Abstract
Understanding how p53 activity is regulated is crucial in elucidating mechanisms of cellular defense against cancer. Genetic data indicate that Mdmx as well as Mdm2 plays a major role in maintaining p53 activity at low levels in nonstressed cells. However, biochemical mechanisms of how Mdmx regulates p53 activity are not well understood. Through identification of Mdmx-binding proteins, we found that 14-3-3 proteins are associated with Mdmx. Mdmx harbors a consensus sequence for binding of 14-3-3. Serine 367 (S367) is located within the putative binding sequence for 14-3-3, and its substitution with alanine (S367A) abolishes binding of Mdmx to 14-3-3. Transfection assays indicated that the S367A mutation, in cooperation with Mdm2, enhances the ability of Mdmx to repress the transcriptional activity of p53. The S367A mutant is more resistant to Mdm2-dependent ubiquitination and degradation than wild-type Mdmx, and Mdmx phosphorylated at S367 is preferentially degraded by Mdm2. Several types of DNA damage markedly enhance S367 phosphorylation, coinciding with increased binding of Mdmx to 14-3-3 and accelerated Mdmx degradation. Furthermore, promotion of growth of normal human fibroblasts after introduction of Mdmx is enhanced by the S367 mutation. We propose that Mdmx phosphorylation at S367 plays an important role in p53 activation after DNA damage by triggering Mdm2-dependent degradation of Mdmx.
The major role of the p53 tumor suppressor protein in preventing tumorigenesis is well established (20). Approximately half of human cancers harbor deletions or inactivating mutations of the p53 gene, and the remainder display thwarted p53 function due to other genetic or functional defects (31, 54). Therefore, inactivation of the p53 pathway seems to be essential for development of cancer.
p53 is maintained at low levels in nonstressed cells to allow normal growth. Once cells are exposed to various forms of stresses, p53 is stabilized and activated as a sequence-specific transcription factor (20). Subsequently, activated p53 turns on multiple target genes, which induce a variety of biological outcomes such as cell cycle arrest or apoptosis (17). It is likely that induction of these target genes collectively protects organisms from developing cancer. Therefore, accurate regulation of p53 activity is crucial to maintain normal growth under nonstressed conditions and to prevent cells from undergoing oncogenic transformation.
A critical regulator of p53 is Mdm2. Mdm2 is the main protein responsible for maintaining p53 activity at a low level in nonstressed cells (15, 19, 36). Mdm2 inactivates p53 by promoting its degradation by functioning as an E3 ubiquitin ligase, stimulating its nuclear export, and blocking its transcriptional activity (22, 30). Interestingly, the mdm2 gene itself is a direct transcriptional target of p53, thus forming a negative feedback loop involving Mdm2 and p53 (5, 56). Mdm2 is overexpressed in a variety of human cancers, and most of these cancers retain wild-type p53. Therefore, the mdm2 gene is regarded as an oncogene that inactivates p53 by its overexpression (30).
Upon activation of p53 by external or internal stimuli, inactivation of Mdm2 plays an important role. Internal stimuli such as activated oncogenes activate p53 via induction of the p14ARF tumor suppressor, and it was shown that p14ARF induces p53 activity by inactivating Mdm2 (44). External stress, such as DNA damage, activates p53 by phosphorylating Mdm2 and inhibiting its activity as well as by directly inducing multiple posttranslational modifications of p53 itself (24, 26, 45, 49). Thus, Mdm2 is responsible for sensing a variety of cellular stresses and delicately modulating p53 activity.
The Mdm2-related protein Mdmx is another key negative regulator of p53 function. The importance of Mdmx on negative regulation of p53 is underscored by the fact that the embryonic lethal phenotype manifested by mdmx-null mice is rescued by p53 deficiency (10, 29, 33). Similar to Mdm2, Mdmx is overexpressed in a wide spectrum of human tumors that retain wild-type p53 (7, 37, 38). Thus, accumulating data indicate that mdmx is another oncogene that inactivates p53 upon overexpression.
Mdmx is related to Mdm2 in its structure and, like Mdm2, can inhibit transcriptional activity of p53 via direct binding at the N-terminal domain of p53 (46). In contrast to Mdm2, the mechanism by which Mdmx inhibits p53 activity is not well understood. Mdmx does not show robust E3 ubiquitin ligase activity toward p53 and is unable to target p53 for ubiquitin-dependent proteolytic degradation (14, 47). It was reported that Mdmx reduces p53 acetylation (7, 39), which may contribute to p53 inhibition by Mdmx.
It was demonstrated that Mdmx regulates Mdm2 as well as p53; Mdmx stabilizes Mdm2 and p53 (12, 14, 43, 47) and augments the ubiquitinase activity of Mdm2 toward p53 (3, 23, 47). On the other hand, a number of reports indicate that Mdmx in turn is regulated by Mdm2 and p53. Activated p53 stimulates Mdmx cleavage by caspase-3 (11), and Mdm2 targets Mdmx for ubiquitination and degradation (8, 16, 32). In addition, Mdm2 and p53 induce nuclear import of Mdmx (12, 21, 28). Thus, intricate feedback loops exist between p53, Mdm2, and Mdmx (15).
In contrast to Mdm2, it is not clear if inactivation of Mdmx is involved in p53 accumulation in response to various stimuli. It was shown that DNA damage induces Mdmx nuclear translocation (21) and Mdm2-dependent degradation of Mdmx (16). However, it remains unclear if nuclear transport or degradation of Mdmx impacts on p53 activity.
In order to gain further insight into the mechanisms of Mdmx regulation, we searched for a novel protein(s) that physically interacts with Mdmx. We found that several isoforms of 14-3-3 proteins bind to Mdmx and that binding of 14-3-3 is regulated by Mdmx phosphorylation at serine 367 (S367). S367 phosphorylation facilitates its degradation by Mdm2, while mutating S367 to alanine enhances the ability of Mdmx to inhibit transcriptional activity of p53 and promotes cell growth of normal fibroblasts. Furthermore, we demonstrate that DNA damage induces S367 phosphorylation and that the induction of the phosphorylation is associated with increased binding of Mdmx to 14-3-3 and shortening of the half-life of Mdmx. We discuss how DNA damage-induced phosphorylation of Mdmx at S367 can contribute to p53 activation.
MATERIALS AND METHODS
Plasmids.
The expression vectors encoding hemagglutinin (HA)-tagged human p53 (pCMV-HA p53), Flag-tagged human Mdm2, His-tagged ubiquitin, pBax-luc, and pAIP1-luc were previously described (30a, 45). To create Flag-tagged human wild-type Mdmx (Flag-Mdmx), full-length human mdmx cDNA was amplified by PCR with primers (5′-ATATGGTACCGTGACATCATTTTCCACCTCT-3′ and 5′-TATAAGCTTTTATGCTATAAAAACCTTAATAA-3′), digested with KpnI and HindIII, and ligated in frame into the simian virus 40-driven vector with Flag tag (35) digested with KpnI and HindIII. The Flag-tagged Mdmx mutants with alanine substitutions (Flag-S367A, Flag-D361A, and Flag-C463A) were created using the Quikchange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The double mutants of Flag-Mdmx (Flag-S367A/D361A and Flag-S367A/C463A) were created by sequentially performing site-directed mutagenesis.
To create pCMV-Myc-Hdm2, full-length human mdm2 cDNA was amplified by PCR with primers (5′-ATAGAATTCTGGTGAGGAGCAGGCAA-3′ and 5′-TATCTCGAGCTAGGGGAAATAAGTTAGCA-3′), digested with EcoRI and XhoI, and inserted in frame into the corresponding sites of pCMV-Myc (Clontech). To create pCMV-HA-14-3-3ɛ, full-length human 14-3-3ɛ cDNA was amplified by PCR with primers 5′-ATAGAATTCTGGATGATCGAGAGGATC and 5′-TATCTCGAGTCACTGATTTTCGTCTTCC-3′, digested with EcoRI and XhoI, and inserted in frame into the corresponding sites of pCMV-HA (Clontech). To create pBabe-hygro-Flag-Mdmx-wt and pBabe- hygro-Flag-Mdmx-S367, Flag-tagged full-length Mdmx or its S367A mutant was amplified by PCR with primers (5′-TATGGATCCGCCGCCACCATGGACT-3′ and 5′-TATGTCGACTTATGCTATAAAAACCTTAATAA-3′) using Flag-Mdmx and Flag-S367 as a template, respectively. Subsequently amplified DNA was digested with BamHI and SalI, and ligated into the corresponding sites of pBabe-hygro.
Antibodies.
