc purification. (A) Scheme of the L1Tc element; the endonuclease (AP), reverse transcriptase (RT), RNase H (RH) (29), and zinc finger (C2H2) domains (21) are separated by thin black lines. The region comprising the C2-L1Tc domain is indicated as a gray box, and the corresponding sequence (the GenBank accession number is indicated at the left of the element) and the Zn finger consensus are shown below the scheme. (B) Coomassie blue staining of one of the preparations of recombinant C2-L1Tc used in NAC activity assays. Lane 1, noninduced TAP-F E. coli cells transformed with the pCAS-C2-L1Tc vector; lane 2, salicylate-induced TAP-F E. coli cells transformed with the pCAS-C2-L1Tc vector; lane 3, C2-L1Tc recombinant protein after Ni2+-NTA chromatography; lane 4, purified C2-L1Tc recovered after Superdex 75 fast protein liquid chromatography.
C2-L1Tc binds to different nucleic acids with different affinities.
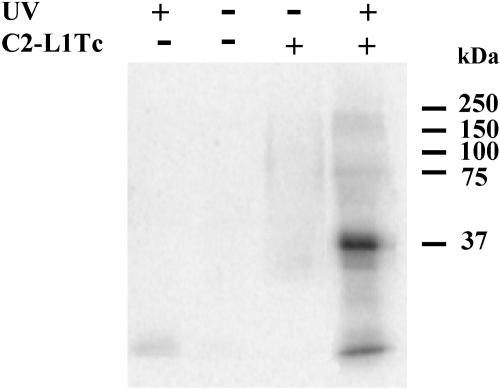
To determine the ability of C2-L1Tc to bind to RNA, a UV cross-linking assay was performed using C2-L1Tc and a 144-nt radiolabeled in vitro-transcribed RNA (144-nt RNA), 77 nt of which are derived from the 5′ end L1Tc and 67 nt of which are derived from the pGEMT vector (Fig. 2). A specific antibody raised against the recombinant protein identified the 37-kDa radiolabeled protein observed in the cross-linked reaction, corroborating that C2-L1Tc is able to bind RNA.
FIG. 2.
UV-induced cross-linking of C2-L1Tc to RNA. Fifty nanograms of 32P-labeled in vitro-transcribed 144-nt RNA was incubated with (+) or without (−) 2 μg of C2-L1Tc and subjected (+) or not subjected (−) to UV irradiation (252 nm) for 5 min. The complexes formed were resolved by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Protein was detected only in the lane preincubated with C2-L1Tc (+) and UV irradiated (+). Background of the radiolabeled RNA exposed to UV light is observed in the lane subjected to UV irradiation (+) incubated without C2-L1Tc (−) addition.
To determine differences in the RNA-binding capacity of C2-L1Tc to different transcripts, two in vitro-transcribed and radiolabeled transcripts of similar size, a 144-nt RNA and a 130-nt RNA not related to L1Tc, were incubated with different amounts of C2-L1Tc and compared by EMSA. In both cases, 0.06 μM of C2-L1Tc was enough to form slowly migrating bands (Fig. 3A). The similar behavior of C2-L1Tc on different transcripts would suggest that C2-L1Tc binds RNA in a sequence-independent manner. That the nature of the slowly migrating bands was due to binding of the RNA by C2-L1Tc was corroborated by the addition of loading dye containing 0.4 mg/ml tRNA and 0.2% SDS to the reaction mixture and by phenol-chloroform extraction to disrupt the RNA-protein interaction. In both cases, the labeled RNA migrated at a position similar to that of the control without protein (data not shown). Therefore, the mobility shift was not due to a conformational change of the RNA but rather to direct interaction of the RNA with C2-L1Tc.
When protein concentration was increased, additional and more abundant complexes of reduced motility were observed. This is probably due to the binding of several C2-L1Tc polypeptides to a single RNA molecule (Fig. 3A). However, it cannot be excluded that the reduced-mobility complexes are due to protein-protein interactions forming higher-molecular-weight complexes. As expected, the same result was obtained when a transcript corresponding to just the pGEMT sequence included in the 144-nt RNA was employed in the EMSA (data no shown).
To further investigate the binding specificity of C2-L1Tc to different nucleic acids, a variety of nucleic acids were examined by competition experiments using EMSA. Thus, the in vitro-transcribed and radiolabeled 130-nt RNA was incubated with the 130-nt RNA or 144-nt RNA unlabeled competitors in the presence of C2-L1Tc at a protein concentration that maintains some RNA in the free form and produces several slowly migrating complexes. Both RNA competitors displaced the radiolabeled RNA, but it required a 10- to 20-times-greater amount of 144-nt RNA than 130-nt RNA to obtain the same degree of competition (Fig. 3B1). Afterward, radiolabeled 130-nt RNA or 144-nt RNA was incubated with cold 144-nt RNA or tRNA as a competitor. As observed in Fig. 3B2 and B3, the amount of tRNA that is required to reduce the amount of the slowly migrating complex formed by the 32P-labeled 130-nt RNA (Fig. 3B2) or the 144-nt RNA (Fig. 3B3) transcript and C2-L1Tc, and thereby increasing free labeled transcript, is in both cases approximately 10 times higher that the amount of the 144-nt transcript necessary to produce the same effect. The 130-nt RNA and the 144-nt RNA adopt several conformations which exhibit a different mobility shift under native conditions. In both cases, C2-L1Tc has a different affinity for each one of them (Fig. 3A). Although it cannot be excluded that C2-L1Tc could have a preference for specific sequences, these data indicate that the RNA-binding capacity of C2-L1Tc depends on the RNA structure.
