Abstract
Null mutations in DNA mismatch repair (MMR) genes elevate both base substitutions and insertions/deletions in simple sequence repeats. Data suggest that during replication of simple repeat sequences, polymerase slippage can generate single-strand loops on either the primer or template strand that are subsequently processed by the MMR machinery to prevent insertions and deletions, respectively. In the budding yeast Saccharomyces cerevisiae and mammalian cells, MMR appears to be more efficient at repairing mispairs comprised of loops on the template strand compared to loops on the primer strand. We identified two novel yeast pms1 alleles, pms1-G882E and pms1-H888R, which confer a strong defect in the repair of “primer strand” loops, while maintaining efficient repair of “template strand” loops. Furthermore, these alleles appear to affect equally the repair of 1-nucleotide primer strand loops during both leading- and lagging-strand replication. Interestingly, both pms1 mutants are proficient in the repair of 1-nucleotide loop mispairs in heteroduplex DNA generated during meiotic recombination. Our results suggest that the inherent inefficiency of primer strand loop repair is not simply a mismatch recognition problem but also involves Pms1 and other proteins that are presumed to function downstream of mismatch recognition, such as Mlh1. In addition, the findings reinforce the current view that during mutation avoidance, MMR is associated with the replication apparatus.
DNA mismatch repair (MMR) contributes to genomic integrity by repairing mismatches generated during replication, by chemical damage, and as “heteroduplex” intermediates during recombination (7, 28, 31, 35, 44). In addition, the MMR system in higher eukaryotes plays a role in response to DNA damage (3, 6, 7, 62). Inherited MMR defects lead to a mutator phenotype, which in humans and mice is associated with increased cancer susceptibility (5, 7, 13, 16, 38, 50). The MMR system of Escherichia coli has been reconstituted in vitro with purified proteins, including the dedicated proteins MutS, MutL, and MutH (44, 56). The MutS protein, a homodimer, first binds the mispair, followed by recruitment of MutL, the endonuclease MutH, the UvrD helicase, four exonucleases, DNA polymerase, and ligase. Together with transient Dam-mediated hemimethylation, these proteins impose strand specificity that leads to specific repair of the newly replicated strand (10, 25, 26, 43, 44, 74).
In the budding yeast Saccharomyces cerevisiae, six MutS homologues (Msh proteins Msh1 to Msh6) and four MutL homologues (Mlh proteins, Mlh1 to Mlh3, and Pms1) function in various MMR transactions (7, 28, 31, 35). Unlike E. coli, the MutS and MutL activities of budding yeast and mammals are each comprised of heterodimers. Mismatches in nuclear DNA replication intermediates are recognized by the Msh2/Msh6 and Msh2/Msh3 heterodimers, which have partial functional overlap (7, 35, 42). Msh2/Msh6 operates in the repair of base-base mispairs and 1-nucleotide “insertion/deletion” loops (28, 32, 41), while Msh2/Msh3 functions in the repair of 1- to 4-nucleotide insertion/deletion loops (28, 32, 41). Similarly for the MutL homologs, Mlh1 forms heterodimers with Pms1, Mlh2, or Mlh3 (19, 28, 30, 49, 72). Genetic studies indicate that the Mlh1/Pms1 heterodimer is the primary MutL activity in MMR-mediated mutation avoidance, whereas the Mlh1/Mlh3 complex plays a minor role in Msh2/Msh3-dependent repair of insertion/deletion loops (19, 24, 30, 49, 53, 54). Current data suggest that initial recognition of the mismatch is by Msh2/Msh6 or Msh2/Msh3, possibly aided by PCNA, which is subsequently joined by Mlh1-Pms1 to form a higher order complex on DNA. This complex is thought to be responsible for directing the downstream and less well characterized MMR events, including strand discrimination, excision, and resynthesis (4, 7, 28, 35, 42).
Although the MutS and MutL proteins must interact during MMR-mediated mutation avoidance, the nature of these and other protein-protein interactions is not clear. Whereas MLH1, PMS1, and MHS2 deletion mutations appear to result in a null, or near null, MMR state for mutation avoidance, specific pms1 or mlh1 mutant alleles might produce a novel mutator phenotype, for example, by differentially impacting interactions with Msh2/Msh3 or Msh2/Msh6. In turn, these types of alleles might provide additional insights into the function of the MutL homologs, including interactions with other MMR factors. Therefore, we have initiated genetic screens for mutations of PMS1 or MLH1 that result in mutational spectra different from the corresponding MMR-null strains. Here, we report the isolation of two novel pms1 alleles, which in contrast to a pms1Δ strain preferentially elevate +1-bp frameshifts in mononucleotide runs, with little or no effect on −1-bp frameshifts. These pms1 alleles are in close proximity, affecting residues near the end of the protein in a previously uncharacterized but highly conserved amino acid motif. Further analyses suggest that these pms1 alleles do not differentially impact MMR during leading- versus lagging-strand replication but, rather, fail to efficiently repair single nucleotide loops arising on the primer strand. The results presented here reinforce the current view that MMR is associated with the replication apparatus.