The anti-Flag antibody (M2) conjugated with agarose and anti-Actin antibody were purchased from Sigma. Anti-14-3-3τ antibody was purchased from Calbiochem. Anti-HA antibody was purchased from Covance. Anti-Flag (M2), anti-Myc tag (9E10), anti-14-3-3ɛ, anti-14-3-3γ, anti-14-3-3 (K-19), and anti-Mdmx antibody (D-19) were purchased from Santa Cruz. Monoclonal anti-Mdmx antibodies 6B1A, 11F4D, and 12G11G were previously described (48). Monoclonal antibodies 3G12 and 7C8 were raised against purified glutathione S-transferase (GST)-Mdmx fusion proteins (K. Okamoto, unpublished data). A mixture of 6B1A, 11F4D, 12G11G, 3G12, and 7C8 was used for Western blot analysis described for Fig. 3B. Anti-LacZ antibody was purchased from Abcam. The anti-phospho-S367 polyclonal antibody was raised against a phosphopeptide corresponding to the sequences that span serine 367 (RTISAPVVRPC [phosphorylated serine is underlined]). The anti-phospho-S367 polyclonal antibody was further purified as described before (51).
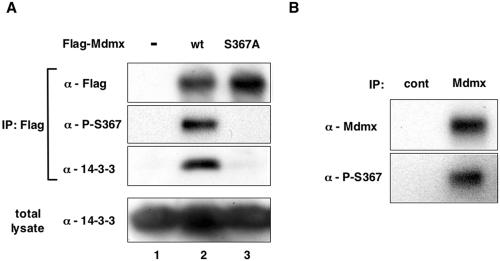
FIG. 3.
S367 of Mdmx is phosphorylated in vivo. (A) WI-38 cells were infected with the control viruses or retroviruses that express wild-type Flag-Mdmx or the S367A mutant. After selection of infected cells by hygromycin resistance, lysates from infected cells were used for immunoprecipitation (IP) with anti-Flag antibody. Total lysates or theanti-Flag immunoprecipitates were then analyzed by Western blot analyses with anti-Flag antibody, anti-phospho-S367 antibody, or an anti-14-3-3 antibody that detects all isoforms (K-19). (B) Lysates from MCF-7 cells were immunoprecipitated with anti-Mdmx antibody (D-19; Santa-Cruz) and analyzed by Western blot analyses with a mixture of monoclonal anti-Mdmx antibodies (see Materials and Methods) or anti-phospho-S367 antibody.
Cells.
BJ and WI-38 cells were obtained from the American Type Culture Collection. p53/Mdmx-deficient mouse embryonic fibroblasts (p53−/−/mdmx−/− MEFs) were previously described (29). CV-1, H1299, and COS-1 cells were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum (GIBCO BRL). WI-38, BJ, and p53−/−/mdmx−/− MEFs were maintained in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% fetal calf serum (GIBCO BRL), 2 mM glutamine, and 0.1 mM β-mercaptoethanol.
DNA transfection.
In DNA transfection experiments using H1299, COS-1, CV-1 and MEF cells, 1 μg of DNA, and 3 μl of Fugene6 reagent (Roche) were introduced per 1.0 × 105 cells. Cells were then incubated for 48 h (COS-1 and CV-1) or 24 h (H1299 and MEFs) before harvest.
Retrovirus-mediated gene transfer.
pBabe-hygro, pBabe-hygro-Flag-Mdmx-wt, or pBabe-hygro-Flag-Mdmx-S367 was used for preparation of amphotropic retroviruses as previously described (25). WI-38 or BJ cells were infected with retroviruses as described before (42), and hygromycin-resistant cells were selected with medium containing 150 μg/ml hygromycin for 4 days. Selected cells were then used for growth assays under the 3T3 protocol.
Purification of Mdmx-binding proteins.
Flag-Mdmx was introduced into COS-1 cells by DNA transfection. For transfection in a large scale, 300 μg of DNA and 900 μl of fugene6 reagent (Roche) were introduced per 3.0 × 107 cells; 48 h after transfection, cells were lysed in MBP buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 0.5% Tween 20, 10% glycerol, and 1 mM EDTA) supplemented with protease inhibitor cocktail (10 μg/ml antipain, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml chymostatin, 10 μg/ml E64, 10 μg/ml phenylmethylsulfonyl fluoride) and phosphatase inhibitor cocktail (PhI) (0.1 mM Na3VO4, 10 mM NaF) for 10 min. Lysates were sonicated, centrifuged twice at 20,000 × g for 10 min, and supernatant were agarose conjugated with anti-Flag antibody for 90 min. Anti-Flag immunoprecipitates were then washed two times with MBP buffer supplemented with protease inhibitor, PhI and 0.8 M NaCl, and four times with MBP buffer supplemented with protease inhibitor and PhI. Subsequently, Flag-Mdmx was eluted with Flag peptide (Sigma), and the eluent were concentrated by acetone precipitation. Flag-tagged proteins and copurified proteins were then separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and visualized by Coomassie staining. Proteins that were bound to Flag-Mdmx were eluted from the gel and used to determine their peptide sequences by mass spectrometry.
Immunoprecipitation and Western blot analysis.
For immunoprecipitation of cell lysates, cells were washed in phosphate-buffered saline and lysed in 200-NP buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5% Nonidet P-40, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol) supplemented with protease inhibitor cocktail and PhI cocktail. In addition, the buffer was supplemented with 1 mg/ml N-methylmaleimide (Sigma), except for experiments described for Fig. 1B and 2B. Lysates were then centrifuged at 20,000 × g for 10 min, and supernatants were incubated with appropriate antibodies in the presence of protein G-Sepharose (Pharmacia) for 2 h. Immunoprecipitates were then washed four times with 200-NP buffer.
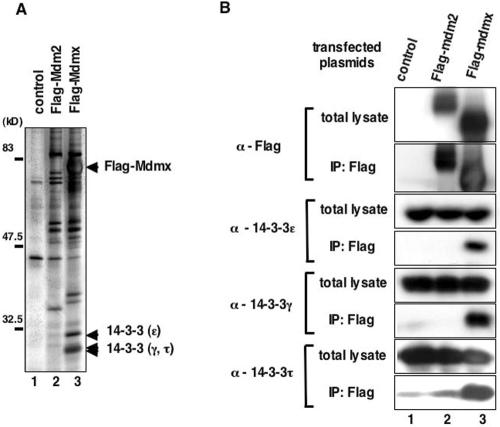
FIG. 1.
14-3-3 proteins bind to Mdmx. (A) Flag-Mdmx, Flag-Mdm2, or a control vector was transfected into COS-1 cells, and lysates prepared from transfected cells were used to purify Flag-tagged proteins on anti-Flag affinity columns as described in Materials and Methods. Polypeptides copurified with Flag-tagged proteins were separated on an SDS-PAGE gel and visualized by silver staining. (B) Polypeptides copurified with Flag-tagged proteins as described for panel A were separated on an SDS-PAGE gel, and Western blot analyses were performed with anti-Flag antibody (M2), or with anti-14-3-3 antibodies which detect the indicated isoforms.
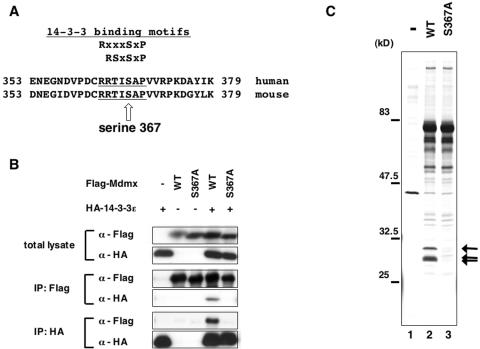
FIG. 2.
Alanine substitution at S367 of Mdmx abolishes binding of 14-3-3 proteins to Mdmx. (A) Mdmx harbors an amino acid sequence that matches known 14-3-3 binding motifs. Previously reported consensus sequences for 14-3-3 binding sites are shown in the upper part of the figure (RXXXSXP and RSXSXP) (9), and aligned with amino acid sequences from human and mouse Mdmx. Amino acid sequences that match 14-3-3 binding motifs are underlined. The putative phosphorylation site (serine 367) is indicated by an arrow. (B) Wild-type Flag-Mdmx, the Flag-S367A mutant Mdmx, or the control vector was transfected into H1299 cells alone or together with HA-14-3-3ɛ. Lysates from transfected cells were used to immunoprecipitate Flag-Mdmx proteins with anti-Flag antibody or HA-14-3-3ɛ proteins with anti-HA antibody. The anti-Flag immunoprecipitates, the anti-HA immunoprecipitates, or total lysates were then analyzed by Western blot analyses with anti-Flag antibody or anti-HA antibody. (C) The control vector, wild-type Flag-Mdmx, or the S367A mutant was transfected into COS-1 cells, and lysates from transfected cells were used to purify Flag-Mdmx on agarose conjugated with anti-Flag antibody as described for Fig. 1A. Proteins copurified with Flag-Mdmx were visualized by silver staining. Polypeptides corresponding to the 14-3-3 proteins are shown by arrows.
