Abstract
The establishment of bacterial communities in two healthy babies was examined for more than the first 10 months of life by monitoring 16S ribosomal DNA (rDNA) diversity in fecal samples by PCR and denaturing gradient gel electrophoresis (DGGE) and by analyzing the sequences of the major ribotypes. DGGE profiles of the dominant populations in the intestines of the infants were obtained by analyzing daily or weekly fecal samples. After delivery, the germfree infant gastrointestinal tracts were rapidly colonized, and the succession of bacteria in each ecosystem was monitored. During the first few days of life the profiles were simple, but they became more complex as the bacterial diversity increased with time in both babies. Clone libraries of amplified 16S rDNA fragments from baby feces were constructed, and these libraries allowed identification of the bacterial types by comparative DNA sequence analysis; the bacteria identified included members of the genera Bifidobacterium, Ruminococcus, Enterococcus, Clostridium, and Enterobacter. Species most closely related to the genera Bifidobacterium and Ruminococcus in particular dominated the intestinal microbiota based on the stability over time and the numbers, as estimated by the intensities of the bands. However, 19 of the 34 cloned rDNA sequences exhibited less than 97% identity with sequences of known bacteria or cloned sequences in databases. This study showed that using PCR-DGGE and 16S rDNA sequence analysis together resulted in a dynamic description of bacterial colonization in the infant intestinal ecosystem and allowed visualization of bacteria that are difficult to cultivate or to detect by other methods.
During the birth process and soon thereafter, bacterial colonization of a previously germfree infant gut intestinal tract begins. Since the pioneering study of Tissier in 1900 (39), several studies have described bacterial succession in this system based on analysis of the microbiota in infants’ stools (reviewed in references 8, 9, and 19). In full-term vaginally delivered infants, colonization starts immediately after delivery, and enterobacteria and streptococci appear in feces. The composition of the gut microbiota is profoundly influenced by the diet of the infant. In breast-fed infants, the microbiota is rapidly dominated by bifidobacteria, and a more diverse microbiota develops only after dietary supplementation commences. In contrast, the intestines of formula-fed infants are colonized by members of a variety of bacterial genera, including enterobacterial genera, Streptococcus, Bacteroides, and Clostridium, in addition to members of the genus Bifidobacterium. This characteristic succession continues until after weaning, when a dense, complex, more stable microbiota becomes established. The previous studies, which have relied almost exclusively on the use of culturing methods, have generated our current understanding of gut microbiology and ecology in infants. Normal colonization by the human intestinal commensal microbes stimulates a range of important functions, from postnatal intestinal maturation to maintenance of the mucosal barrier and nutrient absorption (6, 15). Adequate knowledge of the types of microorganisms, as well as the events that influence the timing of colonization, may provide opportunities to modulate the microbiota when modulation is necessary to enhance these functions.
Gastrointestinal microbial ecology is experiencing a revival because of the development of molecular techniques, particularly techniques based on 16S rRNA genes, that are used to study complex bacterial ecosystems (37, 40). A major proportion of the microbiota in the digestive tract is composed of a large number of diverse anaerobic bacteria that cannot be cultivated on the existing selective or nonselective media. Anaerobic culturing is also slow and limits studies of population dynamics. Therefore, the use of molecular methods to analyze the gastrointestinal ecosystem allows more complete and rapid assessment of the biodiversity in this ecosystem (11, 26, 27, 36, 43). So far, only a limited number of infant microbiota studies have been performed with molecular techniques. Quantitative results that are more accurate than classic plate count results have been obtained by using fluorescent in situ hybridization (FISH) probes targeting RNA of specific groups of bacteria. Using a repertoire of FISH probes, researchers have confirmed the differences in microbiota succession between breast-fed and formula-fed infants (12, 13, 21). However, only bacterial groups recognized by the currently available probes can be detected since the design of probes depends on prior 16S ribosomal DNA (rDNA) sequence information. PCR and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA amplicons from babies’ feces have been used to investigate the contribution of bacteria not detected by culturing to necrotizing enterocolitis in preterm infants (23). Although an association between uncultured bacteria and necrotizing enterocolitis could not be established, a species related to Streptococcus salivarius, which did not survive in a culturable form in feces, was detected in a large proportion of the infants studied.