MATERIALS AND METHODS
Media and growth conditions.
All media were prepared as described (58) except that synthetic medium contained increased leucine (60 mg/liter). Growth and sporulation were at 30°C and at 18°C, respectively. Sporulation of diploid cells and tetrad dissections were performed as previously described (15, 73).
Strain constructions.
Yeast strains used for assaying CAN1 forward mutations or his1-7, his7-2, and hom3-10 reversion are derivatives of a RAD5 CAN1 W303-1B strain (65) (Table 1). The hom3-10 allele was introduced at the HOM3 locus by a two-step recombination procedure using the plasmid pK8 linearized with SpeI (41). Presence of the hom3-10 allele was verified by the inability of the strain to grow on medium lacking threonine. The his1-7 (71) and his7-2 (57) alleles were introduced by a cloning-free, PCR-based allele replacement approach (14). The sequences of primers are the following (with uppercase letter sequences denoting the adaptamer sequences and lowercase letters denoting HIS1 or HIS7 sequences): his1-7 Adap A (5′-AATTCCAGCTGACCACCATGAAattgagagaaaaacgaaggg-3′) and his1-7 Adap B (5′-GATCCCCGGGAATTGCCATGctgacaaatatgctacgaag-3′) for his1-7, and his7-2 Adap A (5′-AATTCCAGCTGACCACCATGgcgtcgggctacaagcgc-3′) and his7-2 Adap B (5′-GATCCCCGGGAATTGCCATGctgtgccaactgaacaggc-3′) for his7-2. Introduction of the mutant allele was verified by the inability of the strain to grow on medium lacking histidine and confirmed by sequencing analysis.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W1558 | MATα ade2-1 CAN1 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 | 77 |
| NEY190 | W1558 hom3-10 | This study |
| NEY186 | NEY190 pms1::TRP1 | This study |
| NEY398 | NEY190 pms1-G882E | This study |
| NEY402 | NEY190 pms1-H888R | This study |
| NEY524 | MATα ade2-1 CAN1 his1-7 hom3-10 leu2-3,112 trp1-1 ura3-1 RAD5; HIS3 his1-7 derivative of NEY190 | This study |
| NEY442 | NEY524 pms1::TRP1 | This study |
| NEY510 | NEY524 pms1-G882E | This study |
| NEY506 | NEY524 pms1-H888R | This study |
| NEY570 | MATaade2-1 CAN1 his7-2 hom3-10 leu2-3,112 trp1-1 ura3-1 RAD5; HIS3 his7-2 derivative of NEY190 | This study |
| NEY662 | NEY570 pms1::TRP1 | This study |
| NEY578 | NEY570 pms1-G882E | This study |
| NEY574 | NEY570 pms1-H888R | This study |
| NEY1106 | NEY570 msh2::LEU2 | This study |
| NEY1100 | NEY570 pms1::TRP1 msh2::LEU2 | This study |
| NEY1110 | NEY570 pms1-G882E msh2::LEU2 | This study |
| NEY1126 | NEY570 pms1-H888R msh2::LEU2 | This study |
| NEY815 | NEY570 pol2-C1089Y | This study |
| NEY819 | NEY570 pms1::TRP1 pol2-C1089Y | This study |
| NEY823 | NEY570 pms1-G882E pol2-C1089Y | This study |
| NEY827 | NEY570 pms1-H888R pol2-C1089Y | This study |
| SJR1973 | MATaade2-101oc his4Δ::LYS2R-10C lys2Δ::hyg trp1Δ ura3-52 | This study |
| NEY702 | SJR1973 pms1::URA3 | This study |
| NEY742 | SJR1973 pms1-G882E | This study |
| NEY744 | SJR1973 pms1-H888R | This study |
| NEY1084 | SJR1973 ogg1::TRP1 | This study |
| NEY1090 | SJR1973 pms1::URA3 ogg1::TRP1 | This study |
| NEY1092 | SJR1973 pms1-G882E ogg1::TRP1 | This study |
| NEY1094 | SJR1973 pms1-H888R ogg1::TRP1 | This study |
| SJR1977 | MATaade2-101oc his4Δ::LYS2F-10C lys2Δ::hyg trp1Δ ura3-52 | This study |
| NEY706 | SJR1977 pms1::URA3 | This study |
| NEY747 | SJR1977 pms1-G882E | This study |
| NEY749 | SJR1977 pms1-H888R | This study |
| NEY1086 | SJR1977 ogg1::TRP1 | This study |
| NEY1088 | SJR1977 pms1::URA3 ogg1::TRP1 | This study |
| NEY1096 | SJR1977 pms1-G882E ogg1::TRP1 | This study |
| NEY1098 | SJR1977 pms1-H888R ogg1::TRP1 | This study |
| AS4 | MATα trp1 arg4 tyr7 ade6 ura3 | 61 |
| NEY630 | AS4 pms1::URA3 | This study |
| NEY673 | AS4 pms1-G882E | This study |
