Abstract
Inactivation of the tumor suppressor p53 by degradation is a mechanism utilized by cells to adapt to endoplasmic reticulum (ER) stress. However, the mechanisms of p53 destabilization by ER stress are not known. We demonstrate here that the E3 ubiquitin-ligase Hdm2 is essential for the nucleocytoplasmic transport and proteasome-dependent degradation of p53 in ER-stressed cells. We also demonstrate that p53 phosphorylation at S315 and S376 is required for its nuclear export and degradation by Hdm2 without interfering with the ubiquitylation process. Furthermore, we show that p53 destabilization in unstressed cells utilizes the cooperative action of Hdm2 and glycogen synthase kinase 3β, a process that is enhanced in cells exposed to ER stress. In contrast to other stress pathways that stabilize p53, our findings further substantiate a negative role of ER stress in p53 activation with important implications for the function of the tumor suppressor in cells with a dysfunctional ER.
The p53 gene encodes for a protein whose loss of function is associated with the majority of human cancers (17). The p53 protein primarily functions as a transcription factor and mediates several biological effects including growth arrest, senescence, and apoptosis in response to diverse forms of stress (50). In the absence of stress, p53 is a short-lived protein whose activity is maintained at low levels. Upon exposure to a variety of stresses, p53 becomes stabilized and accumulates in the nucleus, where it resumes its transcriptional function. The levels and localization of p53 are tightly controlled through several posttranslational mechanisms, including protein stability, phosphorylation, and subcellular localization (3, 42, 46). Although several factors influence p53 function, the Hdm2 (human Mdm2) protein plays an essential role in regulating p53 protein levels in unstressed cells. Hdm2 is a nucleoplasmic and nucleolar RING-finger protein that promotes p53 nuclear export and degradation through its specific E3 ubiquitin ligase activity (2, 40). Other studies have revealed various Hdm2-independent pathways that impinge on p53 turnover, including c-Jun N-terminal kinase (10), COP1 (9), and Pirh2 (32). However, little is known about how and in what cellular context these pathways act.
The current model places both p53 and Hdm2 in an autoregulatory feedback loop where p53 induces the transcription of Hdm2 gene. The Hdm2 protein then binds to and ubiquitylates p53 in the nucleus, a process that allows the nuclear export and cytoplasmic proteasome-dependent degradation of the tumor suppressor (40, 56). The importance of this autoregulatory loop was demonstrated when the lethality of mdm2-null mice was rescued by the deletion of the p53 gene (22, 41). In this model, Hdm2 does not physically shuttle p53 out of the nucleus since the nuclear export sequence (NES) of Hdm2 is not necessary for p53 degradation, as opposed to its RING domain, which is important for p53 ubiquitylation and degradation (5, 11, 43). It has also been proposed that low Hdm2 levels in unstressed cells promote p53 mono-ubiquitylation in the nucleus, which then facilitates p53 nuclear export (30, 35, 65). However, other factors such as p300-CBP can also participate in the polyubiquitylation and degradation of p53 (16). Furthermore, other reports suggested that p53 can also be degraded in the nucleus (54, 64) when Hdm2 levels are elevated (35). The physiological significance of the nuclear degradation is not yet known, but it could represent an important mechanism for the poststress recovery of cells from a p53 response. Several other mechanisms have been described to modulate Hdm2 activity and p53 protein levels, including phosphorylation of Hdm2 and binding to other factors. For example, the tumor suppressor p14ARF and the human ribosomal protein L11 interact with Hdm2 by relocating Hdm2 to the nucleolus, thus reducing p53 ubiquitylation and degradation (38, 47, 67, 68). Although the Hdm2-dependent p53 nuclear export and degradation are critical for p53 function, the molecular mechanism(s) mediating this process is not fully understood and still remains a matter of debate.
Stress of the endoplasmic reticulum (ER) induced by physiological conditions (such as glucose starvation and hypoxia) or by pharmacological agents such as tunicamycin (inhibitor of glycosylation) or thapsigargin (inhibitor of the Ca2+-ATPase in the ER) can lead to accumulation of unfolded proteins and protein aggregates that are detrimental for cell survival (26). Under ER stress, cells initiate adaptive responses by activating specific signaling pathways to limit the accumulation of unfolded proteins. One of them is called the unfolded protein response, in which the transcription of genes encoding ER chaperones and folding catalysts is upregulated to increase protein folding activity (66). Other adaptive responses include the inhibition of protein synthesis as a means to decrease the protein overload in the ER (49) or the elimination of unfolded or misfolded proteins by proteasome-dependent proteolysis (28). If adaptation is not possible, then the stressed cell is eliminated by apoptosis through the activation of the JNK pathway and caspases 7, 12, or 3 (27).
We recently demonstrated a novel mechanism of adaptation of cells to ER stress involving the inactivation of p53 (48). Specifically, we showed that pharmacological or physiological inducers of ER stress prevent the proapoptotic function of p53 by enhancing its nucleocytoplasmic export and degradation (48). This regulation of the tumor suppressor protein requires its phosphorylation within the nuclear localization signal (NLS), which is mediated by the activation of the glycogen kinase 3β (GSK-3β), a protein kinase with pleiotropic effects on tumorigenesis, cell differentiation, and apoptosis (8, 14). However, the molecular mechanisms that modulate the cytoplasmic translocation and destabilization of p53 in ER stressed are poorly understood. We demonstrate here that induction of the cytoplasmic translocation and degradation of p53 by ER stress is mediated by Hdm2 and requires the phosphorylation of the tumor suppressor protein at serine S315 and S376 by GSK-3β. Significantly, the cooperative action of GSK-3β and Hdm2 also occurs in unstressed cells, but it is enhanced in cells subjected to ER stress. Our data reveal a new role for GSK-3β in the regulation of p53 nucleocytoplasmic shuttling and degradation by Hdm2, with possible important implications for therapies aiming at p53 stabilization through the inhibition of the p53-Hdm2-GSK-3β pathway.
MATERIALS AND METHODS
Cell culture and treatments.
Mouse embryonic fibroblasts (MEFs), human embryonic kidney HEK293T cells, and 3T3 cells were maintained in Dulbecco modified Eagle medium plus 10% calf serum (Invitrogen) and antibiotics. A549, WI38, and LNCaP cells were maintained in F12K medium (Cellgro), minimal essential medium, and RPMI 1640 medium, respectively, supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics. Mouse 70Z/3 cells were maintained in RPMI medium supplemented with 15% fetal bovine serum, 50 μM β-mercaptoethanol, and antibiotics. The ER stress inducers tunicamycin (TM; Sigma) and thapsigargin (TG; Sigma), the nucleocytoplasmic transport inhibitor leptomycin B (LMB; Sigma), and the proteasome inhibitor MG132 (Biomol) were dissolved in dimethyl sulfoxide at stock concentrations of 10 mg/ml, 1 mM, 4 μM, and 10 mM, respectively. Cycloheximide and adriamycin (ADR) were dissolved in ethanol at stock concentrations of 100 mg/ml and 1 mM, respectively. All stock solutions were kept in −20°C. The final concentrations of the drugs and the duration of treatments are indicated in the figure legends.
DNA constructs and transfection.
The wild-type (WT) Hdm2 cDNA, the NES Hdmd2 mutant cDNA and the C464A Hdm2 mutant cDNA in pcDNA/3.1 (Neo) vector were previously described (5). The WT and the NES mutant (L348A/L350A) of p53 in the pEGFP/N1 vector were also described previously (5). The green fluorescent protein (GFP)-p53 cDNA bearing the S315A or S376A mutation was generated as described previously (48). WT HA-tagged GSK-3β or the kinase-dead (KD) GSK-3β cDNA in pcDNA/3.1 vector was described previously (55). For transient transfections, the Lipofectamine Plus reagent (Invitrogen) or FuGENE 6 (Roche) were used as recommended by the manufacturer.
Immunoprecipitation and Western blotting.