When the EMSA was performed with radiolabeled 144-nt RNA under the same conditions, but with the addition of either a 77-mer ssDNA or a 3.09-kb dsDNA as a competitor, 2,000 and 250 times more cold ssDNA and dsDNA, respectively, were necessary to reach the same quantity of free labeled RNA as when 1 ng of cold 144-nt RNA was used as the competitor (Fig. 4A). To overcome the possible influence of the nucleic acid size in the C2-L1Tc binding affinity, the same assay was carried out using a cold 144-nt dsDNA or an ssDNA of the same size generated by denaturing the 144-nt dsDNA as a competitor. In this case, the assay was performed at 4°C to avoid the influence of DNA renaturation and structure; this temperature causes a slight change in the electrophoretical motility of the free labeled RNA compared to that observed in the experiments carried out at 37°C. As shown in Fig. 4B, twice as much dsDNA than ssDNA was necessary to obtain the same degree of competition (see the quantity of free labeled RNA in each case), indicating that C2-L1Tc has a higher binding affinity for ssDNA than for dsDNA. Taken together, these data demonstrate that C2-L1Tc binds to RNA > tRNA > ssDNA > dsDNA.
Finally, to investigate the influence of the nucleic acid size on the affinity of C2-L1Tc for nucleic acids, EMSA was carried out using dsDNAs of 144 bp and 3,090 bp as competitors. As observed in Fig. 4C, a lower amount of the high-molecular-weight competitor is required to produce the same degree of competition than of the low-molecular-weight DNA. This finding suggests that C2-L1Tc binds to the DNA with positive cooperativity.
C2-L1Tc promotes annealing of complementary oligonucleotides.
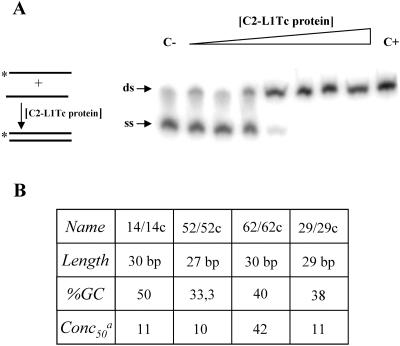
To investigate whether C2-L1Tc has the ability to accelerate annealing of two complementary oligonucleotides, a characteristic of proteins with NAC activity, increasing amounts of C2-L1Tc were incubated with complementary oligonucleotides, one of which was 5′ end radiolabeled. As schematized and shown in Fig. 5A, the C2-L1Tc recombinant protein accelerates the annealing of the 52/52c oligonucleotide pair in a concentration-dependent manner. The annealing assay was also carried out employing the oligonucleotide pairs 29/29c, 14/14c, and 62/62c. The 29/29c pair, used in the NAC activity determination of mouse L1 ORF1p, has no significant sequence similarity with the L1Tc sequence. Primer pairs 14/14c and 62/62c, as the 52/52c primer pair, contain previously identified target site duplications derived from L1Tc insertions into the T. cruzi genome (30). In all cases, C2-L1Tc accelerated DNA duplex formation independently of the oligonucleotide length or GC content (Fig. 5B). Fifty percent of duplex formation is reached with 10 to 11 nM of C2-L1Tc except for the 62/62c pair, where a higher concentration of C2-L1Tc is required. C2-L1Tc annealing activity is slightly lower than that described for mouse L1 ORF1, where 50% of the duplex is generated with less than 10 nM (23). In addition, the retention of single-stranded DNA substrates observed with high concentrations of the murine ORF1 protein (23) was not detected when C2-L1Tc was used.
FIG. 5.
Annealing assays. (A) Scheme of the assay. Asterisks indicate the 32P-labeled strand. Shown are the results obtained when 0.95 nM of the 32P-labeled oligonucleotide (*52) and 1.0 nM of its complementary primer (52c) were used. The triangle indicates increasing amounts of C2-L1Tc as a threefold serial dilution from 0.3 to 635 nM. C− corresponds to the reaction carried out without C2-L1Tc, and C+ indicates the preannealed *52/52c pair, used as a mobility control of the DNA duplex. The arrows indicate the single-stranded (ss) and double-stranded (ds) state of the labeled (*) 52 oligonucleotide. (B) Table summarizing the annealing activity of the C2-L1Tc when primer pairs 14/14c, 52/52c, 62/62c, and 29/29c were used. Oligonucleotide length, percentage of GC content (%GC) in each oligonucleotide, and protein concentration (nM) required to reach 50% of duplex formation (Conc50) are indicated.