| NEY669 | AS4 pms1-H888R | This study |
| PD24 | MATaleu2 ade6 ura3 his4-713 | 11 |
| NEY714 | PD24 pms1::URA3 | This study |
| NEY738 | PD24 pms1-G882E | This study |
| NEY726 | PD24 pms1-H888R | This study |
Strains containing 10C runs were constructed using a derivative of YPH45 (MATa ura3-52 ade2-101oc trp1Δ1) in which the open reading frame of LYS2 was replaced with a hygromycin resistance cassette (lys2Δ::hyg) by transformation with a PCR fragment derived from hphMX4 (20). A wild-type LYS2 gene was then inserted at the HIS4 locus on chromosome III by transformation with a PCR fragment amplified from pDP6 (17) using primers with terminal homology to the HIS4 locus. The primer pair Lys2FHis4F2 (5′-ACTTGGTTGAACAATTGAATGTACCAAAGGAGCGTGTTGTTGTGGAAGAGAACGGTGTTTagaggcatcgcacagttttagc-3′; base pairs complementary to HIS4 sequences are in uppercase while those complementary to LYS2 are in lowercase) and Lys2RHis4R (5′- GTTCGGTTTCCAAGTTAGAAATAATCTACTGGAAATCCTTTGGGATCAACCCAAGCTTACccgaaaagaagctaagtctt-3′) were used to generate a fragment for inserting LYS2 at HIS4 in the forward orientation (his4Δ::LYS2F), with LYS2 transcribed in the same direction as HIS4. The primer pair Lys2Fhis4R2 (5′-GTTCGGTTTCCAAGTTAGAAATAATCTACTGGAAATCCTTTGGGATCAACCCAAGCTTActagaggcatcgcacagttttagc-3′) and Lys2RHis4F (5′-ACTTGGTTGAACAATTGAATGTACCAAAGGAGCGTGTTGTTGTGGAAGAGAACGGTGTTTccgaaaagaagctaagtctt-3′) were used to generate a fragment for inserting LYS2 at HIS4 in the reverse orientation (his4Δ::LYS2R), with LYS2 transcription opposing that of HIS4. In strains with the his4Δ::LYS2F allele, transcription and replication fork movement proceed in the same direction; in strains with the his4Δ::LYS2R allele, the direction of transcription opposes that of replication fork movement.
Mononucleotide run of 10C was inserted into the coding strand of the his4Δ::LYS2 alleles using the delitto perfetto method (63). First, the CORE cassette containing URA3 and Kan was amplified with the primer pair CORE406Lys2F (5′-CGAGCTAGCTGAAAAATTCAAAGTTGCCAAGATCTGGAAAGGACCCCTCgagctcgttttcgacactgg-3′; uppercase letters denote the LYS2 sequence and lowercase letters denote CORE cassette sequence) and CORE406Lys2R (5′-CTCGTCTAATTTGAAATCTTGGTTTTCCAAAAAGGCCAAACGGAACAACTtccttaccattaagttgatc-3′), with the CORE cassette inserted after base pair position 406 relative to the LYS2 start codon. Following transformation with the amplified fragment, Ura+, geneticin-resistant transformants were selected and screened for a Lys− phenotype. The CORE cassette within LYS2 was then replaced with a 240-bp LYS2-10C fragment amplified from SJR1354 (21). Lys+ transformants were selected and screened for loss of CORE sequences (Ura−, geneticin-sensitive phenotype). The presence of the 10C run was confirmed by sequence analysis. Runs are named according to the nucleotides on the coding strand; a LYS2-10C allele thus has a 10C run on the coding strand and the complementary 10G run on the noncoding strand.
PMS1 and OGG1 disruptions were constructed in relevant haploid strains by a PCR-based gene disruption method (2) utilizing TRP1 and URA3 for PMS1 and TRP1 for OGG1 as selective markers and the following primer pairs (uppercase letters indicate sequences complementary to PMS1 or OGG1 sequences and lowercase letters denote selective marker sequences): PMS1-L1 (5′-CGAAAAGAAAAGACGCGTCTCTCTTAATAATCATTATGCGATAAAgagcagattgtactgag-3′) and PMS1-L2 (5′-ATAATGTATTTGTTAATTATATAATGAATGAATATCAAAGCTAGAtgtgcggtatttcacacc-3′; OGG1-L1 (5′-TTTGAAGCGTCCTGATTCATAATTGCGATTTTATTTATCAACCAGgagcagattgtactgag-3′) and OGG1-L2 (5′-TTCGGTCGCGTGCTTTTATCGTGGTATTTACTATGACTTTTTAAGtgtgcggtatttcacacc-3′). Each disruption was confirmed by PCR.