Whole-cell extracts (WCE) were extracted in ice-cold lysis buffer containing 20 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% NP-40, 20% glycerol, 0.2 mM EDTA, 1.5 mM MgCl2, 1 mM dithiothreitol, 100 mM NaF, 20 mM β-glycerophosphate, 50 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 4 μg of aprotinin/ml, 4 μg of pepstatin A/ml, and 4 μg of leupeptin/ml. After incubation on ice for 15 min, the lysates were cleared by centrifugation at 18,000 × g for 15 min. For p53/Hdm2 coimmunoprecipitation, cell extracts (∼3 mg of protein) were immunoprecipitated with an Hdm2 antibody (Ab-1; Oncogene Science) or p53 Ab (DO-1) conjugated to protein G-Sepharose beads (Amersham) as described previously (51). For the detection of p53 ubiquitylation, cells were treated with 10 μM MG132 for 3 h prior to ER stress treatments, and the immunoprecipitation was performed with a polyclonal p53 Ab (FL393) with 200 μg of protein extract. For immunoblotting, WCE containing 50 μg of protein or immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and incubated with the indicated Ab. Abs to p53 (FL-393) and CM-5 were purchased from Santa Cruz, Inc., and Novocastra, respectively; DO-1 (Ab-6) and PAb421 (Ab-1) were from Oncogene Science. The mouse monoclonal Ab to actin was from ICN. The anti-Hdm2 (Ab-1) mouse monoclonal Ab was from Oncogene Science. The monoclonal Ab to androgen receptor (AnR) and polyclonal Ab to p21 were from Santa Cruz, Inc., whereas the anti-GFP rabbit polyclonal Ab was from Promega. All Abs were used at dilutions recommended by the manufacturer. Proteins were visualized by the enhanced chemiluminescence detection method (ECL; Amersham) according to the manufacturer's specification.
Immunofluorescence studies.
The detection of GFP-p53, Hdm2, or p53 by immunofluorescence was performed as previously described (48). GFP-positive and live cells were scored and classified into two groups; the first group with fluorescence predominantly in the nucleus and the second with fluorescence in both the nucleus and cytoplasm. Total green fluorescence in the nucleus and whole cells was quantified by using NIH Image from 200 randomly selected GFP-positive and live cells. For the detection of Mdm2, cells were stained with a 1:100 diluted monoclonal Ab (Ab-4; Oncogene Science), whereas for p53 the cells were stained with a 1:200-diluted anti-p53 rabbit polyclonal Ab (FL393; Santa Cruz). Cells were incubated with primary antibodies for 16 h at 4°C, washed with phosphate-buffered saline, and incubated for 1 h with Alexa Fluor 488-conjugated secondary Ab or Alexa Fluor 546-conjugated secondary Ab (both from Molecular Probes). The nucleus was visualized after staining with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole; Sigma)/ml.
Lentivirus infection and Hdm2 knockdown by shRNA.
For the Hdm2 knock-down experiments, the lentivirus vectors expressing Hdm2 short-hairpin RNA (shRNA) were previously described (24). Lentivirus-containing supernatant was collected 36 h after transfection in HEK293T cells, 0.2 μm-filtered, and snap-frozen at −70°C. A549 cells were infected by retrovirus at low multiplicity (ca. 10% transduction efficiency) by adding 4 mg of Polybrene (Sigma)/ml for 12 h prior to incubation with fresh medium. Cells were selected in puromycin, and polyclonal populations were expanded and analyzed.
RESULTS
Nuclear export is required for the Hdm2-mediated proteasome-dependent degradation of p53 in ER-stressed cells.
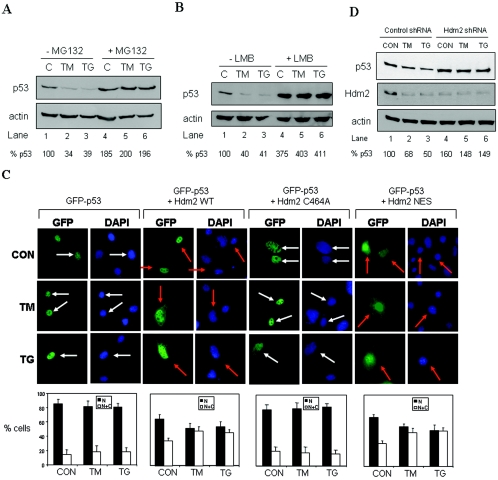
We previously showed that ER stress decreases the half-life of p53 without decreasing its mRNA levels, suggesting an effect of ER stress on p53 degradation (48). We further investigated this phenomenon by verifying the p53 levels in ER-stressed cells in the presence of proteasome inhibitors. To this end, we used A549 cells, which carry a WT p53 allele (51) and respond to ER stress induced by TM or TG treatment (29). We noticed that treatment of A549 cells with the inducers of ER stress led to the downregulation of p53 protein levels, which was blocked when cells were subjected to ER stress in the presence of the proteasome inhibitor MG132 (Fig. 1A). These data suggested that the proteasome degradation machinery is responsible for the decrease of p53 protein levels in ER-stressed cells. Because the proteasome-dependent degradation of p53 has been suggested to be mainly a cytoplasmic process, we next determined whether nuclear export of p53 is a prerequisite for its degradation in response to ER stress. When A549 cells were subjected to ER stress in the presence of LMB, an inhibitor of p53 nuclear export (53, 56), we observed a rescue of p53 protein levels in ER stressed and LMB-treated cells (Fig. 1B). This finding indicated that degradation of p53 in ER-stressed cells requires the nucleocytoplasmic shuttling of the tumor suppressor protein.
FIG. 1.
Hdm2-mediated nucleocytoplasmic shuttling and degradation of p53 in ER-stressed cells. (A and B) A549 cells were pretreated with either 10 μM MG132 (A, lanes 4 to 6) or 4 nM LMB (B, lanes 4 to 6) for 4 h prior to treatment with 1 μM TG (lanes 2 and 5) or 10 μg of TM/ml (lanes 3 and 6) for 2 h. Protein extracts were subjected to Western blotting with anti-p53 (DO-1) Ab (top panel) and anti-actin Ab (bottom panel). The p53 protein levels were normalized to actin levels (% p53) by using Scion Image 2.0 software. (C) Subcellular localization of GFP-p53. GFP-p53 WT cDNA (0.25 μg) was transiently transfected in the absence or presence of an equal amount of Hdm2 WT, Hdm2 NES, or Hdm2 C464A cDNA into 2KO cells. After 24 h, the cells were left untreated or treated with either 10 μg of TM/ml or 1 μM TG for 4 h, and GFP-p53 localization was examined by fluorescence microscopy. The nuclei were visualized by DAPI staining. White arrows indicate nuclear localization of GFP-p53 only, whereas orange arrows indicate both cytoplasmic and nuclear localization. The quantification of GFP-p53 localization is described in Materials and Methods. Cells with predominantly nuclear p53 are represented by the black bars, whereas cells with p53 both in the nucleus and in the cytoplasm are represented by the white bars. Values are means ± the standard deviation (SD) from three experiments. (D) Inhibition of Hdm2 expression by shRNA. A549 cells subjected to Hdm2 silencing by shRNA (see Materials and Methods) were treated with either 10 μg of TM/ml or 1 μM TG for 2 h. Protein extracts (50 μg) were immunoblotted with anti-p53 (DO-1) Ab (top panel), anti-Hdm2 Ab (middle panel), or antiactin Ab (bottom panel).