DGGE and temperature gradient gel electrophoresis (TGGE) (26) are molecular fingerprinting techniques that are being used more frequently in microbial ecology. When combined with sequencing of 16S rDNA clones, they permit determination of the taxonomic affiliations of members of the microbial community. Analyses of amplified 16S rDNA fragments by DGGE and TGGE are rapid and efficient approaches for obtaining profiles of the complex intestinal microbial community structure (2, 43). These methods are especially valuable for monitoring shifts in community structure that occur in response to environmental perturbations, such as diet (29, 32, 38). In the present study, we investigated the feasibility of using PCR-DGGE to monitor rapid changes in the populations in the intestinal tracts of babies. We describe for the first time the succession of bacterial communities in babies’ feces for the first 10 months of life as shown by progressive DGGE profiles. Construction of 16S rDNA clone libraries from fecal samples allowed tentative identification of many dominant species and also revealed numerous rDNA sequences that did not correspond closely to sequences of previously described microorganisms.
MATERIALS AND METHODS
Subjects and preparation of samples.
Informed consent was obtained from the parents of the children used in this study. Two healthy, full-term, unrelated baby boys, designated babies D and L, who were vaginally delivered, participated in this study. Both baby D and baby L were breast fed immediately after birth. Baby D received only breast milk until the weaning period, which began on day 130. Additionally, infant formula was added to the diet starting on day 150 and gradually replaced breast feeding, which was completely withdrawn by day 200. Baby L was breast fed until day 17, when he received the first formula food, and then the proportion of formula milk was progressively increased with time; solid food was given after 3 months.
DNA extraction.
Fresh fecal samples were collected from both babies every day for the first 2 weeks of life and then at least twice monthly for 10 to 12 months. These samples were stored at 4°C until further processing, which occurred within 6 h after defecation. For the daily samples 180 mg (wet weight) was homogenized in 1 ml of TN 150 (10 mM Tris-HCl [pH 8], 150 mM NaCl), and 90 mg of each weekly fecal sample was homogenized. DNA was isolated from the fecal samples essentially as previously described (43). Briefly, 0.5 ml of an homogenized sample was added to 150 μl of acid phenol with 0.3 g of zirconium beads (diameter, 0.1 mm), and the suspension was treated at 5,000 rpm for 3 min with a mini bead beater (Biospec Products, Bartlesville, Okla.). The supernatant was further extracted with phenol-chloroform, treated with DNase-free RNase, and precipitated with ethanol.
PCR amplification for DGGE.
Primers U968-GC-f (5′ CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC) and L1401-r (5′ CGG TGT GTA CAA GAC CC) (25) were used to amplify the V6 to V8 regions of the bacterial 16S rRNA. PCRs were performed with a Taq DNA polymerase kit from Life Technologies (Gaithersburg, Md.). Each 50-μl PCR mixture contained each deoxynucleoside triphosphate at a concentration of 10 mM, 1.25 U of Taq polymerase, 10 μmol of each primer, and 1 μl of a DNA solution. The samples were amplified with a Biometra UNO II (Biometra, Westburg, The Netherlands). The effect of denaturation time was studied to ensure that bacterial DNAs with high G+C contents were amplified by PCR and that the PCR products were subsequently visualized by DGGE. A series of PCRs were performed with DNAs from the two infants by using the same PCR mixture, conditions, and machine. The program was modified so that either a 10-, 30-, or 60-s denaturation step could be used. The following conditions were used: 94°C for 5 min, 35 cycles consisting of 94°C for 10, 30, or 60 s, 56°C for 20 s, and 68°C for 40 s, and finally 68°C for 7 min. The 30- and 60-s denaturation times resulted in identical profiles, while a band at the bottom of the 10-s profile, possibly derived from a member of the genus Bifidobacterium (whose members have high G+C contents [55 to 67%]), was not visible (data not shown). Consequently, the 30-s denaturation time was used in further experiments.
Cloning of the PCR-amplified products.