The alleles pms1-G882E and pms1-H888R were each integrated into the genome by the cloning-free, PCR-based allele integration approach described above (14). Primers used to amplify either pms1-G882E or pms1-H888R are the following (with uppercase letters denoting the adaptamer sequence and lowercase letters for PMS1 sequence): PMS1 C-term Adap A (5′-aattccagctgaccaccatgGAAGACGGTGGGTTACGAAG-3′) and PMS1 C-term Adap B (5′-gatccccgggaattgccatgCAAGCATCTTCAATGCACGAG-3′). The fusion fragments were cotransformed into relevant strains and Ura+ transformants were selected. Subsequent retention of a single copy of the mutant pms1 allele after 5-fluoroorotic acid selection was verified by PCR and sequencing using primers PMS1 C-term Adap A and PMS1 C-term Adap B.
pol2-C1089Y was introduced by a two-step allele replacement method (55). AgeI linearized p173-rsa (33) was transformed into cells, followed by selection on medium lacking uracil. Ura+ transformants were purified and grown in YPD medium (containing yeast extract, peptone, and glucose) and streaked to 5-fluoroorotic acid plates to select for Ura− colonies, which were screened for the pol2-C1089Y mutation by PCR and diagnostic digestion with RsaI.
Plasmid constructions.
Plasmid pRS414-PMS1 was constructed by inserting a 4-kb chromosomal BglII/SalI fragment that contains the 2,715-bp PMS1 open reading frame (ORF) into BglII/SalI-digested pRS416 (URA3-CEN vector [60]). Plasmids used in two-hybrid assays were constructed by inserting coding sequences of MLH1 and PMS1 into pBTM116 and pGAD424, which contain the LexA DNA-binding domain and Gal4 activation domain, respectively (49). The pms1-G882E and pms1-H888R alleles were introduced into the pGAD424-PMS1 plasmid by gap repair (40, 48). Three independent plasmids from each transformation were sequenced to verify the pms1-G882E and pms1-H888R mutations.
Construction of a randomly mutagenized PMS1 library.
PCR mutagenesis of the PMS1 ORF was carried out in two separate reactions (75). The primer pair PMS1-MluI (5′-GCACAGATTAATACCGATTC-3′) and PMS1-BsaBI (5′-GCGTAGAGTATTCCACTGGC-3′) and the pair PMS1-ClaIF (5′-CGCAGAGATTGAGCCAGTTG-3′) and PMS1-ClaIRev (5′-GACGATTGAAGGAGACGCTAG-3′) were used to mutagenize the first approximately one-third (fragment I) and the last approximately one-third (fragment II) of the PMS1 ORF, respectively. The PCR mixture contained the following components: 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3), 0.25 mM MnCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 1 μM concentration of each primer, 5 U of Taq DNA polymerase, and 10 ng of plasmid pRS414-PMS1. The PCR conditions were 5 min at 94°C and 40 cycles of 15 s at 94°C, 15 s at 50°C, and 1 min at 72°C, followed by 10 min at 72°C.
The PCR-mutagenized fragments I and II were gel purified and cotransformed individually into yeast strain NEY186 with pRS416-PMS1 linearized with MluI/BsaBI or ClaI, respectively. Transformants were selected on synthetic complete medium lacking uracil. In vivo homologous gap repair recombination between the PCR fragments and the vector DNA produced a library of mutagenized pms1 alleles on a CEN plasmid. Control transformations with only the gapped vector indicated >95% efficiency in gap repair. A total of 2,000 transformants of each mutagenized fragment were patched onto medium lacking uracil. After 2 days, the patches were replica plated onto synthetic complete medium lacking uracil and arginine but containing canavanine and onto medium lacking uracil and threonine to score mutation at CAN1 and hom3-10, respectively. Plasmid DNAs from two transformants that consistently exhibited a mutator phenotype on canavanine, but not in the hom3-10 reversion assay, were used to retransform NEY186.
Rate measurements and statistical analyses.
The method of the median was used to calculate the mutation rate (37). Data from at least 20 independent cultures (typically four cultures from each of five independent isolates) were used for each rate determination. In all cases, different isolates behaved the same as evidenced by side-by-side comparisons. Briefly, purified colonies were grown in liquid yeast extract-peptone-dextrose medium to saturation. Appropriate dilutions were plated onto complete synthetic medium to determine the viability of the cells. Medium lacking histidine was used to select His+ prototrophs; medium lacking threonine was used to select revertants of hom3-10; medium lacking arginine but containing canavanine was used to select can1 mutants; and medium containing alpha amino-adipic acid (8) was used to select Lys− mutants. Colonies were counted 3 to 5 days after selection plating. Statistical analyses were performed using Prism 3.0 software (GraphPad Software Inc.). The efficiencies of mismatch correction (repair efficiencies) were determined by comparing the rates of instability at the LYS2-10C locus in pms1Δ strains to the rates observed in wild-type, pms1-G882E, and pms1-H888R strains (59). If we denote the rate of instability as Rwt, Rpms1Δ, Rpms1-G882E, and Rpms1-H888R in wild-type, pms1Δ, pms1-G882E, and pms1-H888R strains, respectively, then the repair efficiency is calculated by substracting the mutation rate after repair (Rwt, Rpms1-G882E, or Rpms1-H888R) from mutation rate before repair (Rpms1Δ) and then dividing this difference by the mutation rate before repair (Rpms1Δ).