Given that Hdm2 is a major regulator of p53 protein stability (40), we examined its role in nucleocytoplasmic shuttling and degradation of p53 in response to ER stress. To do so, we utilized p53−/− Mdm2−/− MEFs (23), also known as double-knockout (2KO) cells, in which the subcellular localization of GFP-p53 was determined in coexpression assays with Hdm2 (Fig. 1C). Because the subcellular localization and degradation of p53 are controlled by the intracellular concentration of Hdm2, 2KO cells were transfected with an amount of Hdm2 sufficient to induce the cytoplasmic translocation and degradation of the tumor suppressor protein as recently described (35) (Fig. 1C). In the absence of Hdm2, we noticed that ER stress failed to promote the cytoplasmic translocation of p53. In the presence of Hdm2, we observed that a fraction of GFP-p53 became cytoplasmic, a process that was further enhanced in response to ER stress (Fig. 1C). The ability of Hdm2 to promote the cytoplasmic relocation of GFP-p53 was dependent on its ubiquitin ligase activity since Hdm2 bearing the C464A mutation in the RING finger domain failed to relocate GFP-p53 into the cytoplasm, either in the absence or presence of ER stress (Fig. 1C). In contrast to this, the cytoplasmic relocation of GFP-p53 was induced in the presence of an NES mutant of Hdm2, which retains its ubiquitin ligase activity (5, 11), indicating that nuclear export of p53 proceeds independently of the nuclear export of Hdm2 in unstressed, as well as in ER-stressed cells. Taken together, these findings indicated that p53 ubiquitylation by Hdm2 is a signal for the nuclear export of the tumor suppressor protein. This process, which does not require the nucleocytoplasmic transport of Hdm2, is enhanced in cells subjected to ER stress. To confirm the requirement of Hdm2 for p53 degradation in response to ER stress, we used a viral vector that express double-stranded shRNA to knockdown endogenous Hdm2 (24). We reasoned that if Hdm2 was involved in p53 degradation in ER stressed cells, then its elimination by shRNA should rescue p53 from degradation in response to ER stress. Expression of the shRNA in A549 cells led to >80% decrease of the endogenous Hdm2 protein (Fig. 1D, lanes 4 to 6). When cells were subjected to ER stress (Fig. 1D, lanes 2, 3, 5, and 6), we noticed that downregulation of p53 was not possible in cells with targeted Hdm2 (lanes 5 and 6) as opposed to control cells (i.e., cells infected with control lentiviruses [24]) in which p53 levels were decreased (Fig. 1D, top panel). In these experiments, we noticed that retrovirus infection of A549 cells made them less sensitive to ER stress based on the degree of p53 downregulation (Fig. 1D, lanes 2 and 3) compared to ER-stressed A549 cells without infection (Fig. 1A and B, lanes 2 and 3). These data strongly supported a major role for Hdm2 in p53 degradation in response to ER stress.
Decrease of functional Hdm2 is ER-stressed cells.
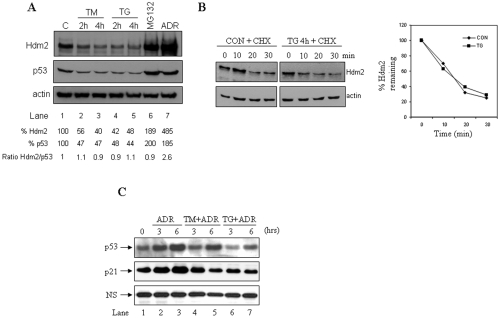
The data presented above also showed that ER stress leads to Hdm2 downregulation (Fig. 1D, middle panel, lanes 2 and 3). To better understand this phenomenon, we first assessed the endogenous Hdm2 protein levels in A549 cells at different time points after ER stress. We observed that Hdm2 protein levels were gradually reduced with the time of the ER stress treatments (Fig. 2A, top panel) concomitant with the downregulation of p53 protein levels (middle panel). In fact, Hdm2 downregulation was proportional to the decrease of p53 protein levels as demonstrated by the constant ratio of Hdm2/p53 before and after the ER stress treatments (Fig. 2A). Furthermore, the half-life of the endogenous Hdm2 in A549 cells was not affected by ER stress (Fig. 2B), indicating the lack of regulation of Hdm2 expression at the posttranslational level. Therefore, inhibition of Hdm2 expression by ER stress could be exerted either at the transcriptional, posttranscriptional or translational level through p53-dependent and/or independent mechanisms. However, ER stress affects the induction of p53-dependent genes as we have previously reported (48). For example, induction of the cdk inhibitor p21 in human diploid WI38 cells upon DNA damage by adriamycin (ADR) was inhibited when cells were exposed to ER stress (Fig. 2C). Furthermore, p21 induction in mouse pre-B 70Z/3 cells upon gamma irradiation was impaired after treatment of cells with TG (data available upon request). Moreover, upregulation of p21 in NIH 3T3 cells exposed to ADR was significantly reduced after treatment with inducers of ER stress for long periods of time (data available upon request). In all of these experiments, p21 expression levels correlated with p53 levels, indicating that regulation of p21 by ER stress was dependent on p53 response. Taken together, these data provide some evidence for a defective transcriptional response of p53 in ER-stressed cells that could account, at least in part, for the reduced Hdm2 levels in cells subjected to ER stress.
FIG. 2.
Inhibition of Hdm2 expression in ER-stressed cells. (A) Downregulation of p53 and Hdm2 proteins by ER stress. A549 cells were left untreated or treated with either 10 μg of TM/ml (lanes 2 and 3) or 1 μM TG (lanes 4 and 5) for the indicated times. Cells were also treated for 4 h with 10 μM MG132 (lane 6) or 1 μM ADR (lane 7) to stabilize p53 (controls). Protein extracts (50 μg) were subjected to immunoblotting with anti-p53 (DO-1) Ab (top panel), anti-Hdm2 Ab (middle panel), or antiactin Ab (bottom panel). Hdm2 and p53 protein levels were normalized to actin by using Scion Image 2.0 software. (B) Hdm2 stability in ER-stressed cells. A549 cells were untreated or treated with 1 μM TG for 4 h, followed by a 40-μg/ml cycloheximide chase for the indicated time. Protein extracts (50 μg) were subjected to Western blotting with anti-Hdm2 Ab (top panel) and antiactin Ab (bottom panel). The exposures shown are different so that the p53 levels in control (CON) and TG-treated cells for time zero (lanes 1 and 5) are equal. Hdm2 levels were normalized to actin levels by using Scion Image 2.0 software, and the ratio in unstressed (control) and ER-stressed cells (TG) plotted against the time is shown. (C) WI-38 cells were treated with 1 μM (adriamycin) ADR alone or with either 10 μg of TM/ml or 1 μM TG for the indicated times. Protein extracts (50 μg) were used for immunoblotting with an anti-p53 rabbit polyclonal Ab (top panel) or with an anti-p21 polyclonal Ab (middle panel). A nonspecific (NS) band was used as a loading control (bottom panel).
Control of p53/Hdm2 interaction and p53 ubiquitylation in ER-stressed cells.
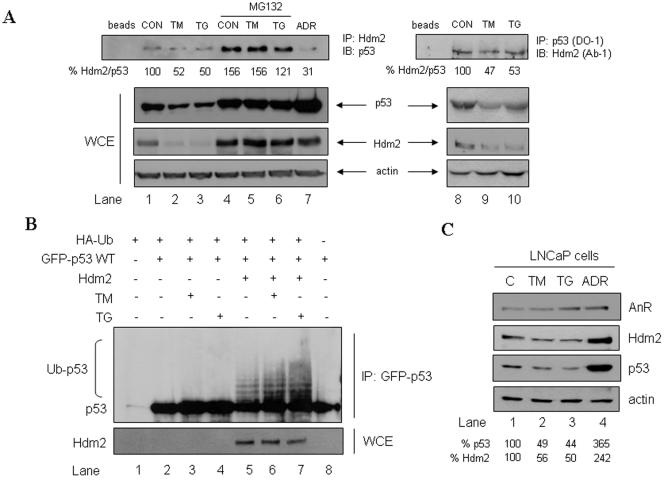
We further investigated the mechanisms of p53 degradation by Hdm2 in ER-stressed cells. First, we examined the interaction between the two proteins in A549 cells subjected to ER stress (Fig. 3A). The formation of Hdm2/p53 complex was detected by immunoprecipitation with either anti-Hdm2 Ab (lanes 1 to 7) or anti-p53 Ab (lanes 8 to 10), followed by immunoblotting with either anti-p53 Ab or anti-Hdm2 Ab, respectively. Using both approaches we measured a partial (∼50%) inhibition of Hdm2/p53 complex formation in cells subjected to ER stress compared to control untreated cells (top panel, compare lanes 2 and 3 with lane 1 and lanes 9 and 10 with lane 8). However, the total protein levels of both Hdm2 and p53 were diminished in cells subjected to ER stress (Fig. 3A, WCE), and as such, this partial inhibition of Hdm2/p53 complex might have been resulted from the different amounts of the proteins subjected to coimmunoprecipitation. To bypass this limitation, the A549 cells were treated with the proteasome inhibitor MG132 to stabilize p53 and prevent the downregulation of Hdm2 prior to exposure to ER stress (lanes 4 to 6). Upon these conditions, coimmunoprecipitation of Hdm2/p53 with anti-Hdm2 Ab showed no differences in the amount of complex formation before and after ER stress (top panel, lanes 4 to 6), indicating that ER stress does not interfere with the intermolecular interaction of the two proteins. On the other hand, DNA damage of A549 cells with ADR led to the dissociation of the Hdm2.p53 complex (lane 7), indicating that these cells are not refractory to signals that modulate the interaction of the two proteins.