Primers 8f (5′ CAC GGA TCC AGA GTT TGA T[C/T][A/C] TGG CTC AG) and 1510r (5′ GTG AAG CTT ACG G[C/T]T ACC TTG TTA CGA CTT) (18) were used for construction of the clone libraries for bacterial 16S rDNA. The following PCR program was used: 94°C for 5 min, 35 cycles consisting of 94°C for 1 min 30 s, 52°C for 30 s, and 68°C for 1 min, and finally 68°C for 7 min. The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and were cloned in Escherichia coli JM109 by using the pGEM-T vector system (Promega, Madison, Wis.). Colonies of ampicillin-resistant transformants were added to 20 μl of Tris-EDTA buffer and boiled for 15 min to lyse the cells. A PCR was performed by using the cell lysates as the template and pGEM-T-specific primers T7 and SP6 to check the sizes of the inserts. The following program was used: 94°C for 3 min, 35 cycles of 94°C for 30 s, 44°C for 30 s, and 68°C for 1.5 min, and finally 68°C for 7 min. The migration profiles of the amplified V6 to V8 regions of the clones were compared to the DGGE profile obtained with the same fecal sample in order to identify the origin of a band. Unique clones that did not correspond to bands in the profiles of the samples were also selected for sequence analysis. Plasmids were purified with a QIAprep Spin miniprep kit (Qiagen) and were used for sequence analysis.
DGGE of PCR amplicons.
PCR fragments, also called amplicons, were separated by DGGE by using the specifications of Muyzer et al. (24) and the Decode system (Bio-Rad Laboratories, Hercules, Calif.), with the following modifications. Polyacrylamide gels (dimensions, 200 by 200 by 1 mm) consisted of 8% (vol/vol) polyacrylamide (ratio of acrylamide-bisacrylamide, 37.5:1) and 0.5× Tris-acetate-EDTA (pH 8.0) (TAE) buffer; 100% denaturing acrylamide was defined as 7 M urea and 40% formamide. The gels were poured from the top by using a gradient maker and a pump (Econopump; Bio-Rad Laboratories) set at a speed of 5 ml per min. We used 40 to 50% gradients to separate products amplified with universal primers. Before polymerization of the denaturing gel (gradient volume, 28 ml), a 7.5-ml stacking gel without denaturing chemicals was added, and the appropriate comb was subsequently inserted. Electrophoresis was performed first for 5 min at 200 V and then for 16 h at 85 V in 0.5× TAE buffer at a constant temperature of 60°C. The gels were stained with AgNO3 as described by Sanguinetti et al. (28).
DNA sequence analysis.
One microgram of a purified plasmid was used to analyze the sequence of a cloned 16S rDNA fragment. Sequencing reactions were performed with a sequencing kit (Amersham Life Sciences, Slough, United Kingdom) according to the manufacturer’s specifications by using IRD-800 5′-labeled primer T7 or Sp6 (Infra-Red Dye 41; MWG-Biotech, Ebersberg, Germany). After addition of 1.8 μl of loading dye fluorescent samples (Amersham, Life Sciences), sequences were automatically analyzed with a LI-COR 4000L DNA sequencer (Li-Cor, Lincoln, Nebr.) and corrected manually. Similarity searches with sequences in the GenBank database were performed by using the BLAST algorithm (3). The 16S rDNA sequences were checked for chimeric constructs by using the CHECK-CHIMERA program of the Ribosomal Database Project (20) and the ARB software package (35).
Nucleotide sequence accession numbers.
The sequences of the fecal rDNA clones obtained from babies D and L (34 sequences that were 983 ± 303 bases long [mean ± standard deviation]) have been deposited in the GenBank database under accession numbers AF253328 to AF253337, AF253339 to AF253343, AF253372, AF253374 to AF253379, AF253381 to AF253390, AF253393, and AF316538.
RESULTS
Molecular analysis of the infant microbiota by DGGE.
To characterize and compare bacterial succession in the large intestines of newborn babies, fecal samples from two healthy babies, babies D and L, were analyzed from birth for more than 10 months by using PCR-DGGE (Fig. 1 and 2). Briefly, the strategy involved extraction of the DNA from fecal samples from the two babies and PCR of a fragment corresponding to region V6 to V8 of the 16S rRNA genes performed with universal bacterial primers. Analysis of PCR products by DGGE results in a fingerprint that represents the diversity of the rDNA nucleotide sequences due to the different bacterial types. The bands in the profile represent most of the dominant microbial populations in the community, and their appearance and disappearance reflect approximate changes in the microbial community composition. The intensity of a band provides a rough estimate of the proportion of the sequence in a sample. In order to identify the species corresponding to certain bands in the DGGE gels, several clone libraries of 16S rDNA sequences (E. coli rDNA nucleotide positions 8 to 1510) were prepared from a number of fecal samples obtained at various ages. The mobilities of the cloned 16S rDNA fragments were compared to the mobilities in the original profiles by DGGE, and the DNA sequences of clones that corresponded to dominant bands or unique clones were determined.