C2-L1Tc stabilizes perfect DNA duplexes.
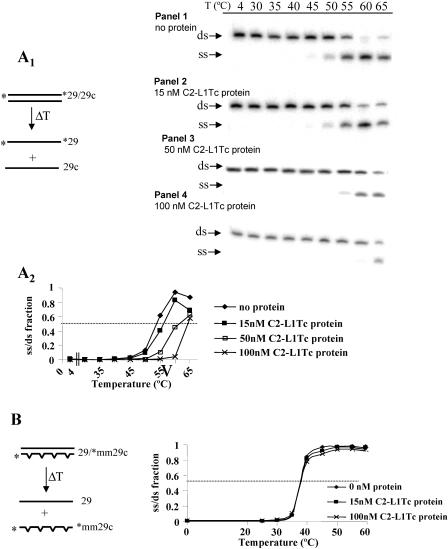
To further determine the NAC activity associated with C2-L1Tc, the effect that C2-L1Tc has on the melting temperature of DNA duplexes was analyzed. Thus, a perfect 29-nucleotide-long preannealed DNA duplex (*29/29c substrate [the asterisk indicates that the oligonucleotide is 32P 5′ end labeled.) was incubated with different concentrations of C2-L1Tc at increasing temperatures. As shown in Fig. 6A1, the addition of C2-L1Tc stabilizes the DNA duplex, preventing DNA denaturation in a concentration-dependent way (Fig. 6A1, panels 2 to 4). With 50 nM and 100 nM of C2-L1Tc, an increase of more than 5°C and 10°C, respectively, in the melting temperature of the perfect duplex was observed (Fig. 6A1, panel 3 and 4, and A2). We next analyzed the effect of C2-L1Tc on the melting temperature of a mismatched duplex. For this purpose, the preannealed 29/*mm29c substrate, which contains four internal mismatches, was incubated with C2-L1Tc at different temperatures (scheme in Fig. 6B). As shown in Fig. 6B, under our experimental conditions, C2-L1Tc has no influence on the melting temperature of this imperfect DNA duplex. In contrast to what was observed for C2-L1Tc, the NAC activity of mouse L1 ORF1 lowered the melting temperature of a mismatched duplex, probably due to its high affinity for ssDNA that allowed it to recognize local single-strand regions in the mismatched duplex (23).
FIG. 6.
Melting assays. (A1) A scheme of the assay is represented at the left of the figure. Asterisks indicate the 32P-labeled strand of the *29/29c duplex. Panels showing duplex disruption, as a function of the temperature (T) (in degrees Celsius), using different concentrations of C2-L1Tc are illustrated on the right. The assay was carried out with 1 nM of 32P-labeled 29/29c duplex without protein (panel 1) or with 15 nM (panel 2), 50 nM (panel 3), or 100 nM (panel 4) of C2-L1Tc for 5 min at the temperatures indicated above the panels. Single-stranded (ss) and double-stranded (ds) DNA motilities are indicated with arrows. (A2) The graph represents the single-stranded/double-stranded fraction of 32P-labeled *29 and 29c primers as a function of temperature and C2-L1Tc concentration. Quantification was performed using Image Quant software (Pharmacia). (B) Scheme of the assay carried out with the mismatched duplex 29/*mm29c. The radiolabeled primer is indicated with an asterisk. The graph represents the single-stranded/double-stranded fraction of the 29/*mm29c duplex as a function of temperature and C2-L1Tc concentration. Quantification was performed using ImageQuant software (Pharmacia).
C2-L1Tc induces generation of the thermodynamically most stable duplex DNA.