FIG. 3.
Physical and functional interactions between p53 and Hdm2 in ER-stressed cells. (A) Physical interaction between Hdm2 and p53 in ER-stressed cells. A549 cells were left untreated (lanes 1, 2, 3, 8, 9, and 10) or pretreated with 10 μM MG132 for 2 h (lanes 4, 5, and 6), followed by treatment with 10 μg of TM/ml (lanes 2, 5, and 9) or 1 μM TG for 4 h (lanes 3, 6, and 10). Cells were also exposed to DNA damage with 1 μM ADR (lane 7), which was used as a control. WCE (50 μg of protein) were used for immunoblotting with anti-p53 (DO-1) Ab (second from the top panel), anti-Hdm2 Ab (third from the top panel), or antiactin Ab (bottom panel). The same protein extracts (3 mg) were subjected to immunoprecipitation with either anti-Hdm2 Ab (lanes 1 to 7) or anti-p53 (DO-1) Ab (lanes 8 to 10) and subsequently to immunoblotting with anti-p53 (DO-1) Ab or anti-Hdm2 Ab, respectively. (B) ER stress enhances the ubiquitylation of p53 by Hdm2. GFP-p53 WT cDNA (0.5 μg) was transiently transfected in the absence or presence of Hdm2 WT cDNA (0.5 μg) and/or HA-Ub cDNA (0.5 μg) into 2KO cells. Cells were treated with 10 μM MG132 for 3 h, prior to ER stress treatment with 10 μg of TM/ml or 1 μM TG for 3 h. Protein extracts (200 μg) were subjected to immunoprecipitation with anti-53 rabbit polyclonal Ab, followed by immunoblotting with anti-p53 (DO-1) Ab. WCE (50 μg of protein) were used for immunoblotting to detect the transfected Hdm2. (C) Specificity of p53 degradation ER stress. LNCaP cells were exposed to ER stress with 10 μg of TM/ml (lane 2) or 1 μM TG (lane 3) or subjected to DNA damage with 1 μM ADR (lane 4) for 2 h. Protein extracts (50 μg) were used for immunoblotting with anti-AnR Ab (top panel), anti-Hdm2 Ab (second from the top), anti-Hdm2 Ab (third from the top), or antiactin Ab (bottom panel).
Given that ER stress does not significantly affect Hdm2/p53 interaction, we next examined whether p53 ubiquitylation by Hdm2 is modulated in ER-stressed cells. To this end, 2KO cells were cotransfected with GFP-p53 and HA-tagged ubiquitin (HA-Ub) in the absence or presence of Hdm2, followed by treatment with inducers of ER stress (Fig. 3B). In the absence of Hdm2, the ubiquitylation of GFP-p53 was undetectable, but it was significantly induced by the presence of Hdm2 (compare lanes 2 and 5). In cells subjected to ER stress, the ubiquitylation of GFP-p53 in the absence of Hdm2 remained undetectable (lanes 3 and 4). However, when Hdm2 was present, GFP-p53 ubiquitylation was enhanced upon treatment with either TM (lane 6) or TG (lane 7) compared to unstressed cells (lane 5). In similar experiments, we found that GFP-p53 ubiquitylation was possible with Hdm2 NES and further enhanced in response to ER stress as opposed to Hdm2 C464A, which was unable to act on GFP-p53 (data not shown). We further wanted to determine whether the induction of Hdm2 activity in ER-stressed cells was specific for p53 or degradation of other Hdm2 substrates was also enhanced in response to ER stress. To this end, we examined the effect of ER stress on AnR protein levels, which is a substrate of Hdm2 (37). As shown in Fig. 3C, treatment of LNCaP cells with ER stress did not induce the degradation of AnR as opposed to p53, which was downregulated by this treatment. These results suggested a specific effect of Hdm2 on p53.
Degradation of p53 by Hdm2 requires phosphorylation of p53 at S315 and S376.
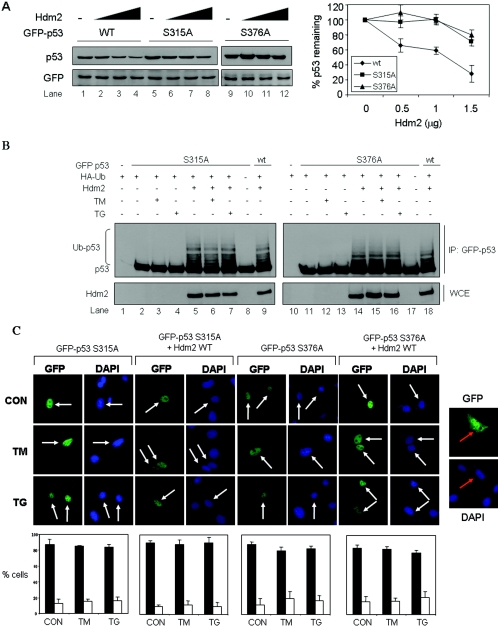
We recently demonstrated that phosphorylation of p53 at S315 and S376 is required for its cytoplasmic relocation in ER-stressed cells (48). Specifically, we showed that both serine residues are constitutively phosphorylated within cells and their phosphorylation is enhanced in response to ER stress (48). To determine the role of these serine residues in the regulation of p53 stability by Hdm2, we assessed the Hdm2-mediated degradation of GFP-p53 bearing either the S315A or S376A mutation. Using transient-expression assays in 2KO cells, we found that GFP-p53-S315A and GFP-p53-S376A were more resistant to Hdm2-mediated degradation compared to GFP-p53 WT (Fig. 4A). This indicated that phosphorylation of S315 and S376 is critical for efficient degradation of p53 by Hdm2. To gain a better mechanistic insight into this process, we next determined the ubiquitylation of GFP-p53-S315A and GFP-p53-S376A by Hdm2 in 2KO cells. We found that ubiquitylation of either GFP-p53-S315A or GFP-p53-S376A was undetectable in the absence of Hdm2 (Fig. 4B, top panel, lanes 2 and 11), but it was induced when Hdm2 was coexpressed (Fig. 4B, top panel, lanes 5 and 14). In the absence of ER stress, ubiquitylation of both GFP-p53 mutants was equal to GFP-p53 WT ubiquitylation (compare lanes 5 and 14 with lanes 9 and 18, respectively). However, treatment with ER stress did not further enhance the ubiquitylation of the GFP-p53 mutants by Hdm2 (Fig. 4B, lanes 6, 7, 15, and 16), as opposed to GFP-p53 WT, whose ubiquitylation was increased (Fig. 3B). These data suggested that S315 and S376 phosphorylation does not interfere with p53 ubiquitylation in unstressed cells but is required for the induction of ubiquitylation of p53 in ER-stressed cells. Since phosphorylation at S315 and S376 is required for the nucleocytoplasmic shuttling of p53 in both unstressed and ER-stressed cells (48), we next examined the effects of Hdm2 on the nuclear export of the GFP-p53 phosphorylation mutants in 2KO cells (Fig. 4C). We found that the distribution of GFP-p53 mutants remained mainly nuclear when expressed alone or together with Hdm2. Furthermore, the GFP-p53 mutants remained nuclear in cells subjected to ER stress. This was different from the regulation of the nucleocytoplasmic regulation of GFP-p53 WT, which was enhanced by the coexpression of Hdm2 and treatment with ER stress inducers (Fig. 2A). Collectively, these data showed that ubiquitylation of p53 is necessary but not sufficient for the nuclear export of the tumor suppressor in unstressed and ER-stressed cells. Furthermore, nuclear export of p53 by Hdm2 requires phosphorylation of p53 at S315 and S376.
FIG. 4.