Two-hybrid analysis.
Protein-protein interactions were assayed by the two-hybrid method as previously described (49).
Sequence analysis of mutants.
Independent isolates for sequence analysis were obtained by streaking individual colonies onto appropriate selective medium. The relevant gene regions were amplified from the yeast genome by “colony PCR” (39). PCR products were sequenced using the following primers: for CAN1, CAN1 For (5′-CTTAACTCCTGTAAAAAC-3′), CAN1 Seq1 (5′-CATTGGCCGCACCAAATGC-3′), CAN1 Seq2 (5′-TTCATCCCTGTTACATCC-3′), CAN1 Seq3 (5′-CCAAATGCAGCAGTAACG-3′), and CAN1 Rev (5′-GAAATGGCGTGGGAATGT-3′); for LYS2, LYS2 5′ (5′-GCTACATATTCGTTACAGC-3′) and LYS2 3′ (5′-GGTCCGCAACAATGGTTACTC-3′); and for his1-7, HIS1 For (5′-CTCCTATTAACGGTTTGAATC-3′) and HIS1 Rev (5′-GAATAAGATAGAACTCTATC-3′).
RESULTS
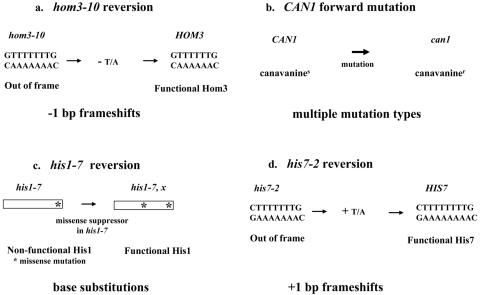
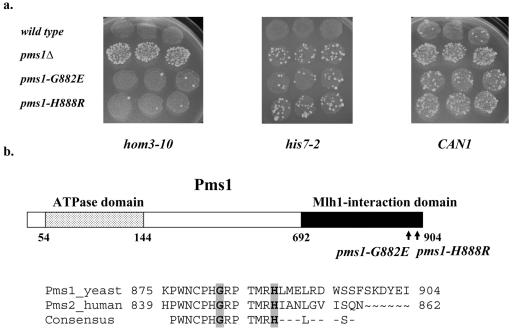
To understand better the role of the Mlh1/Pms1 (MutLα) heterodimer in MMR, we set out to identify alleles of PMS1 that differentially impact MMR-mediated mutation avoidance. Thus, we mutagenized the most conserved regions of the yeast PMS1 ORF and screened for alleles that resulted in a mutator phenotype different from that seen in a pms1Δ strain. A library of randomly mutagenized PMS1 genes in a CEN vector was screened initially in a pms1Δ strain using two mutation assays: reversion of hom3-10 and forward mutation to canavanine resistance at CAN1, which report −1-bp frameshifts in a 7A/7T run and multiple types of mutations, respectively (Fig. 1a and b). Previous work demonstrated that in a pms1Δ strain, reversion of hom3-10 was increased approximately 1,000-fold while forward mutation at the CAN1 locus was increased 30-fold, with more than 60% of the can1 mutations being −1-bp frameshifts in short mononucleotide runs (67). In a pms1Δ strain, elevated mutation levels at hom3-10 and CAN1 can be detected by an increased number of papillae on the appropriate selective media (Fig. 2a). Whereas most mutator strains increased papillation in both assays, two exceptional mutants consistently conferred a strong increase in the number of can1 papillae but no detectable increase in reversion at hom3-10 (Fig. 2a). We did not detect any candidates with the opposite phenotype. To address strain-specific effects, we tested the two pms1 alleles on CEN plasmids in two additional strain backgrounds, GCY35 (45) and AMY125 (59), and observed similar results (data not shown). DNA sequencing analysis of these two unusual pms1 alleles revealed in each strain the presence of a single mutation resulting in a change of glycine to glutamate at residue 882 (pms1-G882E) or histidine to arginine at residue 888 (pms1-H888R) (Fig. 2b).
FIG. 1.
Schematic of four mutator assays. (a) hom3-10 reversion assay measures −1-bp frameshifts in a stretch of 7A/7T bp. (b) CAN1 forward mutation assay detects multiple types of mutations. (c) his1-798 (his1-7) reversion assay measures intragenic missense suppressor mutations near the 3′ end of HIS1. (d) his7-2 reversion assay reports +1-bp frameshifts in a run of 7A/7T bp.
FIG. 2.