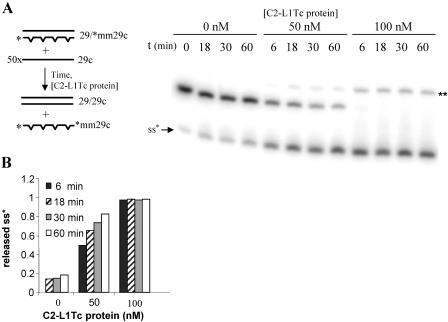
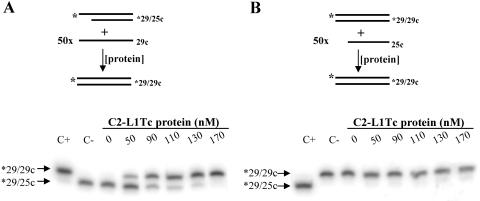
We further explored NAC activities of C2-L1Tc by monitoring whether C2-L1Tc can facilitate strand exchanges that generate the thermodynamically most stable DNA duplex. Two different assays were performed. Thus, preannealed mismatched duplex DNA (29/*mm29c) was incubated in the absence or in the presence of increasing amounts of C2-L1Tc with a 50-fold molar excess of the single-strand complementary perfect primer 29c (scheme in Fig. 7A). As shown in Fig. 7A, the perfect primer displaces the mismatched radiolabeled oligonucleotide from the preannealed mismatched duplex. Thus, a perfect DNA duplex (29/29c) is formed, and the single-strand radiolabeled mismatched oligonucleotide (*mm29c) is liberated. The generation of the most stable DNA duplex (29/29c) depends on the C2-L1Tc concentration. We observed that when 50 nm of C2-L1Tc is used, 80% of perfect DNA duplex is formed in 60 min, while at a 100 nm of the C2-L1Tc, the total release of the mismatched single-strand DNA occurs in less than 6 min after protein addition (Fig. 7B). The ability of C2-L1Tc to facilitate strand exchange was also tested using a DNA duplex formed by a radiolabeled 29-mer oligonucleotide (*29) and a 25-mer complementary primer (25c) which was challenged with a 50-fold excess of ssDNA 29-mer complementary primer (29c) as represented in Fig. 8A. The addition of C2-L1Tc again induces the formation of the most stable dsDNA duplex, *29/29c (Fig. 8A). In contrast, when the preannealed dsDNA *29/29c duplex was challenged with a 50-fold excess of ssDNA oligonucleotide 25c, formation of the *29/25c duplex was not observed (Fig. 8B). Instead, the labeled oligonucleotide remained in the initial *29/29c duplex. In both cases, the *29/29c duplex was the most thermodynamically favored form (Fig. 8A and B). No strand exchange was observed when the preannealed *29/25c complex and 29c primer were incubated with the fraction eluted under similar conditions to those described for C2-L1Tc from extracts of E. coli overexpressing RT-L1Tc protein (data not shown).
FIG. 7.
Strand exchange assay employing mismatched DNA duplexes. (A) Scheme of the strand exchange assay using 1 nM of 29/*mm29c preformed duplex and a 50-fold molar excess of cold 29c primer. Reactions were performed at 37°C using different concentrations of C2-L1Tc and different periods of incubation (as indicated). The single-strand form of the radiolabeled primer (ss*) and the alternative structure that the free *mm29C primer can adopt (**), as previously described (23), are indicated. (B) Quantification of the released single-stranded 32P-labeled *mm29c oligonucleotide was performed using ImageQuant software (Pharmacia).
FIG. 8.
Strand exchange assay employing perfect DNA duplexes. Shown are a scheme and results of the strand exchange assay using preformed perfect duplexes with different numbers of paired bases. A total of 1 nM of preannealed *29/25c (A) or *29/29c (B) DNA duplex was challenged with a 50-fold molar excess of 29c (A) or 25c (B) oligonucleotide and incubated for 15 min with different amounts of C2-L1Tc. *29/29c and *29/25c preformed duplexes were loaded in both cases as size standards.
Mapping C2-L1Tc to determine the region responsible for NAC activity.
Analysis of the C2-L1Tc deduced amino acid sequence revealed positively charged regions flanking the two L1Tc C2H2 motifs and the presence of two RRR stretches located immediately upstream and downstream of the C2H2 motifs (Fig. 9A). An RRRKEK stretch, described as a nuclear localization signal (NLS) and a DNA binding region (6), has been also found upstream of the first C2H2 domain (Fig. 9A). To define the region responsible for the C2-L1Tc NAC activity, strand exchange assays employing perfect DNA duplexes were carried out as described above using synthetic peptides that cover the most relevant regions of C2-L1Tc (Fig. 9A) and C2-L1Tc-derived peptides bearing mutated regions (Table 1). Thus, the peptides tested were as follows: peptide 4821, which covers the two C2-L1Tc C2H2 motifs; peptides 5016 and 10987, which cover the first and second C2H2 motif, respectively, from C2-L1Tc; and peptides 5015 and 5020, which flank the C2H2 motifs and contain the RRR and RRRKEK stretches or one RRR region, respectively. Peptide 4814, which has a positive-charge distribution similar to that of peptide 4821, was employed as a control. The formation and maintenance of the thermodynamically most stable DNA duplex (*29/29c) were induced by all the assayed peptides except for the 4814 control peptide and the 5020 peptide, which did not exhibit NAC activity. The NAC activities of all the assayed peptides are summarized in Table 2. The peptide containing the two zinc fingers with the NLS, peptide 4821, was the most active peptide, showing an NAC activity only three times less than the full protein (Fig. 9B1 and B2). Peptide 5015, which bears an RRR stretch and the NLS and which has an isoelectric point similar to that of the inactive peptide 5020, exhibits an NAC activity seven times lower than the activity of the recombinant protein (Fig. 9A and C1 and Tables 1 and 2). Peptide 5016, which contains the first zinc finger and a partial NLS, was 21 times less active than the intact protein (Fig. 9A and C2 and Tables 1 and 2). To confirm the sequence requirements necessary for optimal NAC activity of C2-L1Tc, some point mutations were produced based on the sequence from peptides 5015 and 5016 (see Table 1 for details). Thus, when the RRR stretch was eliminated from peptide 5015, in spite of maintaining the NLS, the resultant peptide, 5030, is inactive under the experimental conditions employed (Fig. 9C1 and Tables 1 and 2). Deletion of the NLS from peptide 5016, creating peptide 5032, abolishes the NAC activity of the peptide, although the first zinc finger remains intact (Fig. 9C2 and Tables 1 and 2). Substitution of the CCHH motif for SSHH in peptide 5016, i.e., peptide 5031, reduces the NAC activity 1.6 times (Fig. 9C2 and Tables 1 and 2). However, the addition of the full NLS sequence to peptide 5016 produces the 5033 peptide, a very active peptide, which is just 5.7 times less active than C2-L1Tc (Fig. 9C1 and Tables 1 and 2). Thus, it appears that it is necessary for the NAC activity of C2-L1Tc to have the NLS sequence or several residues contained in the NLS motif such as RR or RKEK and at least one zinc finger or a basic region, such as RRR. Moreover, these data indicate that the zinc finger domains are the most relevant domains for NAC activity of C2-L1Tc, although they are by themselves neither essential nor sufficient.