Role of S315 and S376 phosphorylation in subcellular localization and degradation of p53 in ER-stressed cells. (A) Hdm2-dependent degradation of GFP-p53 and its phosphorylation mutants. GFP-p53 WT cDNA or the indicated GFP-p53 mutant cDNAs (0.5 μg) were transiently transfected in 2KO cells in the presence of increasing amounts of Hdm2 WT cDNA (0.5 to 1.5 μg). The GFP cDNA (0.1 μg) was used as an internal control. After 24 h, protein extracts (50 μg) were used for immunoblotting with anti-p53 (DO-1) Ab (top panel) or anti-GFP Ab (bottom panel). The GFP-p53 proteins were normalized to GFP, and their degradation profiles with increasing amounts of Hdm2 are shown. The values in the graphs represent means ± the SD from three independent experiments. (B) Control of ubiquitylation of GFP-p53 phosphorylation mutants by Hdm2 in ER-stressed cells. GFP-p53 cDNA (0.5 μg) bearing the S315A or S376A mutation was transiently transfected in 2KO cells in the absence or presence of Hdm2 WT cDNA (0.5 μg) and HA-Ub cDNA (0.5 μg). The cells were treated with 10 μM MG132 for 3 h, followed by treatment with 10 μg of TM/ml or 1 μM TG for 3 h. The immunoprecipitation of GFP-p53 and immunoblotting were performed as in Fig. 3B. (C) Control of localization of GFP-p53 phosphorylation mutants by Hdm2. GFP-p53 mutant cDNAs (0.25 μg) were transiently transfected in 2KO cells in the absence or presence of Hdm2 WT cDNA (0.25 μg). After 24 h, cells were left untreated or treated with either 10 μg of TM/ml or 1 μM TG for 4 h, followed by the examination of GFP-p53 localization by fluorescence. Cell nuclei were visualized by staining with DAPI. Transfection of GFP cDNA alone was used as a control (right panel). White arrows indicate nuclear localization of GFP-p53 only. Quantification of GFP-p53 localization was performed as described for Fig. 2A and in Materials and Methods.
GSK-3β is involved in Hdm2-mediated p53 cytoplasmic shuttling in ER-stressed cells.
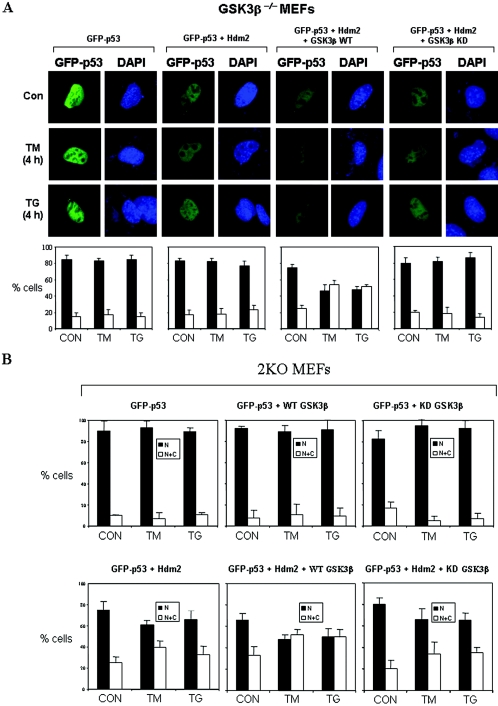
Since p53 phosphorylation at S315 and S376 in ER-stressed cells is mediated by GSK-3β (48), we sought to determine the role of GSK-3β in Hdm2-mediated nuclear export and degradation of the tumor suppressor. To do so, we examined the localization of GFP-p53 in spontaneously immortalized GSK-3β−/− MEFs (20), which were devoid of endogenous p53 (48). In these cells, GFP-p53 exhibited a nuclear localization either in the absence or presence of Hdm2, and it remained nuclear even after treatment with ER stress (Fig. 5A). When the GSK-3β−/− MEFs were transfected with GSK-3β WT and Hdm2, we observed that a fraction of GFP-p53 became cytoplasmic, which was further enhanced in response to ER stress (Fig. 5A). In contrast to GSK-3β WT, the cytoplasmic localization of GFP-p53 by Hdm2 was not induced when the cells were reconstituted with a KD GSK-3β (Fig. 5A). Similar results were obtained when the localization of GFP-p53 was examined in 2KO cells transfected with GSK-3β and Hdm2 (Fig. 5B). Quantification of the subcellular distribution of GFP-p53 showed that cytoplasmic relocation of GFP-p53 was enhanced by GSK-3β WT and impaired by GSK-3β KD. Collectively, these data suggested that GSK-3β is required for the Hdm2-mediated nucleocytoplasmic shuttling of p53.
FIG. 5.
GSK-3β is required for Hdm2-mediated cytoplasmic shuttling of p53 in ER-stressed cells. (A) Subcellular localization of GFP-p53 in GSK-3β−/− MEFs. Cells were transfected with 0.05 μg of GFP-p53 WT cDNA, 0.05 μg of Hdm2 cDNA, and 0.1 μg of either GSK-3β WT or GSK-3β KD cDNA. After 24 h, cells were treated with either 10 μg of TM/ml or 1 μM TG for 4 h and then examined for GFP-p53 localization by fluorescence microscopy. The cell nuclei were visualized by DAPI staining. Quantification of GFP-p53 localization was performed as described in Fig. 2A and Materials and Methods. (B) Subcellular localization of GFP-p53 in 2KO MEFs. Cells were transiently transfected with 0.25 μg of GFP-p53 cDNA in the absence or presence of 0.25 μg of Hdm2 of WT cDNA and 0.5 μg of either GSK-3β WT cDNA or GSK-3β KD cDNA. After 24 h, cells were left untreated or treated with either 10 μg of TM/ml or 1 μM TG for 4 h, followed by examination with fluorescence microscopy. Quantification of the subcellular localization of GFP-p53 was performed as described above.
GSK-3β controls Hdm2-mediated p53 degradation in unstressed and ER-stressed cells.
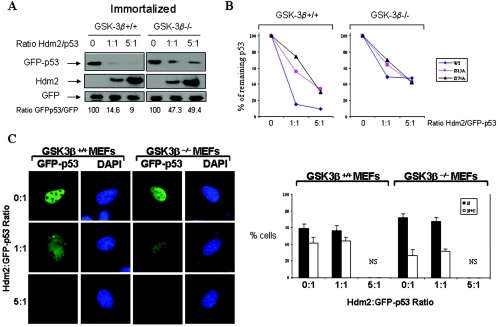
We next wanted to determine the role of GSK-3β in Hdm2-mediated p53 degradation. To this end, spontaneously immortalized GSK-3β+/+ and GSK-3β−/− MEFs were transfected with GFP-p53 WT cDNA and increasing amounts of Hdm2 cDNA, followed by immunoblot analysis for GFP-p53. We found that GFP-p53 WT was more resistant to Hdm2-mediated degradation in GSK-3β−/− MEFs than in GSK-3β+/+ MEFs (Fig. 6A), indicating that p53 degradation by Hdm2 requires GSK-3β. On the other hand, GFP-p53-S315A and GFP-p53-S376A were more resistant to Hdm2-mediated degradation in GSK-3β−/− MEFs and in GSK-3β+/+ MEFs than GFP-p53 WT (Fig. 6B). To confirm the role of GSK-3β in p53 nuclear export and degradation, we determined the localization of GFP-p53 in immortalized GSK-3β+/+ and GSK-3β−/− MEFs transfected with increasing amounts of Hdm2 cDNA (Fig. 6C). We observed that GFP-p53 was mainly nuclear in both GSK-3β+/+ and GSK-3β−/− MEFs. Transfection of cells with equal amounts of Hdm2 and GFP-p53 cDNAs (ratio, 1:1) resulted in the cytoplasmic relocation of GFP-p53 in GSK-3β+/+ MEFs but not in GSK-3β−/− MEFs in which GFP-53 remained nuclear. When the Hdm2/GFP-53 ratio was increased to 5:1, we noticed that the GFP-p53 signal was significantly decreased in both MEF types although GFP-p53 was more resistant to Hdm2-mediated degradation in GSK-3β−/− than in GSK-3β+/+ MEFs (Fig. 6C). From these data, we concluded that the nucleocytoplasmic relocation and subsequent degradation of GFP-p53 by Hdm2 requires GSK-3β.
FIG. 6.