(a) Papillation phenotypes of the wild-type, pms1Δ, pms1-G882E, and pms1-H888R strains. The relative mutator effects at hom3-10, his7-2, and CAN1 alleles in wild-type, pms1Δ, pms1-G882E, and pms1-H888R strains were detected initially by monitoring reversion at hom3-10 and his7-2 and the forward mutation of CAN1 alleles individually by replica plating patches of cells onto appropriate selective media. (b) Pms1 domains and location of altered Pms1 residues. The ATPase domain (residues 54 to 144) and the Mlh1-interacting domain (residues 692 to 904) are indicated by dotted and black boxes, respectively. Also shown are the residues corresponding to the COOH termini of yeast and human homologs. The mutated residues, G882 and H888, are shaded gray. The 13-amino-acid domain, identical between yeast and human homologs, is depicted below the alignment of yeast and human homolog sequences. Numbers correspond to the amino acid position in the protein.
Pms1-G882E and Pms1-H888R interact efficiently with Mlh1 and are expressed at wild-type levels.
Because residues G882 and H888 both lie in the C-terminal region of Pms1, which is essential for interaction with Mlh1 (49), we tested the ability of the mutant proteins to interact with Mlh1 using a two-hybrid assay. As shown in Table 2, both mutant forms of Pms1 interacted efficiently with Mlh1. In addition, we determined the expression levels of Pms1-G882E and Pms1-H888R at the endogenous PMS1 locus. No significant differences between the levels of FLAG-tagged Pms1-G882E, Pms1-H888R, or wild type Pms1 were observed using Western blot analysis (data not shown). Together, comparison of Pms1 protein levels and the two-hybrid assay suggest that the pms1-G882E and pms1-H888R alleles have little if any effect on protein stability or interaction with Mlh1.
TABLE 2.
Mlh1/Pms1 two-hybrid interactions
| Strain | β-Galactosidase activity | Increase (fold) |
|---|---|---|
| pBT+pGAD-PMS1 | 0.18 ± 0.05 | 1 |
| pBT-MLH1 + pGAD-PMS1 | 10.8 ± 1.8 | 60 |
| pBT-MLH1 + pGAD-pms1-G882E | 7.8 ± 0.09 | 43 |
| pBT-MLH1 + pGAD-pms1-H888R | 10.3 ± 0.4 | 57 |
Further analysis of the pms1-G882E and pms1-H888R mutator phenotype.
To confirm the differential effects of pms1-G882E and pms1-H888R in the hom3-10 and CAN1 mutator assays, the plasmid-encoded alleles were introduced at the endogenous PMS1 locus in wild-type strain NEY190. As shown in Table 3, both pms1-G882E and pms1-H888R strains displayed significantly elevated mutation rates at CAN1, although both alleles caused less of a mutator phenotype than seen in a pms1Δ strain (3-fold and 5-fold increases, respectively, versus a 17-fold increase in pms1Δ). In contrast, whereas pms1Δ resulted in a 1,100-fold increase in hom3-10 reversion, the pms1-G882E and pms1-H888R strains displayed only 3- to 4-fold increases in hom3-10 reversion relative to wild type (Table 3).
TABLE 3.
Mutation rates at CAN1, his1-7, hom3-10 and his7-2
| Strain |
CAN1
|
his1-7
|
hom3-10
|
his7-2
|
||||
|---|---|---|---|---|---|---|---|---|
| Mutation rate (10−7)a | Increase (fold)b | Mutation rate (10−7)a | Increase (fold)b | Mutation rate (10−8)a | Increase (fold)b | Mutation rate (10−9)a | Increase (fold)b | |
| PMS1 | 4.3 (3.4-5.2) | 1 | 2.6 (1.1-4) | 1 | 1.01 (0.13-2) | 1 | 5.9 (3.8-8) | 1 |
| pms1Δ | 71 (50-92) | 17 | 23 (14-32) | 9 | 1,170 (400-1947) | 1,100 | 1020 (990-1,050) | 172 |
| pms1-G882E | 13 (8.4-17) | 3 | 4.4 (3.2-5.5) | 1.7 | 4 (3.1-4.9) | 3.7 | 329 (277-381) | 56 |
| pms1-H888R | 20 (17-23) | 4.7 | 8 (4.2-11.7) | 3.1 | 3.6 (2.7-4.5) | 3.4 | 903 (821-984) | 153 |
Values in parentheses are 95% confidence intervals.
Increases are calculated relative to PMS1.
To gain additional insight into the MMR defects, we tested the pms1-G882E and pms1-H888R alleles using his1-798 (his1-7) reversion, which reports intragenic missense suppressor mutations near the 3′ end of HIS1 (Fig. 1) (71). First, the his1-7 allele was integrated and characterized in wild-type and pms1Δ backgrounds. pms1Δ strains displayed a ninefold elevated his1-7 reversion rate compared to the wild type (Table 3). As predicted (71), all 20 revertants from the pms1Δ strain were sequenced and found to be intragenic missense suppressors at HIS1 locus (data not shown). The pms1-G882E strain did not display a significant increase in the base substitution rate. However, the pms1-H888R strain showed a small but significant threefold increase over the wild type in base substitutions (Table 3).