For Western blotting analysis, immunoprecipitates were boiled in Laemmli sample buffer, separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 10% nonfat dried milk in TBST (20 mM Tris-HCl [7.5], 140 mM NaCl, 0.1% Tween 20) and probed with appropriate antibodies. In experiments described for Fig. 6 and 7A, RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholic acid, 1 mM dithiothreitol) was used instead of 200-NP buffer.
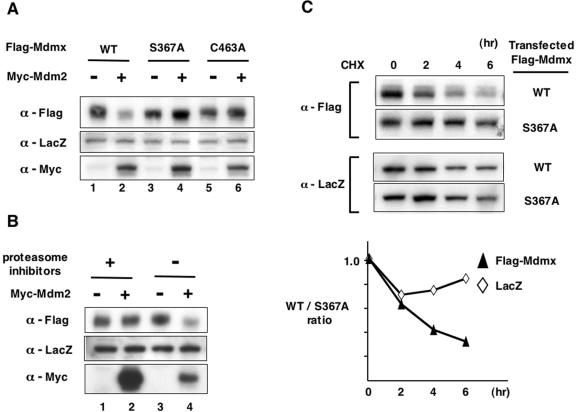
FIG. 6.
S367A is resistant to Mdm2-mediated degradation of Mdmx. (A) We transfected 200 ng of wild-type Flag-Mdmx or its mutants (S367A or C463A) into H1299 cells together with the LacZ expression vector (pCH110), with or without 100 ng of Myc-Mdm2; 24 h after transfection, lysates prepared from transfected cells were used for Western blot analyses with anti-Flag antibody, anti-LacZ antibody, or anti-Myc antibody. (B) Wild-type Flag-Mdmx was transfected into H1299 cells together with the LacZ expression vector, with or without Myc-Mdm2 as described for panel A; 24 h after transfection, cells were incubated with proteasome inhibitors (30 μM ALLN, 50 μM MG132, plus 50 μM LLnL) or dimethyl sulfoxide (DMSO) for an additional 5 h. Western blot analyses were performed as described for panel A. (C) S367A mutation stabilizes Mdmx in the presence of Mdm2. (Upper panel) Wild-type Flag-Mdmx or the S367A mutant was transfected into H1299 cells together with the LacZ expression vector and Myc-Mdm2 as described for Fig. 6A; 24 h after transfection, 50 μg/ml of cycloheximide (CHX) was added to the medium, and cells were harvested at the indicated times. Lysates prepared from cycloheximide-treated cells were used for Western blot analyses with anti-Flag antibody or anti-LacZ antibody. (Bottom panel) Levels of introduced LacZ and Mdmx shown in the upper panel were quantified, and the wild-type/S367A ratio (values from cells transfected with wild-type Mdmx/values from cells transfected with the S367A mutant) was calculated at each time point. Each calculated value was normalized such that the wild-type/S367A ratio at 0 h is presented as 1.0.
FIG. 7.
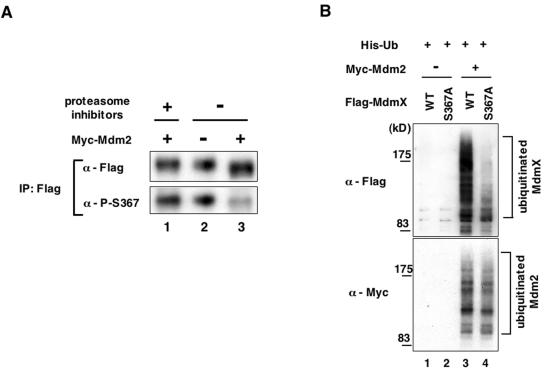
S367 phosphorylation facilitates Mdm2-dependent degradation of Mdmx, and the S367A mutant is resistant to Mdm2-mediated ubiquitination. (A) Mdmx phosphorylated at S367 is preferentially degraded by Mdmx. Wild-type Flag-Mdmx was transfected into H1299 cells alone or together with Myc-Mdm2 as described for Fig. 6B; 24 h after transfection, cells were incubated with proteasome inhibitors as described for Fig. 6 or dimethyl sulfoxide for an additional 5 h. Subsequently, lysates prepared from transfected cells were used to immunoprecipitate Flag-Mdmx with anti-Flag antibody. Approximately equal amounts of immunoprecipitated Flag-Mdmx were used for Western blot analyses with anti-Flag antibody or anti-phospho-S367 antibody. (B) Either wild-type Flag-Mdmx or the S367A mutant was transfected into H1299 cells together with (His)6-tagged ubiquitin (His-Ub), with or without Myc-Mdm2; 24 h after transfection, cells were incubated with a proteasome inhibitor (50 μM MG132) for an additional 3 h. Subsequently, cells were lysed with a buffer containing 8 M urea, and lysates were used to purify His-ubiquitin on Ni-NTA-agarose (QIAGEN). Flag-Mdmx or Myc-Mdm2 conjugated with His-ubiquitin was detected by Western blot analyses with anti-Flag antibody or anti-Myc antibody, respectively.
Cycloheximide chase assay.
H1299 cells were transiently transfected using Fugene 6. 24 h after transfection, culture medium was replaced by growth medium supplemented with cycloheximide (50 μg/ml). Cells were harvested at 0, 2, 4, or 6 h after addition of cycloheximide, and lysates from cycloheximide-treated cells were analyzed by Western blot analyses. Levels of Flag-Mdmx and LacZ were quantified using NIH image software, and the effect of the the S367A mutation on these proteins was calculated as described in the legends.
In vivo ubiquitination assay.
At 24 h after transfection, cells were treated with 50 μM MG132 for 3 h. Lysis of the cells and the purification of His-tagged proteins were performed as described before (8). Purified proteins conjugated with (His)6-ubiquitin were analyzed by Western blot analyses.
In vitro GST pull-down assay.
GST and different GST-14-3-3 fusion proteins were expressed in bacteria and purified on glutathione-Sepharose; 1 μg of GST fusion protein was incubated with lysates prepared from neocarzinostatin-treated or untreated MCF-7 cells. The mixture was rotated for 2 h at 4°C and washed three times with Giordano lysis buffer (27). Proteins copurified on the Sepharose were separated on SDS-PAGE gels and used for Western blot analyses.
RESULTS
14-3-3 proteins associate with Mdmx.
In order to gain insight into the mechanisms by which Mdmx function is regulated, we set out to identify novel Mdmx-binding protein(s). Constructs expressing wild-type Flag-tagged human Mdmx (Flag-Mdmx) or Flag-tagged human Mdm2 (Flag-Mdm2) were transfected into COS-1 cells, and lysates from transfected cells were used to purify Flag-tagged proteins on agarose conjugated with anti-Flag antibody. Subsequently, polypeptides that were copurified with Flag-tagged proteins were separated on an SDS-PAGE gel. Silver staining of the gel revealed three prominent polypeptides (∼30 kDa) that were specifically bound to Mdmx (Fig. 1A).
After large-scale copurification of Mdmx binding proteins, the identity of these polypeptides was determined by mass spectrometry. Analyses of oligopeptides derived from these endogenous proteins revealed that all of these polypeptides belong to a family of 14-3-3 proteins. The 14-3-3 family is composed of seven isoforms in mammals (β, γ, ɛ, σ, ζ, τ, and η), and each member is involved in unique yet overlapping functions in cells (9, 52). The polypeptide that migrated more slowly than the others corresponds to 14-3-3ɛ, while the other two polypeptides that migrated faster as a doublet correspond to 14-3-3γ and 14-3-3τ (Fig. 1A). Binding of these 14-3-3 proteins to Mdmx was further confirmed by Western blot analyses (Fig. 1B).
Inspection of the amino acid sequences of human and mouse Mdmx revealed that they both harbor consensus motifs for binding of 14-3-3 (9, 52) that spans from amino acids 363 to 369 (Fig. 2A). In contrast, Mdm2 does not harbor a 14-3-3 consensus sequence, and failed to bind 14-3-3 proteins (Fig. 1A and 1B).
When we performed Western blot analyses under the same conditions as described for Fig. 1B, we also detected association of Mdmx with 14-3-3β, 14-3-3ζ, and 14-3-3η (data not shown). 14-3-3σ expression was barely detectable, and association of Mdmx with 14-3-3σ was not determined (data not shown). Thus, Mdmx associates with many isoforms of the 14-3-3 family.
Alanine substitution at serine 367 abolishes binding of Mdmx to 14-3-3.