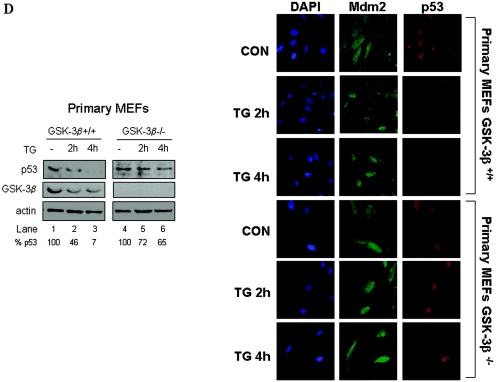
GSK-3β is necessary for p53 degradation in ER-stressed cells. (A) Control of GFP-p53 stability by GSK-3β. GFP-p53 WT cDNA (0.25 μg) was transiently transfected with increasing amounts of Hdm2 cDNA (0.25 to 1.25 μg) into immortalized GSK-3β−/− and GSK-3β+/+ MEFs. The GFP cDNA (0.25 μg) was used as an internal control. The GFP-p53, Hdm2, and GFP protein levels were detected by immunoblotting of 50 μg of protein extracts with antibodies against each protein. The bands were quantified with the Scion Image 2.0 software, and the ratio of GFP-p53 to GFP in each lane is indicated. (B) Hdm2-dependent degradation of GFP-p53 and its phosphorylation mutants. GFP-p53 WT cDNA or the indicated GFP-p53 phosphorylation mutants were cotransfected with Hdm2 WT cDNAs in immortalized GSK-3β−/− and GSK-3β+/+ MEFs as described in panel A. Immunoblot analyses of GFP-p53 and GFP were performed as in panel A. The bands were quantified, and the graphs representing the degradation rates of the GFP-p53 forms plotted against Hdm2/GFP-p53 ratios are shown. (C) Immortalized GSK-3β+/+ and GSK-3β−/− MEFs were transfected with different ratios of GFP-p53 WT to Hdm2 cDNAs as indicated. At 24 h posttransfection, cells were examined for GFP-p53 fluorescence. Cell nuclei were visualized by DAPI staining. The quantification of the subcellular distribution of GFP-p53 was performed as described for Fig. 2A and Materials and Methods. NS, not significant. (D) Regulation of p53 in primary GSK-3β−/− MEFs. MEFs (passage 3) were treated with 1 μM TG for 2 and 4 h, followed by immunoblotting of 50 μg of protein extracts for endogenous p53 (top panel), GSK-3β (middle panel), and actin (bottom panel). Immunostaining of endogenous Mdm2 (green) or p53 (red) in the primary MEFs was performed as described in Materials and Methods. The cell nuclei were detected by DAPI staining.
To further substantiate the role of GSK-3β in p53 degradation, we looked at the endogenous WT p53 in primary GSK-3β+/+ and GSK-3β−/− MEFs. Specifically, we assessed the protein levels and subcellular localization of p53 before and after ER stress treatment with TG (Fig. 6D). We found that p53 protein was more resistant to the inhibitory effects of ER stress in GSK-3β−/− MEFs than in GSK-3β+/+ MEFs demonstrating that GSK-3β is involved in the downregulation of p53 in ER-stressed cells. It is of interest that primary MEFs are more sensitive to downregulation of p53 by ER stress than tumor cells (Fig. 1), indicating differences in ER stress responses between normal and transformed cells. The immunostaining data showed a significant loss of nuclear p53 staining in TG-treated GSK-3β+/+ MEFs as opposed to GSK-3β−/− MEFs. Furthermore, we noticed that in untreated GSK-3β+/+ MEFs a fraction of p53 was cytoplasmic, as opposed to GSK-3β−/− MEFs in which p53 was predominantly nuclear. This result indicated that GSK-3β also regulates the nucleocytoplasmic shuttling of p53 in unstressed cells. On the other hand, Mdm2 localization was both nuclear and cytoplasmic in both MEF types (Fig. 6D). However, treatment with TG decreased the Mdm2 signal GSK-3β+/+ MEFs concomitant with the decrease of p53 signal in these cells. The Mdm2 signal did not significantly change in TG-treated GSK-3β−/− MEFs consistent with the resistance of p53 to ER stress-mediated degradation in these cells. Collectively, these data demonstrated the important role of GSK-3β in localization and degradation of p53.
DISCUSSION
Our findings provide a mechanistic explanation for the higher degradation rates of p53 in ER-stressed cells (48). Specifically, we show that Hdm2-dependent ubiquitylation, nuclear export, and degradation of p53 are all enhanced in ER-stressed cells. At the molecular level, Hdm2 interaction with p53 was not significantly affected by ER stress (Fig. 2), indicating that their interaction is necessary but not sufficient for the enhanced ubiquitylation of the tumor suppressor protein. Our data support the notion that enhanced ubiquitylation of p53 is likely to be mediated by posttranslational modifications of p53 rather than Hdm2. Indeed, our observations showed that the auto-ubiquitylation activity of Hdm2 was not affected in cells subjected to ER stress (data not shown) and that degradation of AnR, which is another substrate of Hdm2 (37), was not induced in response to ER stress (Fig. 3). In fact, our findings support the notion that phosphorylation of p53 is necessary for its enhanced ubiquitylation by Hdm2. That is, ER stress induces p53 phosphorylation at S315 and S376 (48), modifications that are important for the induction of p53 ubiquitylation (Fig. 3) and subsequent degradation (Fig. 4) in response to ER stress. It is possible then that phosphorylation of p53 upon ER stress results in conformational changes that further facilitate its ubiquitylation process by Hdm2. This is reminiscent of the regulation of p53 degradation by phosphorylation at threonine (T) 55, which is mediated by TAF1 and facilitates p53 oligomerization (33), a process required for efficient Hdm2-mediated ubiquitylation and degradation of the tumor suppressor protein (39). The role of S315 in p53 destabilization is further supported by recent findings demonstrating that p53 phosphorylation at this site by Aurora A kinase facilitates Hdm2-mediated ubiquitylation and degradation of p53 (25). Concerning S376, our data are supported by other findings for a negative effect of its phosphorylation on p53 stabilization (6). Interestingly, S376 of p53 was found to be phosphorylated in unstressed cells and its dephosphorylation to be induced upon DNA damage by gamma irradiation, resulting in a higher capacity of p53 to bind DNA (62). Our data show that S315 and S376 phosphorylations are equally important for p53 ubiquitylation, nucleocytoplasmic shuttling, and degradation in response to ER stress (Fig. 4B and C). Since the functional status of p53 is usually determined by its overall phosphorylation status rather than phosphorylation at a single residue (51), it remains possible that phosphorylation at S315 or S376 affects phosphorylation at other sites that also contribute to the inactivation of p53 in ER stress-treated cells.
It is of interest that degradation of either transfected or endogenous p53 still takes place in GSK-3β−/− cells but to a much lesser extent than in GSK-3β+/+ cells (Fig. 6), providing evidence that GSK-3β may not be the only determinant of p53 degradation. One possibility is that GSK-3α also exerts a similar effect on p53 degradation in ER-stressed cells. Also, in A549 cells ER stress had a partial (∼50%) inhibitory effect on p53 protein levels (Fig. 1), which raises the interesting question what of makes the other 50% of p53 resistant to ER stress. It is possible that upon ER stress only a fraction of p53 is modified by phosphorylation from GSK-3β, thus leading to the partial destabilization of p53. Another interpretation may have to do with the differential sensitivity of cells to ER stress since destabilization of p53 is greater in primary MEFs (Fig. 6D) than in the A549 tumor cells (Fig. 1). Furthermore, ER stress may modulate p53 through recently identified pathways that control its stability. For example, recent findings demonstrated that p53 degradation is inhibited by the interaction of Hdm2 with the ribosomal proteins L11 and L23 (7, 21, 38). Although ER stress signals to the translational ribosomal machinery through the activation of the eIF2α kinase PERK (49), it is not presently known whether Hdm2 interaction with either L11 and/or L23 is affected by ER stress and whether this type of stress can cause perturbations in ribosomal biogenesis that signal to Hdm2 and p53. Other control mechanisms might implicate the Yin Yang 1 (YY1) protein, which has recently been shown to facilitate Hdm2-mediated ubiquitylation of p53 (15, 57). A link between YY1 and ER stress might be implied by the ability of YY1 to induce the expression of grp78 gene, which is also transcriptionally induced in response to ER stress (36). Moreover, YY1 can enhance the transactivation capacity of ATF6, a protein involved in the transcriptional induction of many genes in response to ER stress (34). Another tentative link between p53 degradation and ER stress may be provided by the function of Rad23, a protein implicated in the stimulation of ER-associated protein degradation pathway (59). Specifically, human Rad23 (HR23) exhibits a dual function in p53 degradation by preventing the deubiquitylation of p53 and delivering the ubiquitylated protein to the proteasome machinery for degradation (12). Furthermore, HR23 interacts with p300/CBP resulting in the inhibition of the protein stability and transcriptional activity of p53 (69). In addition to ubiquitylation, conjugation of both p53 and Hdm2 by the ubiquitin-like protein NEDD8 (neddylation) has recently been shown to interfere with the biological functions of both proteins (63). Nevertheless, neddylation of p53 is not affected by treatment of cells with inducers of ER stress (data not shown), thus excluding the possibility for a regulatory role of this modification in p53 function in response to ER stress.