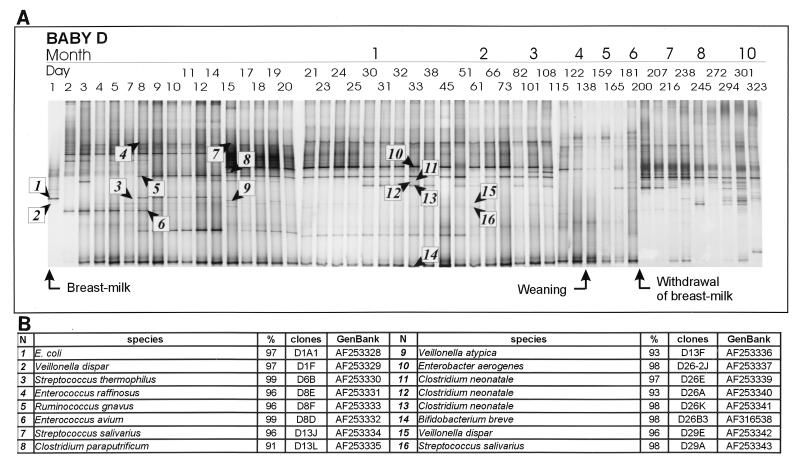
FIG. 1.
(A) PCR-DGGE profiles representing the bacterial diversity in baby D generated from samples taken from birth to 323 days. Both the day and the approximate time of the month of sampling are indicated. Changes in the diet are indicated by arrows. The bands identified from the 16S rDNA clone libraries are numbered and are indicated by arrowheads; these bands are described in the table in panel B. (B) Closest relative as determined by comparative sequence analysis, level of identity with this relative, clone designation, and accession number for each band identified in panel A.
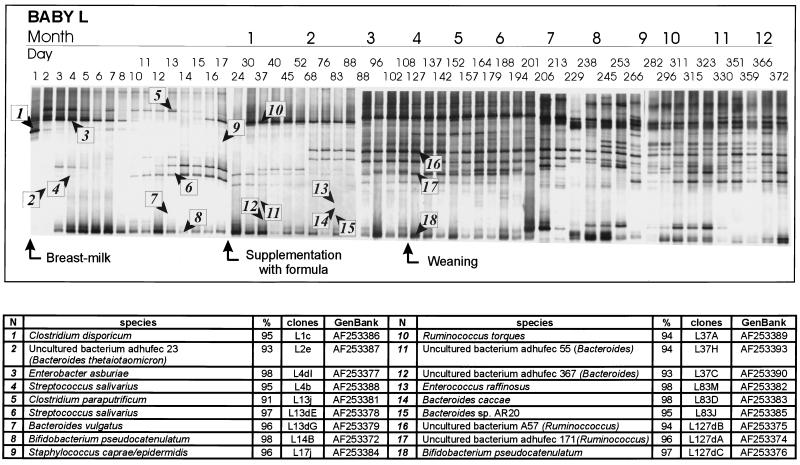
FIG. 2.
(A) PCR-DGGE profiles representing the bacterial diversity in baby L generated from samples taken from birth to 372 days. Both the day and the approximate time of the month of sampling are indicated. Changes in the diet, such as addition of formula milk, are indicated by arrows. The bands identified from the 16S rDNA clone libraries are numbered and are indicated by arrowheads; these bands are described in the table in panel B. (B) Closest relative as determined by comparative sequence analysis, level of identity with this relative, clone designation, and accession number for each band identified in panel A.
The first observation made with the DGGE profiles obtained for the two babies (Fig. 1 and 2) was the low diversity of the 16S rDNA sequences present in the feces during the first few weeks. Only a few different bands were present; the number varied from one or two major bands at birth (day 1 for babies D and L) to a maximum of six bands that were visible after 1 month. The most noticeable feature of the profiles was the appearance and disappearance of bands, especially during the first month or two. Several bands detected in the first days of life were rapidly replaced by different bands. Some bands were dominant for only a day, while others disappeared after several days; sometimes bands reappeared, but more often they did not.
Bacterial succession in baby D.