It is well documented that, in many cases, the conserved serine residue of the 14-3-3-binding consensus sequence (Fig. 2A) is phosphorylated in vivo, and that 14-3-3 binds preferentially to target proteins that are phosphorylated at this serine residue (9, 52). Thus, serine phosphorylation at the 14-3-3 binding site regulates functions of the target proteins by facilitating association between 14-3-3 and its targets.
In order to determine if binding of 14-3-3 to Mdmx is dependent on integrity of the conserved serine residue (S367) at the 14-3-3 binding consensus site, we created a mutant form of Mdmx in which S367 is substituted by alanine (S367A) (Fig. 2A). We transfected either wild-type Flag-Mdmx or its S367A mutant alone or together with HA-tagged 14-3-3ɛ (HA-14-3-3ɛ) into cells and determined if the S367A mutation inhibits interaction between Mdmx and 14-3-3 by Western blot analyses. As expected, the S367A mutation abolished binding of Flag-Mdmx to HA-14-3-3ɛ (Fig. 2B). Next we transfected either wild-type Flag-Mdmx or the S367A mutant into COS-1 cells, and examined if the the S367A mutation affects the association between transfected Mdmx and endogenous 14-3-3 proteins. As already presented in Fig. 1A, silver staining of proteins copurified with Mdmx demonstrated that wild-type Mdmx was capable of binding to endogenous 14-3-3 proteins (Fig. 2C, lane 2). In contrast, the S367A mutant was defective in binding to these proteins (Fig. 2C, lane 3). These data indicate that the S367A mutation abolishes binding between Mdmx and 14-3-3 proteins.
S367 is phosphorylated in vivo.
Abolishment of Mdmx-14-3-3 interaction by mutation of S367 strongly suggests that this residue is phosphorylated in vivo. Therefore, we raised an antibody that specifically reacts with phosphorylated S367 (anti-P-S367). We validated specificity of the antibody by its loss of reactivity with Mdmx by the S367A mutation (Fig. 3A) and after phosphatase treatment of immunoprecipitated Mdmx (see Fig. S1 in the supplemental material). Using anti-P-S367, we determined if S367 of Mdmx is phosphorylated by Western blot analyses. We found that, after retrovirus-mediated transfer of wild-type Flag-Mdmx into WI-38 cells, the ectopically expressed Mdmx was phosphorylated at S367 and bound to 14-3-3 (Fig. 3A, lane 2). In contrast, neither S367 phosphorylation nor interaction with 14-3-3 was observed after introduction of the S367A mutant (Fig. 3A, lane 3).
In addition to WI-38, we also used two different cell lines, COS-1 and CV-1, to examine whether S367 is phosphorylated after introduction of Mdmx into cells by DNA transfection. In both cell lines, we found that wild-type Mdmx was phosphorylated on S367, and that the S367A mutation abolished both S367 phosphorylation and binding to 14-3-3 (data not shown). Thus, in three different cells, S367 is phosphorylated after exogenous introduction of Mdmx, and the S367A mutation abolishes both S367 phosphorylation and binding of Mdmx to 14-3-3. These data indicate that, like other 14-3-3 binding proteins, serine phosphorylation at the binding consensus sequence induces binding of 14-3-3 to Mdmx. Of note, aspartate mutation at S367 (S367D), as well as S367A, abolished binding of Mdmx to 14-3-3 (see Fig. S2 in the supplemental material). Presumably conformational change rather than a mere increase of negative charge at the 367 residue is necessary for binding of 14-3-3 to Mdmx phosphorylated at S367.
In order to determine if endogenous Mdmx is phosphorylated at S367 in vivo, we examined S367 phosphorylation in MCF-7 cells, which express high levels of endogenous Mdmx (7). Western blot analyses of anti-Mdmx immunoprecipitates indicated that S367 of endogenous Mdmx was phosphorylated in MCF-7 (Fig. 3B). Therefore, at least a fraction of S367 is phosphorylated at S367 under normal growth conditions.
The S367A mutation of Mdmx augments cooperative repression of p53-dependent transcriptional activation by Mdm2 and Mdmx.
It was reported that Mdmx inhibits transcriptional activation by p53 (14, 46). In order to evaluate the potential effects of Mdmx S367 phosphorylation on transcriptional activation by p53, we determined the effect of either wild-type Mdmx or the S367A mutant on transactivation of the p53-responsive promoters (Fig. 4). Because it was reported that Mdm2 cooperates with Mdmx to inhibit transcriptional activity of p53 (12), we initially performed the transfection experiments both in the presence and the absence of Mdm2 in p53-deficient cells (H1299).
FIG. 4.
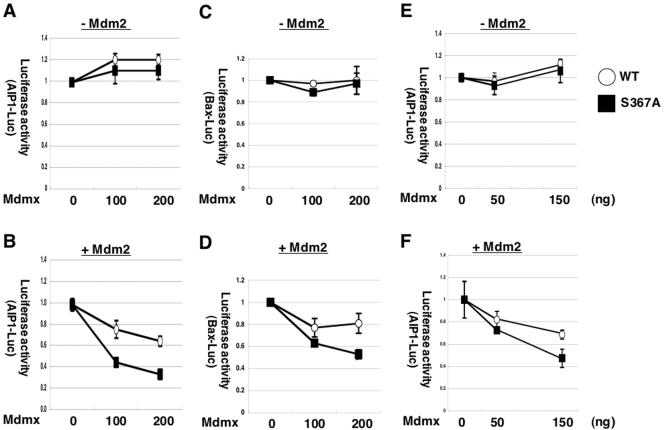
Cooperative repression of p53 transcriptional activity by Mdmx and Mdm2 is augmented by the S367A mutation. H1299 cells (A, B, C, and D) or p53−/−/Mdmx−/− MEFs (E and F) were transfected with HA-p53, Renilla luciferase control vector, and firefly luciferase vector linked to the aip1 (A, B, E, and F) or the bax (C and D) promoter, together with (B, D, and F) or without (A, C, and E) Myc-Mdm2. Increasing amounts of wild-type Flag-Mdmx or the S367A mutant were cotransfected as indicated at the bottom of each graph. In experiments with Mdm2 cotransfection (B, D, and F), 100 ng (B and D) or 200 ng (F) of Myc-Mdm2 was introduced. In all experiments, 50 ng of HA-p53 was transfected and the total amount of DNA was adjusted to 2 μg with pBluescript plasmid (Stratagene); 24 h after transfection, cells were lysed and luciferase activity was measured using the dual-luciferase assay system (Promega). Mean values (±standard deviation) from three independent experiments were determined. Basal promoter activity expressed in the absence of HA-p53 was measured and subtracted in each experiment. Values presented were calculated as follows: value from cells transfected with indicated amount of Mdmx/value from cells transfected without Mdmx.
In order to clearly demonstrate the potential cooperative effect of Mdm2 and Mdmx on p53 activity, we determined the maximum amounts of transfected Mdm2 that did not cause significant reduction of p53 activity in the absence of transfected Mdmx. We found that cotransfection of p53 together with moderate excess amounts of Mdm2 (p53 to Mdm2, 1:2) caused only marginal reduction (11%) of luciferase activity from the p53-responsive promoter (aip1) in H1299 cells. The amounts of Mdm2 thus determined were used in the cotransfection experiments.
In the presence of cotransfected Mdm2, we observed moderate reduction of luciferase activity from two p53-responsive promoters (the aip1 and the bax promoters) by wild-type Mdmx in H1299 cells. Notably, inhibition of luciferase activity by Mdmx was enhanced approximately by twofold when the S367A mutation was introduced (Fig. 4B and D), indicating that the S367A mutation augments the ability of Mdmx to repress transactivation by p53.
In contrast, in the absence of cotransfected Mdm2, we did not observe a significant reduction of luciferase activity from the aip1 or the bax promoter either by wild-type Mdmx or the S367A mutant under the same experimental conditions (Fig. 4A and C). These data indicate that Mdm2 and Mdmx cooperatively repress transcriptional activity of p53, and that the S367A mutation augments such cooperative repression of p53.
Next, we examined the effect of the S367A mutation in cells that are deficient in Mdmx or Mdm2. After cotransfection under conditions similar to those described for Fig. 4A and B, we measured luciferase activity from the aip1 promoter in p53−/−/mdmx−/− MEFs or p53−/−/mdm2−/− fibroblasts. Again we observed that the S367A mutation enhanced functional cooperation between Mdmx and Mdm2 to repress transactivation by p53 in p53−/−/mdmx−/− MEFs (Fig. 4E and F) and p53−/−/mdm2−/− fibroblasts (data not shown). Thus, enhancement of Mdmx function by the S367A mutation is not mediated by augmenting the activity of endogenous wild-type Mdmx or endogenous Mdm2, and the S367A mutant per se possesses higher activity to repress p53 function.