Our data demonstrate that p53 phosphorylation at S315 and S376 mediated by GSK-3β (49) serves as a signal that is crucial not only for the ubiquitylation but also for the cytoplasmic relocation and degradation of p53. These effects of GSK-3β on p53 take place in unstressed cells but are accelerated in cells subjected to ER stress (for a model of p53 regulation by ER stress, see Fig. 7). Indeed, the loss of GSK-3β impeded p53 nuclear export (Fig. 5) and increased p53 stability (Fig. 6) in the absence of ER stress. Moreover, endogenous WT p53 is more stable in primary GSK-3β−/− than GSK-3β+/+ MEFs in response to ER stress, a finding consistent with a higher nuclear presence of p53 in GSK-3β−/− MEFs exposed to ER stress (Fig. 6D). The immunostaining data show a decrease in endogenous Mdm2 levels concomitant with the downregulation of endogenous p53 in response to ER stress. This regulation is more apparent in GSK-3β+/+ MEFs than in GSK-3β−/− MEFs. These data provide strong evidence that GSK-3β plays a major role in p53 nucleocytoplasmic transport and degradation. The molecular event(s) that lead to activation of GSK-3β upon ER stress are currently under investigation. One possibility is that GSK-3β becomes activated by phosphorylation by another kinase(s) that is induced in response to ER stress (18). Such phosphorylation events may be important in determining the substrate specificity of GSK-3β. Given the pleiotropic effects of GSK-3β in cell signaling (45), it is conceivable to speculate that the ability of the kinase to regulate p53 function may tightly be dependent on signaling induced by a specific type of stress. For example, induction of GSK-3β by ER stress leads to the downregulation of p53 protein (48; this study), whereas activation of GSK-3β by a specific type of genotoxic stress is associated with p53 activation (58, 60, 61).
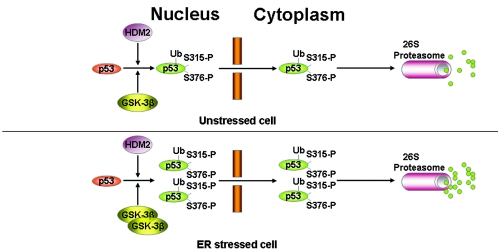
FIG. 7.
Schematic model of p53 regulation by Hdm2 in unstressed and ER-stressed cells. In unstressed cells, the nucleocytoplasmic transport of p53 is mainly mediated by Hdm2. We show here that phosphorylation of p53 at S315 and S376 by GSK-3β does not interfere with the ubiquitylation process but is required for the cytoplasmic relocation of p53 by Hdm2. Cytoplasmic p53 is then prone to degradation by the 26S proteasome pathway. When cells are exposed to ER stress, the nuclear fraction of GSK-3β becomes activated leading to the induction of p53 phosphorylation at S315 and S376 (49). As shown in the present study, these phosphorylation events enhance the ubiquitylation of p53 by Hdm2 and the cytoplasmic relocation of the tumor suppressor. As a result, p53 degradation rates in the cytoplasm are induced.
The role of ER stress in tumor development is strongly implied by the transcriptional induction and expression of glucose-regulated proteins (Grps) or ER chaperones that confer a survival advantage to the stressed cell. Whereas in normal cells the induction of Grps is associated with tissue preservation or organ protection, in cancer cells the induction of Grps can facilitate tumor progression and drug resistance (31). Given the apoptotic functions of p53, its inactivation in ER-stressed cells could certainly provide cells with a growth advantage and promote the transformation process. In fact, our previous findings demonstrated an inhibitory effect of ER stress on p53-depenent apoptosis (49). A physiological inducer of ER stress is hypoxia (29), a condition that is common in solid tumors. Although some reports have shown hypoxia to induce p53 (1, 13), other reports have provided evidence to the contrary (45, 53). This discrepancy can be explained not only by the differences in experimental conditions but also by the pleiotropic effects that are associated with hypoxia, including nutrient deprivation or low pH (acidosis) (19). The latter interpretation is supported by findings demonstrating the downregulation of p53 in hypoxic cells that are free from acidosis (52). Furthermore, recent data also showed the lack of p53 accumulation in hypoxic cells upon conditions that prevented acidosis and maintained nutrient levels (44). These and our data are consistent with a model where induction of the ER stress response in hypoxic tumors is associated with the downregulation of p53. Such an induction of ER stress in the microenvironments of solid tumors may have detrimental effects on therapies aiming at the activation of endogenous p53 and may provide an explanation for tumor resistance to radio/chemotherapies. Understanding the mechanisms of downregulation of p53 by Hdm2 in ER-stressed cells may be useful for the design of strategies leading to the activation of p53 in tumors with deregulated ER (i.e., hypoxic tumors). Tumors that retain WT p53 could be sensitized to p53-mediated apoptosis by inhibiting the expression of Hdm2 or blocking p53/Hdm2 interaction with synthetic peptides. Such approaches alone or in combination with novel strategies that utilize membrane-associated Grps for the destruction of tumors with increased levels of ER stress (4) may prove promising for therapeutic intervention.
Acknowledgments
We thank J. Woodgett for GSK-3β WT cDNA, KD GSK-3β cDNA, and GSK-3β knockout MEFs; S. Boyd and T. Jacks for the GFP-p53 WT, Hdm2 WT, Hdm2 NES, and Hdm2 C464A plasmids; M. Treier for HA-ubiquitin vector; S. Jones for 2KO MEFs; R. Iggo for Hdm2 shRNA vectors; and M. Triffiro for LNCaP cells and androgen receptor Ab.
D.B. is a research student of the Terry Fox Foundation through an award from the National Cancer Institute of Canada (NCIC) and a recipient of the CIHR Cancer Consortium Training Grant Studentship Award. This study was supported by a grant from the NCIC to A.E.K.