In breast-fed baby D, the first dominant band in the DGGE profile had a sequence related to the E. coli sequence (clone 1). In the clone bank obtained for day 1 another clone, whose amplicon was not visible in the DGGE profile, was most closely related to Veillonella dispar (clone 2). Sequences corresponding to bifidobacteria (clone 14) produced dominant bands at the bottom of the profile due to their high G+C contents just 3 days after birth. The bifidobacterial bands remained some of the most intense bands in all of the DGGE profiles during the first 6 months of baby D’s life. In the clone bank obtained for day 33, a sequence related to Bifidobacterium breve (clone 14) was identified, but other bifidobacterial species or strains that produced bands at similar positions may also have been present at different times. Until day 8, bands related to Enterococcus (clones 4 and 6) and Bifidobacterium dominated the profile, and in the days that immediately followed these bands were joined by bands for streptococci (clones 3 and 7), ruminococci (clone 5), and clostridia (clone 8). An Enterobacter aerogenes (clone 10) band appeared on day 15. A faint band in the DGGE profile corresponding to Veillonella atypica (clone 9) was detected on day 15. Around day 30 several bands corresponding to one or more species whose closest relatives were Clostridium neonatale appeared (clones 11 to 13). Subsequently, until the age of about 3.5 months (101 days), the baby’s microbiota consisted predominantly of Bifidobacterium, Clostridium, Enterobacter, and Ruminococcus species. Remarkably, a significant decrease in the intensities of bands corresponding to Enterobacter, Clostridium, and Ruminococcus species was observed from day 115 to 122. The DGGE profile was reduced to essentially one main band, corresponding to bifidobacteria, on day 122.
Following weaning of baby D, which began on day 130, the DGGE profile remained unstable (days 159 to 200). It is noteworthy that just after the end of breast feeding, which was on day 198, the band corresponding to bifidobacteria disappeared momentarily. The intensity of this band never recovered in the period from 7 to 10 months. The DGGE profiles began to stabilize again on day 200 and gradually became more diverse, and more bands appeared.
Bacterial succession in baby L.
After birth, baby L was breast fed until day 17, after which an increasing quantity of formula milk was added to his diet. The profile shifted during the first 3 months after birth. Two strong bands that were most closely related to a Clostridium species (clone 1) and Enterobacter asburiae (clone 3) were present on the first day of life. On the second day a dominant band corresponding to a Clostridium paraputrificum 16S rDNA sequence (clone 5) appeared, while the band corresponding to the first Clostridium species became less intense. The amplicons related to clostridia and E. asburiae were joined by a band at the position for bifidobacteria on day 3, and this band rapidly became dominant. A weak band originating from an uncultured Bacteroides species (clone 2) and a band corresponding to S. salivarius (clone 4) were also detected in the clone banks during the first few days. Thus, for the first 2 weeks, bacteria belonging to the genera Bifidobacterium, Enterobacter, Clostridium, and Streptococcus dominated. By day 16 an intense band corresponding to Ruminococcus torques (clone 10) was present, and this species, Bifidobacterium pseudocatenulatum (clone 8), and S. salivarius (clone 6) dominated the profile. As breast feeding was gradually replaced by formula milk, the profiles became more complex, and bands corresponding to other bacteria distantly related to uncultured bacteria (whose nearest relatives were Ruminococcus; clones 16 and 17) became more dominant. This pattern continued when solid food was introduced into the infant diet during month 4. Several other clones detected in the clone banks had sequences related to sequences of members of the genera Bacteroides (clones 7, 11, 14, and 15) and Enterococcus, but presumably lower numbers of these species were present and so bands were not apparent. From 4 to 6 months the profile remained quite stable despite the introduction of solid food. From 6 to 9 months some more pronounced shifts in the profiles were observed, and after 9 months the profiles stabilized again until monitoring was stopped after 1 year.
Comparison of the DGGE profiles of the two babies.
There were several similar features in the profiles of the two babies. Both profiles indicated that there was early colonization by bifidobacterial species, which were detected on the third or fourth day of life. Amplicons related to bifidobacteria were most dominant in breast-fed baby D during the first 6 months, while in baby L, who was fed both human and formula milk, the amplicons related to bifidobacteria were less intense. Besides amplicons related to Bifidobacterium, other amplicons related to fermentative bacteria that produce acids, such as streptococci and enterococci, were found in both babies mainly during the breast-feeding period. Amplicons related to Ruminococcus, Clostridium, or Enterobacter were also identified in the samples from both babies, and the times of appearance varied. For baby L at about 3 months amplicons related to Ruminococcus or close relatives of this genus appeared, and the bands were stronger than the bands for the bifidobacteria. The species present in the microbiota of baby D at 3 months were more diverse; bifidobacteria clearly dominated, but strong bands corresponding to clostridia, enterobacters, and ruminococci were also present. Bands corresponding to these organisms dominated the profiles of baby D for several months.