Next, we tested the pms1 alleles using the his7-2 reversion assay, which reports +1-bp frameshifts in a run of 7A/7T bp (Fig. 1). With the his7-2 assay, the pms1Δ strain displayed a 172-fold increase in +1 frameshifts compared to a wild-type strain (Table 3). Strikingly, in contrast to their behavior with hom3-10 reporter, both pms1-G882E and pms1-H888R were similar to pms1Δ with the his7-2 reporter. The reversion rate for his7-2 was elevated 56- and 153-fold in the pms1-G882E and pms1-H888R strains, respectively (Table 3).
Based on rate analyses with four mutator reporters (CAN1, hom3-10, his1-7, and his7-2), the pms1-G882E and pms1-H888R alleles both appeared to preferentially elevate +1-bp frameshifts in mononucleotide runs. To confirm this interpretation, 20 to 30 can1 mutants from each strain were sequenced. Strikingly, in pms1-G882E and pms1-H888R strains, approximately 40% of the can1 mutations were +1-bp frameshifts, whereas in wild-type and pms1Δ strains, this percentage was only 5% (Table 4).
TABLE 4.
Spectra of can1 mutations
| Strain | CAN1 mutation rate (10−7)a | No. of base substitutions/ no. of mutants sequenced (fold increase)b | No. of frameshifts/no. of mutants sequenced (fold increase)b
|
|
|---|---|---|---|---|
| −1 bp | +1 bp | |||
| PMS1 | 4.3 (3.4-5.2) | 15/20 (1.0) | 4/20 (1.0) | 1/20 (1.0) |
| pms1Δ | 71 (50-92) | 13/27 (11) | 13/27 (41) | 1/27 (13) |
| pms1-G882E | 13 (8.4-17) | 14/30 (1.9) | 4/30 (2.0) | 12/30 (24) |
| pms1-H888R | 20 (17-23) | 15/30 (3.1) | 2/30 (1.6) | 13/30 (41) |
Values in parentheses are 95% confidence intervals.
Increases are calculated relative to PMS1.
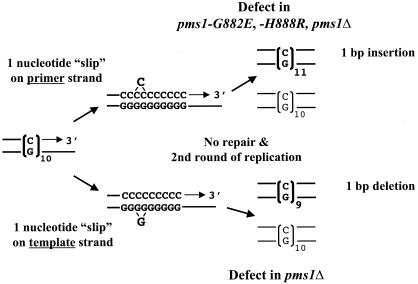
Taken together, the most notable finding from the mutation analyses is that pms1-G882E and pms1-H888R alleles increase +1-bp frameshifts in mononucleotide repeats to an extent comparable to that associated with a pms1Δ allele, while having little or no effect on −1-bp frameshifts and no or little effect on base substitutions. The preferential increase in the +1-bp frameshifts is in contrast to pms1Δ or other MMR-null strains, which generally show a greater increase in −1-bp frameshifts in mononucleotide reporters (21). Our findings suggest that the pms1-G882E and pms1-H888R alleles are preferentially defective in repairing single-strand loops arising on the primer strand during DNA replication (Fig. 3).
FIG. 3.
DNA polymerase slippage model for instability in mononucleotide runs. Following a transient dissociation of the primer and template strand during DNA replication, the strands can reanneal in misaligned configuration, resulting either in a displaced single-strand loop on the primer (upper) or the template (lower) strand. If the resulting mismatches are not corrected before the next round of replication, the mispaired loops will give rise to unit size insertions or deletions, depending on whether the unpaired loop was in the primer or the template, respectively.
pms1-G882E and pms1-H888R mutator effects are similar during leading- and lagging-strand synthesis.
The mutation data presented above suggest that the pms1-G882E and pms1-H888R alleles differentially affect the repair of +1-bp versus −1-bp frameshift intermediates, with a much greater defect in the repair of mispairs due to 1-nucleotide loops on the primer strand than 1-nucleotide loops on the template strand. However, because these data were derived using different genes at different positions in the yeast genome, the mutation pattern might instead reflect sequence effects (e.g., more efficient repair of extra A than of extra T) and/or differences in the repair of errors generated during leading- versus lagging-strand synthesis.
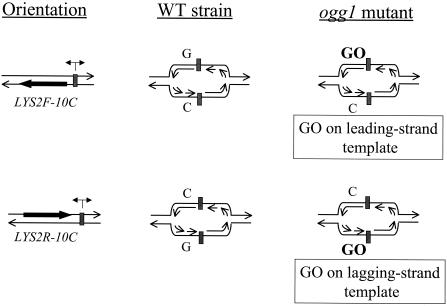
A more direct comparison of the repair efficiencies of +1 versus −1 frameshift intermediates requires the use of a single mutational target for both types of events. Therefore, we constructed strains with an in-frame 10C run on the coding strand of the LYS2 gene (LYS2-10C). As shown previously, a 10N run is sufficiently long to insure that the majority of lys2 forward mutations occur within the run (21, 29, 66). Whether a given sequence resides on the leading- or lagging-strand template during replication was controlled by positioning the LYS2-10C allele at the HIS4 locus on chromosome III, which is replicated from ARS306 >90% of the time (46, 76). We define the forward (LYS2F) orientation as that in which the transcriptional machinery and replication fork move in the same direction; in the reverse (LYS2R) orientation, they converge (Fig. 4). To change the location of a given run on the leading- versus lagging-strand template, we inverted the entire LYS2-10C gene. As illustrated in Fig. 4, the 10C run is on the lagging-strand template in strains containing the LYS2F-10C allele but on the leading-strand template in strains containing LYS2R-10C.