TABLE 2.
Nucleic acid chaperone activity of C2-L1Tc- and C2-L1Tc-derived peptides in strand exchange assaya
| Peptide name | pI | Conc50 (μM) |
|---|---|---|
| C2-L1Tc | 8.70 | 0.07 |
| 4821 | 8.93 | 0.25 |
| 10987 | 7.06 | 2.9 |
| 5020 | 12.18 | NO |
| 5015 | 11.27 | 0.51 |
| 5030 | 9.30 | NO |
| 5016 | 8.68 | 1.5 |
| 5031 | 9.31 | 2.4 |
| 5032 | 6.99 | NO |
| 5033 | 9.61 | 0.4 |
| 4814 | 9.07 | NO |
The name, theoretical isoelectric point (pI), determined at the Expasy website (http://us.expasy.org) according to the method of Bjellqvist et al. (2), and concentration of C2-L1Tc and synthetic peptides required for reaching 50% of the *29/29c duplex formation (Conc50) are summarized. These data are the averages of at least three different experiments. Although a slight variability was observed between them, the relative activity of the different peptides was invariable. NO, no observed activity.
DISCUSSION
The T. cruzi L1Tc element has two C2H2 type zinc finger domains (21) similar to the TFIIIA transcription factor motifs at its carboxy-terminal end. The ORF1-derived protein from Drosophila I-factor LINE, which possesses a region with CCHC cysteine motifs, has NAC activity (9). The mouse LINE-1 ORF1 and the ORF1 from Saccharomyces cerevisiae Ty1 LTR retrotransposon also code for a protein with NAC activity, although they do not share recognizable similarity with gag proteins, except that they present a high content of positively charged amino acids (8, 23). All of these proteins exhibit nucleic acid binding capacity in a sequence-independent manner (8, 9, 17).
In this study, using an EMSA, we show that C2-L1Tc binds both RNA and DNA where several C2-L1Tc-RNA complexes of different sizes are observed. C2-L1Tc does exhibit affinity preferences for particular nucleic acids. Thus, experiments using different types of nucleic acid competitors indicate that C2-L1Tc binds RNA better than tRNA and that it also discriminates between ssDNA and dsDNA. Moreover, C2-L1Tc presents different binding affinities for particular conformations of RNAs. These data also suggest that C2-L1Tc binds DNA with positive cooperativity. The chaperone activity of C2-L1Tc and its stronger affinity for RNA may be implicated in the transposition mechanism of the L1Tc, which transposes via an RNA intermediate. C2-L1Tc could mediate the production of a ribonucleoprotein particle composed of the L1Tc RNA intermediate and the proteins encoded by the LINE which constitute the transposable machinery. Thus, C2-L1Tc could transport L1Tc mRNA into the nucleus for reverse transcription and integration of the newly synthesized element. The ability of the protein encoded by the ORF1 from mammalian L1 and the I-factor gag-like protein to bind to nucleic acids has suggested a role for these proteins in ribonucleoprotein particle formation (16, 32).
Here, we demonstrate that C2-L1Tc has NAC activity and shares some NAC properties with murine ORF1p. However, slight differences are observed between both NAC proteins. While C2-L1Tc accelerates annealing of complementary oligonucleotides at all concentrations tested, the murine ORF1p at a high concentration retains DNA in the single-stranded form in annealing assays (23). This is probably due to the C2-L1Tc binding affinity which is only two times higher for ssDNA than for dsDNA, while murine ORF1p showed a binding affinity 100-fold higher for ssDNA than for dsDNA. In addition, C2-L1Tc does not affect melting temperature of mismatched duplexes, while ORF1p lowers the melting temperature of a mismatched duplex (23). This can be explained by the stronger affinity of ORF1p for ssDNA, a property that could allow ORF1p to recognize the internal mismatches of the preformed mismatched duplexes as ssDNA and to retain it in the single-stranded form (16).