DISCUSSION
Although numerous studies of microbial invasion of the human intestine at birth have been performed and fluctuation of the fecal populations of newborn babies has been well established, little is known about bacterial stability from day to day. This is mainly due to the use of traditional bacteriological techniques, particularly cultivation on selective media combined with a plethora of biochemical and microscopic analyses, that has led essentially to specific studies of a few cultivable genera. In the present study, we demonstrated that PCR-DGGE can be used to monitor the successive populations in the intestinal tracts of babies without prior knowledge of the microbiota.
Several studies have demonstrated that the DGGE and TGGE techniques are effective tools for investigating the diversity of the intestinal ecosystem, which has been shown to be complex, and these studies have revealed that the microbiota of each individual is unique (40, 43). However, these studies were studies of reasonably well-established intestinal communities of adults or children that were relatively stable over time. More recently, DGGE was used to monitor the fecal bacterial communities in pigs of different ages, and the results showed that there were unique fecal bacterial populations that were stable over time (33). In the present study we used samples obtained on the first day of life of each baby and subsequent daily and weekly samples to generate DGGE profiles that clearly demonstrated the microbial succession of the dominant microbiota. The most obvious example was the appearance of bifidobacteria after just a few days on a milk diet. Furthermore, the introduction of solid food and the withdrawal of breast milk from the diet of baby D coincided with major shifts in the profiles.
Bands in the DGGE profiles were identified by analyzing part of the 16S rDNA sequence and comparing the sequences with sequences in databases. Usually, a 2.5 to 3.0% difference in 16S rRNA sequences indicates separate species status. In this study the levels of similarity ranged from 91 to 99%; 19 of 34 clones exhibited levels of similarity of less than 97%, and 5 clones exhibited levels of similarity of 97%. Thus, 56% of the sequences were derived from species not identified previously in the human intestinal microbiota. It is possible that the levels of similarity will change somewhat when the complete sequences are compared, although Zoetendal et al. (43) found the same closest relatives and the same levels of similarity with partial and complete sequences.
Culturing studies have indicated that in general infants are initially colonized by enterobacteria and gram-positive cocci, which are thought to create a reduced environment favorable for the establishment of Bacteroides, Bifidobacterium, and Clostridium by day 7 (reviewed in references 9 and 19). The microbiota of infants fed only breast milk becomes dominated by bifidobacteria during the first week, and there is a concomitant decrease in members of the Enterobacteriaceae (41). The introduction of solid foods into the diets of breast-fed babies results in a major change in the microbiota. In contrast, within the first few days, the microbiota of formula-fed infants becomes more diverse, with a longer presence and higher counts of members of the Enterobacteriaceae and enterococci; subsequently, species of bifidobacteria, Bacteroides, clostridia, enterococci, and streptococci dominate (34, 41). The identities of many of the dominant bands in the DGGE profiles were consistent with some of these patterns of succession for the infants. For example, in baby D E. coli was the initial colonizer, and disappearance of this organism was followed by the appearance of high numbers of bifidobacteria, as indicated by the strong band intensity.
However, several deviations from the usual trends were also observed. The first most dominant colonizer of baby L was a member of the clostridia. Thus, aerobic bacteria are not always the only founders of the intestinal microbiota. Nevertheless, cultivation methods have shown that infants generally excrete clostridia during the first week of life, and species related to C. paraputrificum, as detected in this study, are common. Higher numbers are usually isolated subsequently from bottle-fed babies (7, 31, 34), but certain species have been found frequently or at high levels in breast-fed babies (4, 22). Clostridium spp. are important human pathogens, particularly in infants, in which certain species have been associated with various gastrointestinal disorders (1) and have been implicated in the pathogenesis of necrotizing enteritis (5). In our study, we found sequences of Clostridium corresponding to strong bands in the breast-fed baby but not in the weaned infants. Curiously, in the profiles for baby D, three bands (clones 11 to 13) that were probably derived from the same species or even the same strain of C. neonatale appeared on day 30. The presence of heterogeneous 16S rRNA genes (i.e., 16S rRNA genes that exhibit small variations in the genome of a strain) can result in several bands in a DGGE or TGGE profile (25, 29).