S367A mutant is resistant to Mdm2-dependent degradation of Mdmx.
Enhanced repression of p53 by the S367A mutant prompted us to investigate its biochemical mechanism. In order to determine if the S367A mutation affects levels of introduced p53, Mdm2, or Mdmx, we first performed DNA transfection under the experimental conditions described for Fig. 4B and 4D in which the effect of S367 mutation on p53 was clearly observed, and determined levels of these proteins in H1299 cells.
In agreement with published data, levels of introduced Mdm2 increased after introduction of wild-type Mdmx (Fig. 5A, lanes 1 and 2). However, the S367A mutation did not cause a further increase the levels of Mdm2 (Fig. 5A, lanes 2 and 3). Likewise, the S367 mutation did not affect p53 levels (Fig. 5A, lanes 2 and 3). Therefore, it is not likely that enhanced activity of Mdmx by the S367A mutation depends on changes in the levels of Mdm2 or p53.
FIG. 5.
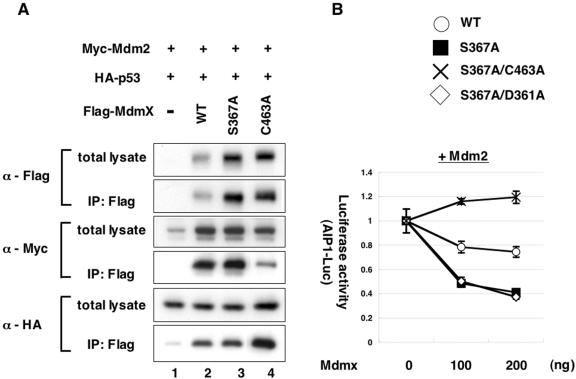
Characterization of the Mdmx mutants. (A) H1299 cells were transfected with 100 ng of Myc-Mdm2 and 50 ng of HA-p53 together with 200 ng of wild-type Flag-Mdmx, the indicated Flag-Mdmx mutant, or the control vector. Lysates from transfected cells were used for immunoprecipitation (IP) with anti-Flag antibody, and the anti-Flag immunoprecipitates or total lysates were analyzed by Western blot analyses with anti-Flag antibody, anti-Myc antibody, or anti-HA antibody. (B) H1299 cells were cotransfected as described for Fig. 4. Increasing amounts of wild-type Flag-Mdmx or indicated Mdmx mutant were transfected together with Myc-Mdm2, HA-p53, Renilla luciferase control vector, and firefly luciferase vector linked to the aip1 promoter. Normalized values are calculated from luciferase activity and presented as described for Fig. 4.
In contrast, levels of Mdmx with the S367A mutation were higher than wild-type Mdmx (Fig. 5A, lanes 2 and 3), suggesting that increased levels of Mdmx may explain the stronger inhibition of the p53 activity by the S367A mutant. Because the effect of the S367A mutation on transcriptional activity of p53 can be observed only if Mdm2 is cointroduced (Fig. 4), we examined whether the increased levels of Mdmx by the S367A mutation are caused by coexpression of Mdm2. We determined levels of wild-type Mdmx and the S367A mutant in either the presence or the absence of cotransfected Mdm2. Mdm2 introduction caused reduction of wild-type Mdmx (Fig. 6A, lanes 1 and 2), whereas the S367A mutant was resistant to the Mdm2-dependent decrease (Fig. 6A, lanes 3 and 4). The resistance of the S367A mutant to Mdm2-mediated inhibition of Mdmx was also demonstrated in p53−/−mdmx−/− MEFs (see Fig. S3 in the supplemental material). Thus, the effects of the S367A mutation on levels of Mdmx parallel its effects on the transactivating function of p53, strongly suggesting that the resistance of the S367A mutant to inhibition by Mdm2 is responsible for the increased ability of the mutant to repress p53.
The RING finger domain of Mdmx is responsible for its binding to Mdm2 (43, 50) and for its ubiquitination and degradation by Mdm2 (8, 23, 32). We examined if integrity of this domain is necessary for Mdm2-dependent inhibition of Mdmx in our experimental conditions. Mutation at C463, in which cysteine is substituted by alanine (C463A), compromises the integrity of the RING finger domain of Mdmx (23). Upon testing the mutant Mdmx (C463A), we found that it markedly reduced the affinity of Mdmx for Mdm2 (Fig. 5A, lane 4), and completely neutralized the ability of the S367A mutant to cooperate with Mdm2 to inhibit transcriptional activity of p53 (Fig. 5B). We also found that the C463A mutant was resistant to Mdm2-dependent inhibition (Fig. 6A, lanes 5 and 6), Thus, an intact Mdmx RING finger domain, which is needed for the Mdmx/Mdm2 interaction, is required for Mdm2-dependent inhibition of Mdmx.
S367A mutant is resistant to Mdm2-mediated proteasomal degradation.
It was previously demonstrated that Mdm2 promotes Mdmx ubiquitination and targets it for proteasomal degradation (8, 16, 32). In order to determine if the reduction of levels of wild-type Mdmx by Mdm2 presented in Fig. 6A is mediated via proteasomal degradation, we introduced wild-type Mdmx alone or together with Mdm2 in either the absence or the presence of proteasome inhibitors. As already demonstrated in Fig. 6A, Mdm2 showed reduced levels of wild-type Mdmx in the absence of proteasome inhibitors (Fig. 6B, lanes 3 and 4). In contrast, reduction of wild-type Mdmx by Mdm2 was blocked in their presence (Fig. 6B, lanes 1 and 2), indicating that inhibition of wild-type Mdmx by Mdm2 is mediated by proteasome-mediated degradation.
It was previously shown that the half-life of wild-type Mdmx is markedly decreased in the presence of Mdm2 (8). In order to confirm that Mdm2 promotes degradation of wild-type Mdmx, the stability of introduced Mdmx was determined by a cycloheximide chase assay in the presence of Mdm2 (Fig. 6C). Either wild-type Flag-Mdmx or the S367A mutant was transfected into H1299 cells together with Mdm2 and lacZ, and transfected cells were further incubated with cycloheximide for 0 to 6 h. Subsequently, levels of Flag-Mdmx and LacZ in transfected cells before and after treatment with cycloheximide were determined by Western blot analyses to evaluate the stability of Flag-Mdmx and LacZ (8).
The results indicate clearly that the levels of wild-type Mdmx decreased more rapidly than those of the S367A mutant, while the stability of LacZ (Fig. 6C) as well as Mdm2 (data not shown) was similar in these transfections. Combined with the results described in Fig. 6A and 6B, these results indicate that the S367A mutation protects Mdmx from Mdm2-dependent proteasomal degradation.
Mdmx phosphorylated at S367 is preferentially degraded by Mdm2.
The resistance of the S367A mutant to Mdm2-dependent degradation suggests that S367 phosphorylation facilitates degradation of Mdmx by Mdm2. In order to examine if Mdm2 preferentially targets Mdmx phosphorylated at S367 for degradation, we introduced wild-type Mdmx alone or together with Mdm2 into H1299 cells, and determined if the fraction of Mdmx phosphorylated at S367 relatively decreased in the presence of Mdm2 (Fig. 7A). Approximately equal amounts of immunoprecipitated Flag-Mdmx were used to compare the extent of S367 phosphorylation. Concomitantly with degradation of Mdmx by Mdm2 (Fig. 6A and 6B), the fraction of Mdmx phosphorylated at S367 decreased markedly (Fig. 7A, lanes 2 and 3). Furthermore, proteasome inhibitors blocked Mdm2-dependent reduction of a fraction of Mdmx phosphorylated at S367 (Fig. 7A, lanes 1 and 3). These results indicate that Mdmx phosphorylated at S367 is a preferred target for Mdm2-dependent proteasomal degradation. Taken together, our data demonstrate that S367 phosphorylation promotes Mdm2-dependent degradation of Mdmx.
S367A Mdmx mutant is resistant to Mdm2-dependent ubiquitination.
Next, we attempted to determine whether the S367A mutation inhibits Mdm2-dependent degradation of Mdmx by blocking Mdmx ubiquitination by Mdm2. Either wild-type or the S367A mutant versions of Flag-tagged Mdmx were introduced into H1299 cells together with Myc-Mdm2, and the extent of Mdmx ubiquitination was determined by Western blot analyses. In order to facilitate detection of ubiquitinated Mdmx, (His)6-tagged ubiquitin was cotransfected. Subsequently we purified His-ubiquitin from lysates prepared from transfected cells and subjected them to Western blot analyses, probing with anti-Flag antibodies (Fig. 7B, upper panel). In the presence of transfected Mdm2, we observed polyubiquitination of wild-type Mdmx that was detected as slower-migrating forms of Mdmx (Fig. 7B, lane 3). In contrast, we did not observe significant polyubiquitination of the S367 mutant under the same conditions (Fig. 7B, lane 4). In the absence of transfected Mdm2, neither wild-type Mdmx nor the S367A mutant was ubiquitinated (Fig. 7B, lanes 1 and 2). We also observed the inhibition of polyubiquitination of Mdmx by S367 mutation if we examined anti-Flag immunoprecipitates after the cotransfection of Flag-Mdmx, Myc-Mdm2, and His-ubiquitin (see Fig. S4 in the supplemental material). Thus, the S367 mutation blocks Mdm2-mediated ubiquitination of Mdm2. Note that ubiquitination of Mdm2 itself was not significantly affected by the S367A mutation, as revealed by Western blotting of Myc-Mdm2 conjugated with His-ubiquitin (Fig. 7B, lower panel).