C2-L1Tc may neutralize the negative charge of the DNA phosphodiester backbone, facilitating association of DNA strands, stabilizing the formed duplex by charge shielding, and increasing the melting temperature of perfect duplex when complementary primers are used. Moreover, C2-L1Tc allows the formation of the most stable duplex, promoting the exchange of strands when both 32P29-25c and 29-32Pmm29 preformed duplex and an excess of single-stranded complementary 29c are used, even though it has no effect on the melting temperature of 29-32Pmm29 mismatched duplexes in melting assays. This result suggests that C2-L1Tc produces the strand exchange by means of specific interactions with nucleic acids and by a transient base-pair destabilization, thereby allowing a subsequent partial base pairing and displacement of the less homologous primer. An ability to destabilize the helical structure of DNA by altering the cooperativity of the helix-coil transition has been described previously for the nucleocapsid (NC) protein of human immunodeficiency virus type 1 (HIV-1) (41).
The existence of NAC activity associated with C2-L1Tc, I-factor ORF1, and L1 ORF1p reinforces the implication of an NAC activity in the transposition mechanism of non-LTR retrotransposons. During TPRT, when the chromosomal DNA is cleaved at the target site, an NAC could promote strand exchange and stabilize the priming between the L1Tc mRNA poly(A) tail and the T-rich region located at the cleaved target site. This pairing allows to the 3′ end of the cleaved target site to act as a primer for reverse transcription and first-strand cDNA synthesis. Subsequently, NAC activity could facilitate the strand exchange required by TPRT to synthesize second-strand DNA as well. This strand exchange would imply template switching from the 3′ end of synthesized first-strand cDNA to the cleaved target of the positive DNA strand, which would function as a primer for the second-strand cDNA synthesis (22, 23). As recently described for some lentiviruses, the non-LTR NAC activity may also cooperate with the RNase H activity encoded by the element in order to unblock the 5′ end of the generated cDNA, allowing a more efficient transfer of the second strand (35).
Using peptides covering the most relevant regions of the C2-L1Tc, we have shown that the two C2H2 cysteine motifs and the C2H2 motif upstream region are involved in NAC activity in strand exchange experiments. Peptide 4821, which covers both C2H2 motifs and NLS (RRRKEK), is the most active of all the assayed peptides (protein concentration required to reach 50% of duplex formation [Conc50], 0.25 μM), being only 3.5 times less active than C2-L1Tc (Conc50, 0.07 μM). It is followed by the 5033 peptide (Conc50, 0.4 μM), which bears the NLS and the first C2H2 domain. Partial (RR) or complete deletion of the NLS, peptides 5016 and 5032, produces, respectively, a decrease (Conc50, 1.5 μM) and the complete loss of the NAC activity in spite of the presence of the first Zn finger. Peptide 10987, which covers the second C2H2 motif and which has an isoelectric point similar to that of the inactive peptide 5032, shows some NAC activity (Conc50, 2.9 μM). Interestingly, the second zinc finger contains a diarginine motif that is not present in the first zinc finger. Different contributions of the zinc finger domains to NAC activity have also been observed in the NC HIV protein (41). Substitution of the CCHH motif for SSHH in peptide 5016, creating peptide 5031, reduces but does not destroy the NAC activity (Conc50, 2.4 μM). The third more active peptide is peptide 5015 (Conc50, 0.51 μM), which bears an RRR stretch and the NLS. However, deletion of the RRR stretch from this peptide to create the 5030 peptide eliminates the NAC activity, although the NLS is maintained. On the other hand, peptide 5020, which maps downstream of the zinc fingers, loses NAC activity in spite of containing RRR. It has been described that while deletion of the Zn fingers in NC HIV has little or no effect on the viral RNA-annealing activity of the NAC protein, the deletion of short peptides containing basic residues flanking Zn fingers leads to a complete loss of this activity (10). Interestingly, there are conserved basic residues in the ORF1 proteins from human, rabbit, rat, and mouse L1 elements, where no zinc finger motifs have been observed; substitution of two arginines with alanines in the human L1 element reduces retrotranposition at least 100-fold (25).
Taking all of these results together and considering that all tested peptides show lower activity than the intact protein, we suggest that the Zn fingers and the basic residues flanking C2-L1Tc cooperate in the NAC activity of the protein. For C2-L1Tc NAC activity, it is essential to have the presence of both a basic stretch such as RR or RKEK accompanied by at least one zinc finger or by another basic region, such as RRR. Moreover, these data indicate that the zinc finger domains, though relevant, are neither essential nor sufficient to generate the thermodynamically most stable DNA duplex under the tested conditions. They are probably required for some aspects of nucleic acid chaperone function as described previously for HIV-1 NC (14). In HIV-1 NC, changes in the structure of the zinc fingers reduce the cooperativity of the helix-coil transition, thereby decreasing in vitro chaperone activity (41).