Another difference between our study and cultivation studies was the lack of Bacteroides as part of the dominant microbiota. There are several possible reasons for this. First, Bacteroides may have been present but at low levels. Interestingly, a recent enumeration of cells in feces of 12 breast- or formula-fed babies by FISH performed with specific probes indicated that a high bifidobacterial count was associated with a low Bacteroides count and vice versa (12). As the bands corresponding to bifidobacteria were very strong in both babies’ profiles in this study, perhaps the proportion of Bacteroides was correspondingly low. Several 16S rDNA sequences related to Bacteroides were identified in the clone libraries for baby L but not in the clone libraries for baby D. Before introduction of formula into the diet of baby L, a Bacteroides vulgatus-like sequence was detected; this species is commonly isolated from infant feces (4, 22). Clone libraries from day 2, 37, and 83 samples also contained Bacteroides-related sequences. It is noteworthy that clone libraries do not necessarily reflect bacterial diversity due to lysis, PCR biases, and random selection of 16S rDNA sequences (10, 43). The Bacteroides-like clones and some clones representing other genera could not be matched with dominant bands in the profiles. The absence of dominant Bacteroides bands in the DGGE profiles might have been due to the presence of small amounts of numerous different species whose sequences migrated differently. In such a case, the sequences could not be observed as bands in DGGE profiles if their individual levels were lower than the detection limit, although some of them could be selected by cloning. Another explanation is that the Bacteroides sequences formed diffuse bands in DGGE gels due to unclear melting domains, which would have hampered their detection (30). Previous studies demonstrated that Bacteroides fragilis and closely related uncultured Prevotella species from feces form fuzzy bands during DGGE (42).
Perhaps the most remarkable difference between the composition of the fecal flora detected by DGGE and the composition of the fecal flora detected other methods is the presence of strong bands corresponding to ruminococcal species in both babies’ feces. Especially in baby L, bands of tentative Ruminococcus-like species dominated the profiles for many months. As reviewed by Mackie et al. (19), who grouped results from at least 10 studies based on cultivation of the microbiota of babies, Ruminococcus strains are usually present at low (≤5 log10 CFU/g) and moderate (6 to 8 log10 CFU/g) levels in feces from infants (before and after 1 month, respectively). It is possible that the nonpathogenic status of ruminococci has caused this genus to be overlooked and that development of selective media and 16S rRNA probes has received insufficient attention. Strains of Ruminococcus, as well as some bifidobacteria, have been defined as the major producers of extracellular glycosidases that degrade ABH and Lewis blood group active oligosaccharide chains of mucin (16, 17). The resulting mono- and disaccharides are utilized by larger fecal populations that cannot degrade these chains (16). The enzymatic potential of ruminococci suggests that these organisms have an essential or substantial role in the intestinal ecosystem or host.
In conclusion, we demonstrate that PCR-DGGE is a powerful tool for monitoring the effects of diet, environment, or host on succession in the infant gut ecosystem. The recent development and application of genus-specific primers for Bifidobacterium and Lactobacillus spp. in combination with DGGE expand the potential of this technique for specific populations (14, 29). Furthermore, the novel sequences generated during this study should facilitate isolation of the corresponding bacteria and aid in the design of new probes for quantification to assess the contributions of these organisms to the intestinal microbiota.
Acknowledgments
We thank the two families who provided fecal samples for this study. We are also grateful to Hans G. H. J. Heilig and Gael Erauso for excellent technical advice and to Wilma M. Akkermans-van Vliet for helpful technical assistance.
This study was supported in part by the EU-FAIR PROBDEMO (grant FAIR-CT96-1028) and EU-FAIR FLORA (grant FAIR-CT97-3035) projects.