Taken together, our results show that S367 phosphorylation facilitates Mdm2-dependent ubiquitination and degradation of Mdmx.
S367 phosphorylation is induced by DNA damage.
The stimulating effect of the S367A mutation on Mdmx-dependent inhibition of p53 activity strongly suggests that S367 phosphorylation may function to activate p53. The major stress known to induce p53 activity in cells is DNA damage. Of note, it was recently demonstrated that DNA damage profoundly affects the stability and subcellular localization of Mdmx (16, 21). Therefore, we examined if treatment of cells with DNA damaging agents can induce S367 phosphorylation in MCF-7 cells, which express high levels of endogenous Mdmx. Since our data indicate that Mdmx phosphorylated at S367 is targeted for proteasomal degradation, MCF-7 cells were pretreated with MG132 to block potential degradation of the phosphorylated Mdmx. Subsequently the treated cells were exposed to a variety of DNA-damaging agents, and lysates prepared from exposed cells were used to determine if S367 phosphorylation was induced.
Treatment of cells with gamma irradiation or etoposide caused rapid and drastic induction of S367 phosphorylation (Fig. 8B; see also Fig. S5 in the supplemental material). Treatment with adriamycin or UV also induced S367 phosphorylation, although the induction was weaker and slower than that caused by gamma irradiation or etoposide (Fig. 8B; see also Fig. S5 in the supplemental material). Thus, a variety of DNA lesions induce S367 phosphorylation of endogenous Mdmx.
FIG. 8.
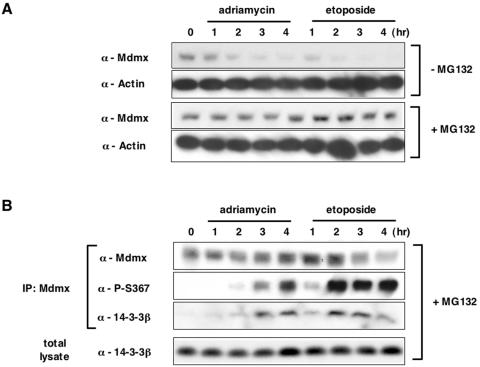
Induction of S367 phosphorylation after DNA damage is associated with increased binding of 14-3-3 to Mdmx and accelerated Mdmx degradation. (A) MCF-7 cells were preincubated with dimethyl sulfoxide (−MG132) or 20 μM MG132 (+MG132) for 1 hour and then incubated with 3 μM adriamycin or 20 μM etoposide for the indicated periods. Subsequently lysates prepared from the drug-treated cells were used for Western blot analyses with anti-Mdmx antibody (D-19; Santa-Cruz) or anti-actin antibody. (B) MCF cells were preincubated with MG132 and exposed to DNA-damaging compounds as described in A. Cell lysates prepared from the drug-treated cells were used for immunoprecipitation with anti-Mdmx antibody (D-19). The anti-Mdmx immunoprecipitates or total lysates were used for Western blot analyses with the anti-Mdmx antibody (D-19), the anti-P-S367 antibody, or anti-14-3-3β antibody (Santa Cruz).
Induction of S367 phosphorylation after DNA damage is associated with increased binding of Mdmx to 14-3-3.
The data presented in Fig. 2 and 3 indicate that S367 phosphorylation of Mdmx facilitates binding of 14-3-3 to Mdmx. Therefore, we next examined if the increased binding of 14-3-3 to Mdmx are associated with induction of S367 phosphorylation observed after DNA damage. Treatment of MCF-7 cells with adriamycin or etoposide induced association of 14-3-3 with Mdmx in accordance with induction of S367 phosphorylation (Fig. 8B). Thus, induction of S367 phosphorylation after DNA damage is associated with increased binding of Mdmx to 14-3-3. Furthermore, in vitro GST pulldown assays with various GST-14-3-3 proteins clearly indicated that 14-3-3 preferentially interacted with Mdmx proteins from lysates prepared from MCF-7 cells after treatment with neocarzinostatin, a compound that causes double-strand breaks in DNA (4) (see Fig. S6 in the supplemental material). p53 also bound slightly better to 14-3-3 after DNA damage, in accordance with previously published results (55). Therefore, DNA damage induces enhanced binding of Mdmx to 14-3-3 both in vivo and in vitro.
Induction of S367 phosphorylation after DNA damage is associated with shortening of the half-life of Mdmx.
The results described in Fig. 6 and 7 indicate that S367 phosphorylation triggers Mdm2-dependent proteasomal degradation of Mdmx. Therefore, we next attempted to determine whether induction of S367 phosphorylation seen after DNA damage is associated with enhanced degradation of Mdmx. Remarkably, treatment of MCF-7 cells with adriamycin or etoposide reduced levels of Mdmx in the absence of MG132, while the proteasome inhibitor blocked such reduction (Fig. 8A). It is likely that reduction of Mdmx levels after etoposide treatment was caused by enhanced degradation of Mdmx, since etoposide treatment reduced stability of Mdmx (see Fig. S7 in the supplemental material).
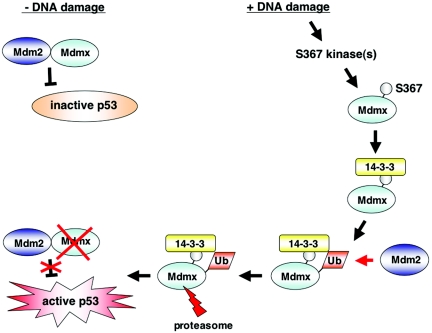
Taken together, treatment of cells with DNA damage induces S367 phosphorylation, binding 14-3-3 to Mdmx, and degradation of Mdmx with a similar kinetics. Our data support the model that induced S367 phosphorylation triggers Mdmx degradation via facilitation of binding of 14-3-3 to Mdmx (see Fig. 10).
FIG. 10.
Model of the molecular mechanism of p53 activation by S367 phosphorylation. In cells with low levels of activity of the S367 kinase(s), Mdm2 and Mdmx cooperatively repress transcriptional activity of p53. After DNA damage induces activation of the S367 kinase(s) and S367 phosphorylation, 14-3-3 binds to Mdmx. Binding of 14-3-3 to Mdmx induces ubiquitination of Mdmx by Mdm2. Ubiquitination of Mdmx causes its proteasomal degradation and leads to p53 activation.
Expression of the S367A mutant causes accelerated cell growth of normal human fibroblasts.
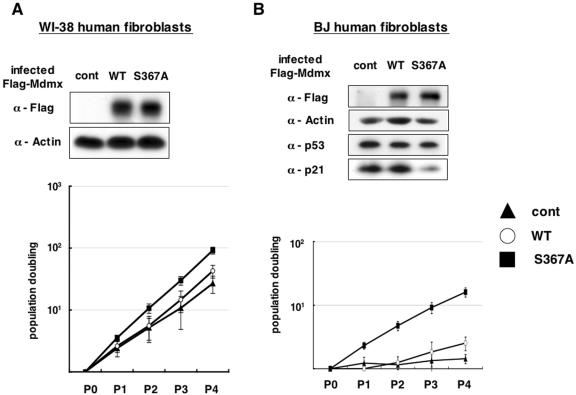
Enhanced suppression of transcriptional activity of p53 by the S367A mutation suggested possibility that the S367A mutation may give growth advantages to cells that harbor wild-type p53. To test this, we infected normal human fibroblasts (young WI-38 and presenescent BJ cells) with either control retroviruses or retroviruses expressing either wild-type or S367A mutant Mdmx (Fig. 9).
FIG. 9.
Introduction of the S367A mutant into normal human fibroblasts causes accelerated cell growth. Young WI-38 cells (A) and presenescent BJ cells (B) were infected with retroviruses that express wild-type Flag-Mdmx (circles), the S367A mutant (squares), or the control retroviruses (triangles). After selection of infected cells by hygromycin resistance, cells were grown under the 3T3 protocol. Cells were split every 3 days, and cell numbers were calculated and plotted on a logarithmic scale. Cell counting started at the end of drug selection. Mean values (±standard deviation) from three independent experiments are presented to evaluate growth of the infected cells. The accumulated number of population doublings is shown on a log scale. Lysates were prepared from infected cells after drug selection, and Western blot analyses were performed with anti-Flag antibody, antiactin antibody, anti-p53 antibody, and anti-p21 antibody.