To our knowledge, this is the first description of NAC activity mediated by a protein containing C2H2 zinc finger motifs. C2H2 motifs similar to those of the TFIIIA transcription factor family have also been described in some non-LTR elements (5, 12). Zinc finger proteins are involved in many fundamental cellular processes such as replication and repair, translation, programmed cell death, and metal regulation (18). Two proteins of T. cruzi that share zinc finger motifs with a diverse range of RNA-binding proteins have been described. These proteins form complexes with synthetic oligoribonucleotides and are implicated in the control of trypanosome differentiation (26).
Since an NAC activity is essential for transposition of mobile retroelements which code for it (7, 25), the presence of this activity in L1Tc confers an especial relevance to the element due to its autonomy in terms of mobility and its potential biological function in the T. cruzi genome. L1Tc, the best-represented LINE in T. cruzi, is present in most, if not all, of the chromosomes from the analyzed T. cruzi strains (29). Mobile elements expand the host genomes and generate a high degree of plasticity to the genome with regard to both structure and function, improving the adaptability of the host organisms. In spite of the fact that it is not clearly understood which are the events that may have shaped the current architecture of trypanosomatid genomes through evolution, there are several lines of evidence that indicate that retrotransposons play an essential role in genome organization and may be one of the molecular means that the parasite has to survive in the human host (11).
Acknowledgments
We are grateful to M. E. Patarroyo, E. Torres, and F. Guzman for peptide synthesis and purification. We thank M. Caro and D. Branciforte for technical assistance and A. Berzal and his team for assistance in RNA transcription assays.
This work was supported by FIS 01/3148 and RICET C03-04 from Fondo de Investigación Sanitaria, MSC, and BMC2003-00834 from Plan Nacional I+D+I (MEC), Spain. S.R.H. was supported by an MEC predoctoral fellowship (FPU), J.L.G.-P. was supported by a Fundación Ramón Areces predoctoral fellowship, and S.L.M. was supported by NIH grant GM 40367.
REFERENCES
- 1.Barroso-del Jesus, A., M. Tabler, and A. Berzal-Herranz. 1999. Comparative kinetic analysis of structural variants of the hairpin ribozyme reveals further potential to optimize its catalytic performance. Antisense Nucleic Acid Drug Dev. 9:433-440. [DOI] [PubMed] [Google Scholar]
- 2.Bjellqvist, B., G. J. Hughes, C. Pasquali, N. Paquet, F. Ravier, J. C. Sanchez, S. Frutiger, and D. F. Hochstrasser. 1993. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023-1031. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bringaud, F., J. L. Garcia-Perez, S. R. Heras, E. Ghedin, N. M. El-Sayed, B. Andersson, T. Baltz, and M. C. Lopez. 2002. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 124:73-78. [DOI] [PubMed] [Google Scholar]
- 5.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 6.Cokol, M., N. Rajesh, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1(5):411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristofari, G., and J. L. Darlix. 2002. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acids Res. Mol. Biol. 72:223-268. [DOI] [PubMed] [Google Scholar]
- 8.Cristofari, G., D. Ficheux, and J. L. Darlix. 2000. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 275:19210-19217. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, A., E. Hartswood, T. Paterson, and D. J. Finnegan. 1997. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 16:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rocquigny, H., C. Gabus, A. Vincent, M. C. Fournie-Zaluski, B. Roques, and J. L. Darlix. 1992. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 89:6472-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donelson, J. E. 1996. Genome research and evolution in trypanosomes. Curr. Opin. Genet. Dev. 6:699-703. [DOI] [PubMed] [Google Scholar]
- 12.Eickbush, T. H. 2002. R2 and related site-specific non-long terminal repeat retrotransposons, p. 813-835. In N. Craig, R. Craggie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 13.Garcia-Perez, J. L., C. I. Gonzalez, M. C. Thomas, M. Olivares, and M. C. Lopez. 2003. Reverse transcriptase activity in a protein encoded by the non-LTR retrotransposon L1Tc from Trypanosoma cruzi. Cell. Mol. Life Sci. 60:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J., T. Wu, B. F. Kane, D. G. Johnson, L. E. Henderson, R. J. Gorelick, and J. G. Levin. 2002. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 76:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houghten, R. A. 1985. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc. Natl. Acad. Sci. USA 82:5131-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolosha, V. O., and S. L. Martin. 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94:10155-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolosha, V. O., and S. L. Martin. 2003. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J. Biol. Chem. 278:8112-81127. [DOI] [PubMed] [Google Scholar]
- 18.Krishna, S. S., I. Majumdar, and N. V. Grishin. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31:532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 20.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 21.Martin, F., C. Marañon, M. Olivares, C. Alonso, and M. C. Lopez. 1995. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 247:49-59. [DOI] [PubMed] [Google Scholar]
- 22.Martin, S. L., D. Branciforte, D. Keller, and D. L. Bain. 2003. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. USA 100:13815-13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, J. V., and N. Gilbert. 2002. Mammalian LINE-1 retrotransposons and related elements, p. 836-869. In N. Craig, R. Craggie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 25.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 26.Morking, P. A., B. M. Dallagiovanna, L. Foti, B. Garat, G. F. Picchi, A. C. Umaki, C. M. Probst, M. A. Krieger, S. Goldenberg, and S. P. Fragoso. 2004. TcZFP1: a CCCH zinc finger protein of Trypanosoma cruzi that binds poly-C oligoribonucleotides in vitro. Biochem. Biophys. Res. Commun. 319:169-177. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira, N. 1987. Biological and molecular aspects of Trypanosoma cruzi, p. 125-134. In M. E. Patarroyo, J. B. Zabriskie, and D. Pizano-Salazar (ed.), Modern biotechnology and health: perspectives for the year 2000. Academic Press, London, United Kingdom.