REFERENCES
- 1.Ahtonen, P., O.-P. Lehtonen, P. Kero, E. Eerola, and K. Hartiala. 1994. Clostridium perfringens in stool, intrapartum antibiotics and gastrointestinal signs in a neonatal intensive care unit. Acta Paediatr. 83:389–390. [DOI] [PubMed] [Google Scholar]
- 2.Akkermans, A. D. L., E. G. Zoetendal, C. F. Favier, G. H. J. Heilig, W. M. Akkermans-van Vliet, and W. M. de Vos. 2000. Temperature and denaturing gradient gel electrophoresis analysis of 16S rRNA from human faecal samples. Biosci. Microflora 19:93–98. [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 4.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975–986. [DOI] [PubMed] [Google Scholar]
- 5.Blakey, J. L., L. Lubitz, N. T. Campbell, G. L. Gillam, R. F. Bishop, and G. L. Barnes. 1985. Enteric colonization in sporadic neonatal necrotizing enterocolitis. J. Pediatr. Gastroenterol. Nutr. 4:591–595. [DOI] [PubMed] [Google Scholar]
- 6.Cebra, J. J. 1999. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69:1046S–1051S. [DOI] [PubMed] [Google Scholar]
- 7.Cooperstock, M., L. Riegle, C. W. Woodruff, and A. Onderdonk. 1983. Influence of age, sex, and diet on asymptomatic colonization of infants with Clostridium difficile. J. Clin. Microbiol. 17:830–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooperstock, M. S., and A. J. Zedd. 1983. Intestinal flora of infants, p.79–99. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 9.Dai, D., and W. A. Walker. 1999. Protective nutrients and bacterial colonization in the immature human gut. Adv. Pediatr. 46:353–382. [PubMed] [Google Scholar]
- 10.Felske, A., H. Rheims, A. Wolterink, E. Stackebrandt, and A. D. L. Akkermans. 1997. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983–2989. [DOI] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variation of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen, H. J. M., A. C. M. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klein, J. G. Bondels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61–67. [DOI] [PubMed] [Google Scholar]
- 13.Harmsen, H. J. M., A. C. M. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523–4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 15.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. [DOI] [PubMed] [Google Scholar]
- 16.Hoskins, L. C. 1993. Mucin degradation in the human gastrointestinal tract and its significance to enteric microbial ecology. Eur. J. Gastroenterol. Hepatol. 5:205–212. [Google Scholar]
- 17.Hoskins, L. C., M. Agustines, W. B. McKee, E. T. Boulding, M. Kriaris, and G. Niedermeyer. 1985. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J. Clin. Investig. 75:944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, Chichester, United Kingdom.
- 19.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S–1045S. [DOI] [PubMed] [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, F., S. A. H. Savage, A. M. Parrett, G. Gramet, J. Doré, and C. Edwards. 2000. Dynamics of colonisation of the colon in breast-fed infants. Reprod. Nutr. Dev. 40:180. [Google Scholar]
- 22.Mevissen-Verhage, E. A. E., J. H. Marcelis, M. N. de Vos, W. C. M. Harmsen-van Amerongen, and J. Verhoef. 1987. Bifidobacterium, Bacteroides, and Clostridium spp. in fecal samples from breast-fed and bottle-fed infants with and without iron supplement. J. Clin. Microbiol. 25:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millar, M. R., C. J. Linton, A. Cade, D. Glancy, M. Hall, and H. Jalal. 1996. Application of 16S RNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J. Clin. Microbiol. 34:2506–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. de Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbaum, V., and D. Riesner. 1987. Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem. 26:235–246. [DOI] [PubMed] [Google Scholar]
- 28.Sanguinetti, C. J., E. Dias Neto, and A. J. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:914–921. [PubMed] [Google Scholar]
- 29.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1989. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simhon, A., J. R. Douglas, B. S. Drasar, and J. F. Soothill. 1982. Effect of feeding on infants’ faecal flora. Arch. Dis. Child. 57:54–58. [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167–179. [DOI] [PubMed] [Google Scholar]
- 33.Simpson, J. M., J. V. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark, P. L., and A. Lee. 1982. The microbial ecology of the large bowel of breast-fed and formula fed infants during the first year of life. J. Med. Microbiol. 15:189–203. [DOI] [PubMed] [Google Scholar]
- 35.Strunk, O., and W. Ludwig. 1995. ARB—a software environment for sequence data. Department of Microbiology, Technical University of Munich, Freising, Germany.
- 36.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannock, G. W. 1999. Analysis of the intestinal microflora: a renaissance. Antonie Leeuwenhoek 76:265–278. [PubMed] [Google Scholar]
- 38.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tissier, H. 1900. Recherches sur la flore intestinale des nourrissons (état normal et pathologique). G. Carre and C. Naud, Paris, France.
- 40.Vaughan, E. E., F. Schut, H. G. H. J. Heilig, E. G. Zoetendal, W. M. de Vos, and A. D. L. Akkermans. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1:1–12. [PubMed] [Google Scholar]
- 41.Yoshioka, H., K. Iseki, and K. Fugita. 1983. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 72:317–321. [PubMed] [Google Scholar]
- 42.Zoetendal, E. G. 2001. Molecular characterization of bacterial communities in the human gastrointestinal tract. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 43.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]