In both fibroblast populations, comparison of cells infected with control viruses and wild-type Mdmx indicated that introduction of wild-type Mdmx gave only marginal growth advantage to these cells. In contrast, WI-38 cells expressing the S367A mutant grew significantly faster than those introduced with wild-type Mdmx or the control viruses, while the effect of the S367A mutation was striking in presenescent BJ cells (Fig. 9B). Remarkably, levels of p21, one of a key transcriptional target of p53, were clearly reduced by the S367A mutation, while p53 levels were not significantly affected (Fig. 9B). Our data thus indicate that the S367A mutation promotes cell growth in normal cells, probably by inhibiting p53 function.
DISCUSSION
Genetic data indicate that Mdmx functions to keep p53 activity at a low level in cells (10, 29, 33). Accordingly, introduction of Mdmx in cells suppresses the transactivating function of p53 (46). However, how Mdmx functions to inhibit p53 remains obscure, and little is known about how its activity is regulated. In an attempt to identify new Mdmx-interacting proteins that may be linked to regulation of Mdmx function, we found that several isoforms of 14-3-3 (γ, τ, and ɛ) bind Mdmx, and that their association with Mdmx is dependent on Mdmx phosphorylation at S367.
Through analyses of S367 phosphorylation, several new findings on Mdmx regulation are presented in this paper. First, S367 phosphorylation facilitates Mdm2-dependent ubiquitination and degradation of Mdmx. Second, the S367A mutation enhances the ability of Mdmx to cooperate with Mdm2 to repress transcriptional activity of p53. Third, S367 phosphorylation is induced after DNA damage. Finally, introduction of the S367A mutant into normal fibroblasts causes enhanced cell growth. Based on these data, we propose a model for the mechanism by which S367 phosphorylation of Mdmx regulates the trans-activating function of p53 (Fig. 10).
Our model depicts how a complicated network formed between Mdm2, Mdmx, and p53 is regulated by S367 phosphorylation. When Mdmx is not phosphorylated at S367, Mdm2 is not capable of targeting it for ubiquitination and degradation. This allows Mdmx to stably exist in cells and, in collaboration with Mdm2, to suppress p53 function. Once S367 is phosphorylated after DNA damage, Mdmx becomes a substrate for Mdm2-mediated ubiquitination and degradation. Thus, p53 is released from suppression by Mdmx and activated.
Mdm2 targets both Mdmx and p53 for degradation. Interestingly, efficient targeting of both substrates by Mdm2 is controlled by phosphorylation of its substrates. Ubiquitination and degradation of p53 by Mdm2 are inhibited by phosphorylation at the N-terminal domain of p53 (1, 36), while Mdm2-dependent ubiquitination and degradation of Mdmx are facilitated by S367 phosphorylation of Mdmx, as presented in this paper. Thus, in both cases, phosphorylation of the substrates of Mdm2 leads to p53 activation. p53 phosphorylation at the N-terminal domain is induced by DNA damage via activation of ATM/ATR kinases and Chk1/Chk2 kinases (6, 13, 40). Interestingly, S367 phosphorylation induced after DNA damage is blocked by caffeine, an inhibitor of ATM/ATR kinases (41) (see Fig. S8 in the supplemental material). Of note, Pereg et al. recently demonstrated that, after DNA damage, phosphorylation of Mdmx is induced at multiple sites, including S367 (S342, S367, and S403), and that one of the phosphorylation sites, S403, is a direct target of ATM kinase (34). Therefore, it is possible that the ATM/ATR kinase pathway plays a major role in phosphorylation of the N-terminal domain of both p53 and Mdmx at S367.
At this moment, the molecular mechanism of how S367 phosphorylation facilitates Mdm2-mediated ubiquitination is not clear. We observed that wild-type Mdmx and the S367 mutant showed a similar affinity for Mdm2 in transfected cells in the presence of MG132 (data not shown). Therefore, it is not likely that the resistance of the S367A mutant to Mdm2-mediated ubiquitination is due to reduced interaction between Mdm2 and the S367A mutant.
Because a direct outcome of S367 phosphorylation is likely to be binding of 14-3-3 proteins to Mdmx, it is likely that such binding facilitates Mdmx ubiquitination by Mdm2. 14-3-3 proteins bind a multitude of functionally diverse signaling proteins, and play critical roles in signal transduction pathways and cell cycle progression (53). One of the many functions attributed to 14-3-3 is shuttling of its target proteins between the cytoplasm and nucleus (9, 52, 53). In fact, it was demonstrated that Mdmx shuttles between these cellular compartments, and that Mdm2 can recruit Mdmx into the nucleus (12, 21, 28). Therefore, it may be possible that degradation of Mdmx is regulated through its translocation between the cytoplasm and the nucleus.
Recently it was demonstrated that Mdmx can be cleaved by caspase between amino acids 358 and 361, and it was proposed that this cleavage plays a functional role in p53 regulation (11). Since the caspase cleavage site is located in the vicinity of S367, we examined if caspase cleavage of Mdmx is regulated by S367 phosphorylation. The D361A mutation, which renders Mdmx resistant to caspase cleavage (11), did not inhibit the ability of the S367A mutant to enhance Mdmx-dependent inhibition of p53 (Fig. 5B). This is in agreement with the previous report that the D361A mutation does not affect Mdm2-dependent degradation of Mdmx (8). Thus, it is not likely that caspase cleavage of Mdmx is involved in regulation of Mdmx degradation by Mdm2.
It was reported that p14ARF stimulates Mdm2-dependent Mdmx ubiquitination and degradation by the proteasome (32). Interestingly, a splicing variant of Mdmx which lacks the Mdmx central domain shows increased sensitivity toward Mdm2- mediated ubiquitination and degradation (8). Thus, stimulation of Mdm2-dependent ubiquitination and degradation of Mdmx are caused by either S367 phosphorylation, binding of p14ARF, or deletion of the central domain. It may be possible that these effectors share the same molecular basis in facilitating Mdmx ubiquitination. Given that 14-3-3 functions as an adapter molecule (53) and S367 phosphorylation is required for interaction between Mdmx and 14-3-3, 14-3-3 binding may also regulate association between Mdmx and p14ARF or a protein(s) that interacts with the central domain of Mdmx. Detailed analyses of these observations may shed light on the mechanisms by which S367 phosphorylation and 14-3-3 binding facilitate Mdm2-dependent ubiquitination and degradation ofMdmx.
In line with our finding that the S367A mutation enhances the ability of Mdmx to suppress p53, we showed that S367A causes enhanced growth of normal cells in culture. Interestingly, the effect of the mutant is more striking in presenescent BJ fibroblasts that show retarded cell growth (Fig. 9B). It was reported that increased activity of p53 is associated with cell senescence (2, 18). Therefore, it may be possible that p53 is more active in presenescent BJ cells, and the potential effect of the S367A mutant to inhibit p53 becomes more pronounced as cells enter the presenescent stage and show higher levels of p53 activity. In fact, the S367A mutation decreases levels of p21, a p53 transcriptional target that plays a crucial role in cell cycle progression (Fig. 9B). Thus, the S367A mutation may affect Mdmx function by modulating transcriptional activity of p53 in BJ cells.
In summary, our data strongly suggest that Mdmx phosphorylation at S367 induced after DNA damage plays an important role in p53 activation. In future it will be very interesting to determine the precise mechanism for regulation of S367 phosphorylation in response to a variety of cellular stresses that are known to activate p53.
Supplementary Material
Acknowledgments
The original human Mdm2 and Mdmx clones are kind gifts from Donna George and Steven Berberich, respectively. We are indebted to Jiangdong Chen, Christian Gaiddon, and Ari Elson for providing us with the wild-type Flag-tagged Mdm2 expression vector, the HA-tagged p53 expression vector, and the GST-14-3-3 expression vectors, respectively. The His-ubiquitin expression plasmid is a kind gift from Dirk Bohmann. We thank Gigi Lozano for providing us p53/Mdm2-deficient fibroblasts. We also thank Tomomi Shinozaki for her experimental assistance at the initial part of the project.