- 28.Olivares, M., C. Alonso, and M. C. Lopez. 1997. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J. Biol. Chem. 272:25224-25228. [DOI] [PubMed] [Google Scholar]
- 29.Olivares, M., J. L. Garcia-Perez, M. C. Thomas, S. R. Heras, and M. C. Lopez. 2002. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J. Biol. Chem. 277:28025-28030. [DOI] [PubMed] [Google Scholar]
- 30.Olivares, M., M. C. Thomas, A. Lopez-Barajas, J. M. Requena, J. L. Garcia-Perez, S. Angel, C. Alonso, and M. C. Lopez. 2000. Genomic clustering of the Trypanosoma cruzi nonlong terminal L1Tc retrotransposon with defined interspersed repeated DNA elements. Electrophoresis 21:2973-2982. [DOI] [PubMed] [Google Scholar]
- 31.Olivares, M., M. C. Thomas, C. Alonso, and M. C. López. 1999. The L1Tc, long interspersed nucleotide elements from Trypanosoma cruzi, encodes a protein with 3′ phosphatase and 3′ phosphodiesterase enzymatic activities. J. Biol. Chem. 274:23883-23886. [DOI] [PubMed] [Google Scholar]
- 32.Ostertag, E. M., and H. H. Kazazian, Jr. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35:501-538. [DOI] [PubMed] [Google Scholar]
- 33.Puentes, F., F. Guzman, V. Marin, C. Alonso, M. E. Patarroyo, and A. Moreno. 1999. Leishmania: fine mapping of the leishmanolysin molecule's conserved core domains involved in binding and internalization. Exp. Parasitol. 93:7-22. [DOI] [PubMed] [Google Scholar]
- 34.Requena, J. M., M. C. López, and C. Alonso. 1996. Genomic repetitive DNA elements of Trypanosoma cruzi. Parasitol. Today 12:279-283. [DOI] [PubMed] [Google Scholar]
- 35.Roda, R. H., M. Balakrishnan, M. N. Hanson, B. M. Wohrl, S. F. Le Grice, B. P. Roques, R. J. Gorelick, and R. A. Bambara. 2003. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J. Biol. Chem. 278:31536-31546. [DOI] [PubMed] [Google Scholar]
- 36.Sarin, V. K., S. B. Kent, J. P. Tam, and R. B. Merrifield. 1981. Quantitative monitoring of solid-phase peptide synthesis by the ninhydrin reaction. Anal. Biochem. 117:147-157. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, M. C., J. L. García-Pérez, C. Alonso, and M. C. López. 2000. Molecular characterization of KMP11 from Trypanosoma cruzi: a cytoskeleton-associated protein regulated at translational level. DNA Cell Biol. 19:47-57. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez, M., C. Ben-Dov, H. Lorenzi, T. Moore, A. Schijman, and M. J. Levin. 2000. The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. USA 97:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickstead, B., K. Ersfeld, and K. Gull. 2003. Repetitive elements in genomes of parasitic protozoa. Microbiol. Mol. Biol. Rev. 67:360-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilhelm, M., and F. X. Wilhelm. 2001. Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci. 58:1246-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, M. C., R. J. Gorelick, and K. Musier-Forsyth. 2002. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl. Acad. Sci. USA 99:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, C., S. Sridhara, J. C. A. Bardwell, and U. Jakob. 2000. Heating greatly speeds Coomassie blue staining and destaining. BioTechniques 28:426-432. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization. 1995. Twelfth Programme Report of the UNPD/World Bank/W.H.O. Special Program for Research and Training in Tropical Diseases, p. 125-126. In World Health Organization (ed.), W.H.O.—tropical disease research. World Health Organization, Geneva, Switzerland.16227955