REFERENCES
- 1.Alarcon, R., C. Koumenis, R. K. Geyer, C. G. Maki, and A. J. Giaccia. 1999. Hypoxia induces p53 accumulation through MDM2 downregulation and inhibition of E6-mediated degradation. Cancer Res. 59:6046-6051. [PubMed] [Google Scholar]
- 2.Alarcon-Vargas, D., and Z. Ronai. 2002. p53-Mdm2: the affair that never ends. Carcinogenesis 23:541-547. [DOI] [PubMed] [Google Scholar]
- 3.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 4.Arap, M. A., J. Lahdenranta, P. J. Mintz, A. Hajitou, A. S. Sarkis, W. Arap, and R. Pasqualini. 2004. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell 6:275-284. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 6.Chernov, M. V., L. J. Bean, N. Lerner, and G. R. Stark. 2001. Regulation of ubiquitination and degradation of p53 in unstressed cells through C-terminal phosphorylation. J. Biol. Chem. 276:31819-31824. [DOI] [PubMed] [Google Scholar]
- 7.Dai, M. S., S. X. Zeng, Y. Jin, X. X. Sun, L. David, and H. Lu. 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24:7654-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doble, B. W., and J. R. Woodgett. 2003. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116:1175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86-92. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, S. Y., V. Adler, T. Buschmann, Z. Yin, X. Wu, S. N. Jones, and Z. Ronai. 1998. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 12:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 12.Glockzin, S., F. X. Ogi, A. Hengstermann, M. Scheffner, and C. Blattner. 2003. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol. Cell. Biol. 23:8960-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graeber, T. G., J. F. Peterson, M. Tsai, K. Monica, A. J. J. Fornace, and A. J. Giaccia. 1994. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 14:6264-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimes, C. A., and R. S. Jope. 2001. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog. Neurobiol. 65:391-426. [DOI] [PubMed] [Google Scholar]
- 15.Gronroos, E., A. A. Terentiev, T. Punga, and J. Ericsson. 2004. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 101:12165-12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2:405-415. [DOI] [PubMed] [Google Scholar]
- 17.Hainaut, P., and M. Hollstein. 2000. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 77:81-137. [DOI] [PubMed] [Google Scholar]
- 18.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 19.Hockel, M., and P. Vaupel. 2001. Biological consequences of tumor hypoxia. Semin. Oncol. 28:36-41. [PubMed] [Google Scholar]
- 20.Hoeflich, K. P., J. Luo, E. A. Rubie, M. S. Tsao, O. Jin, and J. R. Woodgett. 2000. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature 406:86-90. [DOI] [PubMed] [Google Scholar]
- 21.Jin, A., K. Itahana, K. O'Keefe, and Y. Zhang. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24:7669-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 23.Jones, S. N., A. T. Sands, A. R. Hancock, H. Vogel, L. A. Donehower, S. P. Linke, G. M. Wahl, and A. Bradley. 1996. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc. Natl. Acad. Sci. USA 93:14106-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaeser, M. D., S. Pebernard, and R. D. Iggo. 2004. Regulation of p53 stability and function in HCT116 colon cancer cells. J. Biol. Chem. 279:7598-7605. [DOI] [PubMed] [Google Scholar]
- 25.Katayama, H., K. Sasai, H. Kawai, Z. M. Yuan, J. Bondaruk, F. Suzuki, S. Fujii, R. B. Arlinghaus, B. A. Czerniak, and S. Sen. 2004. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 36:55-62. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman, R. J., D. Scheuner, M. Schroder, X. Shen, K. Lee, C. Y. Liu, and S. M. Arnold. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell. Biol. 3:411-421. [DOI] [PubMed] [Google Scholar]
- 28.Kostova, Z., and D. H. Wolf. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koumenis, C., C. Naczki, M. Koritzinsky, S. Rastani, A. Diehl, N. Sonenberg, A. Koromilas, and B. G. Wouters. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, Z., K. V. Ferry, M. A. Diamond, K. E. Wee, Y. B. Kim, J. Ma, T. Yang, P. A. Benfield, R. A. Copeland, and K. R. Auger. 2001. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 276:31357-31367. [DOI] [PubMed] [Google Scholar]
- 31.Lee, A. S. 2001. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem. Sci. 26:504-510. [DOI] [PubMed] [Google Scholar]
- 32.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 33.Li, H. H., A. G. Li, H. M. Sheppard, and X. Liu. 2004. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol. Cell 13:867-878. [DOI] [PubMed] [Google Scholar]
- 34.Li, M., P. Baumeister, B. Roy, T. Phan, D. Foti, S. Luo, and A. S. Lee. 2000. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302:1972-1975. [DOI] [PubMed] [Google Scholar]
- 36.Li, W. W., Y. Hsiung, Y. Zhou, B. Roy, and A. S. Lee. 1997. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Mol. Cell. Biol. 17:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, H. K., L. Wang, Y. C. Hu, S. Altuwaijri, and C. Chang. 2002. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21:4037-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577-587. [DOI] [PubMed] [Google Scholar]
- 39.Maki, C. G. 1999. Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J. Biol. Chem. 274:16531-16535. [DOI] [PubMed] [Google Scholar]
- 40.Michael, D., and M. Oren. 2003. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13:49-58. [DOI] [PubMed] [Google Scholar]
- 41.Montes, d. O. L., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 42.O'Brate, A., and P. Giannakakou. 2003. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist. Update 6:313-322. [DOI] [PubMed] [Google Scholar]
- 43.O'Keefe, K., H. Li, and Y. Zhang. 2003. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 23:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan, Y., P. R. Oprysko, A. M. Asham, C. J. Koch, and M. C. Simon. 2004. p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene 23:4975-4983. [DOI] [PubMed] [Google Scholar]
- 45.Patel, S., B. Doble, and J. R. Woodgett. 2004. Glycogen synthase kinase-3 in insulin and Wnt signalling: a double-edged sword? Biochem. Soc. Trans. 32:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pluquet, O., and P. Hainaut. 2001. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 174:1-15. [DOI] [PubMed] [Google Scholar]
- 47.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 48.Qu, L., S. Huang, D. Baltzis, A. M. Rivas-Estilla, O. Pluquet, M. Hatzoglou, C. Koumenis, Y. Taya, A. Yoshimura, and A. E. Koromilas. 2004. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan, K. M., A. C. Phillips, and K. H. Vousden. 2001. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13:332-337. [DOI] [PubMed] [Google Scholar]
- 51.Saito, S., H. Yamaguchi, Y. Higashimoto, C. Chao, Y. Xu, A. J. Fornace, Jr., E. Appella, and C. W. Anderson. 2003. Phosphorylation site interdependence of human p53 posttranslational modifications in response to stress. J. Biol. Chem. 278:37536-37544. [DOI] [PubMed] [Google Scholar]
- 52.Schmaltz, C., P. H. Hardenbergh, A. Wells, and D. E. Fisher. 1998. Regulation of proliferation-survival decisions during tumor cell hypoxia. Mol. Cell. Biol. 18:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneiderhan, N., A. Budde, Y. Zhang, and B. Brune. 2003. Nitric oxide induces phosphorylation of p53 and impairs nuclear export. Oncogene 22:2857-2868. [DOI] [PubMed] [Google Scholar]
- 54.Shirangi, T. R., A. Zaika, and U. M. Moll. 2002. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 16:420-422. [DOI] [PubMed] [Google Scholar]
- 55.Stambolic, V., and J. R. Woodgett. 1994. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem. J. 303(Pt. 3):701-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sui, G., E. B. Affar, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, S. R. Grossman, and Y. Shi. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859-872. [DOI] [PubMed] [Google Scholar]
- 58.Turenne, G. A., and B. D. Price. 2001. Glycogen synthase kinase3 beta phosphorylates serine 33 of p53 and activates p53's transcriptional activity. BMC Cell Biol. 2:12. http://www.biomedcentral.com/1471-2121/2/12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van, L. T., A. J. van der Eb, and C. Terleth. 2002. A role for Rad23 proteins in 26S proteasome-dependent protein degradation? Mutat. Res. 499:53-61. [DOI] [PubMed] [Google Scholar]
- 60.Watcharasit, P., G. N. Bijur, L. Song, J. Zhu, X. Chen, and R. S. Jope. 2003. Glycogen synthase kinase-3β (GSK3β) binds to and promotes the actions of p53. J. Biol. Chem. 278:48872-48879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watcharasit, P., G. N. Bijur, J. W. Zmijewski, L. Song, A. Zmijewska, X. Chen, G. V. Johnson, and R. S. Jope. 2002. Direct, activating interaction between glycogen synthase kinase-3β and p53 after DNA damage. Proc. Natl. Acad. Sci. USA 99:7951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterman, M. J., E. S. Stavridi, J. L. Waterman, and T. D. Halazonetis. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19:175-178. [DOI] [PubMed] [Google Scholar]
- 63.Xirodimas, D. P., M. K. Saville, J. C. Bourdon, R. T. Hay, and D. P. Lane. 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118:83-97. [DOI] [PubMed] [Google Scholar]
- 64.Xirodimas, D. P., C. W. Stephen, and D. P. Lane. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 65.Yu, Z. K., R. K. Geyer, and C. G. Maki. 2000. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 19:5892-5897. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, K., and R. J. Kaufman. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279:25935-25938. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., G. W. Wolf, K. Bhat, A. Jin, T. Allio, W. A. Burkhart, and Y. Xiong. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 69.Zhu, Q., G. Wani, M. A. Wani, and A. A. Wani. 2001. Human homologue of yeast Rad23 protein A interacts with p300/cyclic AMP-responsive element binding (CREB)-binding protein to down-regulate transcriptional activity of p53. Cancer Res. 61:64-70. [PubMed] [Google Scholar]