This work is supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Y.T and K.O.), a Grant-in-Aid for Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor and Welfare, Japan (Y.T.), a Grant-in-Aid from the Tokyo Biochemical Research Foundation (Y.T.), Research Grants from the Princess Takamatsu Cancer Research Fund and Takeda Science Foundation (Y.T.), and the Program for Promotion of Fundamental Studies in Health Sciences of Organization for Pharmaceutical Safety and Research of Japan (Y.T.). The work was also supported by NCI grant 87497 (to C.P). Work in the laboratory of Y.S. is supported by the A-T Children's Project, the A-T Medical Research Foundation, and the National Institute of Neurological Disorders and Stroke (NS31763).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Atadja, P., H. Wong, I. Garkavtsev, C. Veillette, and K. Riabowol. 1995. Increased activity of p53 in senescing fibroblasts. Proc. Natl. Acad. Sci. USA 92:8348-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badciong, J. C., and A. L. Haas. 2002. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J. Biol. Chem. 277:49668-49675. [DOI] [PubMed] [Google Scholar]
- 4.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 5.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspari, T. 2000. How to activate p53. Curr. Biol. 10:R315-317. [DOI] [PubMed] [Google Scholar]
- 7.Danovi, D., E. Meulmeester, D. Pasini, D. Migliorini, M. Capra, R. Frenk, P. de Graaf, S. Francoz, P. Gasparini, A. Gobbi, K. Helin, P. G. Pelicci, A. G. Jochemsen, and J. C. Marine. 2004. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol. 24:5835-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Graaf, P., N. A. Little, Y. F. Ramos, E. Meulmeester, S. J. Letteboer, and A. G. Jochemsen. 2003. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J. Biol. Chem. 278:38315-38324. [DOI] [PubMed] [Google Scholar]
- 9.Dougherty, M. K., and D. K. Morrison. 2004. Unlocking the code of 14-3-3. J. Cell Sci. 117:1875-1884. [DOI] [PubMed] [Google Scholar]
- 10.Finch, R. A., D. B. Donoviel, D. Potter, M. Shi, A. Fan, D. D. Freed, C. Y. Wang, B. P. Zambrowicz, R. Ramirez-Solis, A. T. Sands, and N. Zhang. 2002. mdmx is a negative regulator of p53 activity in vivo. CancerRes 62: 3221-3225. [PubMed] [Google Scholar]
- 11.Gentiletti, F., F. Mancini, M. D'Angelo, A. Sacchi, A. Pontecorvi, A. G. Jochemsen, and F. Moretti. 2002. MDMX stability is regulated by p53-induced caspase cleavage in NIH3T3 mouse fibroblasts. Oncogene 21: 867-877. [DOI] [PubMed] [Google Scholar]
- 12.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277:19251-19254. [DOI] [PubMed] [Google Scholar]
- 13.Iliakis, G., Y. Wang, J. Guan, and H. Wang. 2003. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene 22:5834-5847. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juven-Gershon, T., and M. Oren. 1999. Mdm2: the ups and downs. Mol. Med. 5:71-83. [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai, H., D. Wiederschain, H. Kitao, J. Stuart, K. K. Tsai, and Z. M. Yuan. 2003. DNA damage-induced MDMX degradation is mediated by MDM2. J. Biol. Chem. 278:45946-45953. [DOI] [PubMed] [Google Scholar]
- 17.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 18.Kulju, K. S., and J. M. Lehman. 1995. Increased p53 protein associated with aging in human diploid fibroblasts. Exp. Cell Res. 217:336-345. [DOI] [PubMed] [Google Scholar]
- 19.Lane, D. P., and P. A. Hall. 1997. MDM2-arbiter of p53's destruction. Trends Biochem. Sci. 22:372-374. [DOI] [PubMed] [Google Scholar]
- 20.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 21.Li, C., L. Chen, and J. Chen. 2002. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol. Cell. Biol. 22:7562-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302:1972-1975. [DOI] [PubMed] [Google Scholar]
- 23.Linares, L. K., A. Hengstermann, A. Ciechanover, S. Muller, and M. Scheffner. 2003. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. USA 100:12009-12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCurrach, M. E., and S. W. Lowe. 2001. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 66:197-227. [DOI] [PubMed] [Google Scholar]
- 26.Meek, D. W. 2004. The p53 response to DNA damage. DNA Repair (Amsterdam) 3:1049-1056. [DOI] [PubMed] [Google Scholar]
- 27.Meulmeester, E., R. Frenk, R. Stad, P. de Graaf, J. C. Marine, K. H. Vousden, and A. G. Jochemsen. 2003. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol. Cell. Biol. 23:4929-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Migliorini, D., D. Danovi, E. Colombo, R. Carbone, P. G. Pelicci, and J. C. Marine. 2002. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J. Biol. Chem. 277:7318-7323. [DOI] [PubMed] [Google Scholar]
- 29.Migliorini, D., E. L. Denchi, D. Danovi, A. Jochemsen, M. Capillo, A. Gobbi, K. Helin, P. G. Pelicci, and J. C. Marine. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 22:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momand, J., H. H. Wu, and G. Dasgupta. 2000. MDM2-master regulator of the p53 tumor suppressor protein. Gene 242:15-29. [DOI] [PubMed] [Google Scholar]
- 30a.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 31.Olivier, M., R. Eeles, M. Hollstein, M. A. Khan, C. C. Harris, and P. Hainaut. 2002. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19:607-614. [DOI] [PubMed] [Google Scholar]
- 32.Pan, Y., and J. Chen. 2003. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parant, J., A. Chavez-Reyes, N. A. Little, W. Yan, V. Reinke, A. G. Jochemsen, and G. Lozano. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29:92-95. [DOI] [PubMed] [Google Scholar]
- 34.Pereg, Y., D. Shkedy, P. de Graaf, E. Meulmeester, M. Edelson-Averbukh, M. Salek, S. Biton, A. F. Teunisse, W. D. Lehmann, A. G. Jochemsen, and Y. Shiloh. 2005. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc. Natl. Acad. Sci. USA 102:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pochampally, R., B. Fodera, L. Chen, W. Lu, and J. Chen. 1999. Activation of an MDM2-specific caspase by p53 in the absence of apoptosis. J. Biol. Chem. 274:15271-15277. [DOI] [PubMed] [Google Scholar]
- 36.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 37.Ramos, Y. F., R. Stad, J. Attema, L. T. Peltenburg, A. J. van der Eb, and A. G. Jochemsen. 2001. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res 61:1839-1842. [PubMed] [Google Scholar]
- 38.Riemenschneider, M. J., R. Buschges, M. Wolter, J. Reifenberger, J. Bostrom, J. A. Kraus, U. Schlegel, and G. Reifenberger. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59:6091-6096. [PubMed] [Google Scholar]
- 39.Sabbatini, P., and F. McCormick. 2002. MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol. 21:519-525. [DOI] [PubMed] [Google Scholar]
- 40.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 41.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59:4375-4382. [PubMed] [Google Scholar]
- 42.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 43.Sharp, D. A., S. A. Kratowicz, M. J. Sank, and D. L. George. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274:38189-38196. [DOI] [PubMed] [Google Scholar]
- 44.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 45.Shinozaki, T., A. Nota, Y. Taya, and K. Okamoto. 2003. Functional role ofMdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 22:8870-8880. [DOI] [PubMed] [Google Scholar]
- 46.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Laar, P. Dekker, M. Bazuine, R. C. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 47.Stad, R., N. A. Little, D. P. Xirodimas, R. Frenk, A. J. van der Eb, D. P. Lane, M. K. Saville, and A. G. Jochemsen. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stad, R., Y. F. Ramos, N. Little, S. Grivell, J. Attema, A. J. van Der Eb, and A. G. Jochemsen. 2000. Hdmx stabilizes Mdm2 and p53. J. Biol. Chem. 275:28039-28044. [DOI] [PubMed] [Google Scholar]
- 49.Stommel, J. M., and G. M. Wahl. 2005. A new twist in the feedback loop: stress-activated MDM2 destabilization is required for p53 activation. Cell Cycle 4:411-417. [DOI] [PubMed] [Google Scholar]
- 50.Tanimura, S., S. Ohtsuka, K. Mitsui, K. Shirouzu, A. Yoshimura, and M. Ohtsubo. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447:5-9. [DOI] [PubMed] [Google Scholar]
- 51.Taya, Y., K. Nakajima, K. Yoshizawa-Kumagaye, and K. Tamai. 2003. Generation and application of phospho-specific antibodies for p53 and pRB, p. 17-26. In W. S. El-Deiry (ed.), Tumor suppressor genes: regulation, function, and medical applications. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 52.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20:6331-6338. [DOI] [PubMed] [Google Scholar]
- 53.van Hemert, M. J., H. Y. Steensma, and G. P. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23:936-946. [DOI] [PubMed] [Google Scholar]
- 54.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 55.Waterman, M. J., E. S. Stavridi, J. L. Waterman, and T. D. Halazonetis. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat.Genet. 19:175-178. [DOI] [PubMed] [Google Scholar]
- 56.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.