FIG. 4.
Schematic representation of replication forks emerging from ARS306. The location of ARS306 is depicted as a gray box. The coding (nontranscribed) LYS2 strand is illustrated as a thick black arrow, whereas the bi-directional arrow above ARS306 indicates the directionality of replication from ARS306. The forward (LYS2F) orientation is defined as that in which the transcriptional machinery and replication fork move in the same direction; in the reverse (LYS2R) orientation, they converge. The 10G run is on the leading-strand template whereas the 10C run is on the lagging-strand template in strains containing the LYS2F-10C allele. With the LYS2R-10C, the 10G run is on the lagging-strand template, and the 10C run is on the leading-strand template.
For each LYS2-10C allele, we determined the forward mutation rate and mutational spectrum in PMS1, pms1Δ, pms1-G882E, and pms1-H888R backgrounds (Table 5). In the wild type, the forward mutation rates for the LYS2F-10C and LYS2R-10C alleles were similar, and an approximately 10:1 bias for +1 events was observed, suggesting less efficient repair of +1 than of −1 frameshift intermediates. In spite of this apparent bias, however, it should be noted that the MMR efficiency for both +1 and −1 intermediates exceeded 95%. The forward mutation rates were elevated approximately 100-fold in the pms1Δ strains, with −1 events outnumbering +1 events approximately 2:1. These data from PMS1 and pms1Δ backgrounds agree well with observations reported previously for strains containing the same LYS2-10C alleles at the LYS2 locus in a different strain background (21). Although the forward LYS2 mutation rates in the pms1-G882E and pms1-H888R strains were comparable to the pms1Δ strain rates, the distributions of +1 versus −1 events within the 10C/10G runs were not. There was a very strong bias for +1 events (39:1), which is the reverse of that seen in the pms1Δ strains, but very similar to the bias in the PMS1 strains. The C/G runs of the LYS2-10C alleles thus behave similarly to the A/T runs of the hom3-10 and his7-2 alleles and support the conclusion that the primary MMR defect conferred by the pms1-G882E and pms1-H888R alleles is in the repair of primer strand loop mispairs (Fig. 3).
TABLE 5.
Forward mutation in LYS2-10C strains
| Strain | 10G lagging-strand template (LYS2R-10C)
|
10G leading-strand template (LYS2F-10C)
|
||||||
|---|---|---|---|---|---|---|---|---|
| Rate (10−6)a | Increase (fold)b | No. of frameshiftsc
|
Rate (10−6)a | Increase (fold)b | No. of frameshiftsc
|
|||
| −1 bp | +1 bp | −1 bp | +1 bp | |||||
| PMS1 OGG1 | 9.5 (7.3-11.6) | 1 | 1/10 | 9/10 | 19.2 (14-23) | 1 | 1/10 | 8/10 |
| pms1Δ | 1934 (886-2,981) | 203 | 17/20 | 2/20 | 4,524 (1,797-7,251) | 235 | 12/19 | 7/19 |
| pms1-G882E | 302 (153-451) | 32 | 0/20 | 20/20 | 1,316 (600-2,032) | 68 | 1/19 | 18/19 |
| pms1-H888R | 693 (350-1,037) | 73 | 0/20 | 20/20 | 1,651 (811-2,490) | 86 | 0/20 | 19/20 |
| ogg1Δ | 91 (12-170) | 9.5 | 0/12 | 7/12 | 96.5 (70-123) | 5 | 0/12 | 8/12 |
| pms1Δ ogg1Δ | 11,950 (4,310-28,220) | 1,258 | 8/13 | 5/13 | 16,570 (7,190-25,940) | 863 | 8/13 | 4/13 |
| pms1-G882E ogg1Δ | 5,587 (1,770-9,390) | 588 | 1/13 | 11/13 | 20,470 (14,360-26,570) | 1,066 | 0/13 | 13/13 |
| pms1-H888R ogg1Δ | 8,420 (5,800-11,040) | 886 | 0/13 | 11/13 | 29,100 (8,010-50,180) | 1,516 | 0/13 | 11/13 |
Values in parentheses are 95% confidence intervals.
Increases are calculated relative to PMS1.
Number/total number of mutants sequenced.
Although the above data demonstrate that the inefficiency in primer strand loop repair conferred by the pms1-G882E and pms1-H888R alleles is not sequence specific, the analysis did not address the possibility of differential effects during leading- versus lagging-strand synthesis. Addressing the leading/lagging issue requires knowledge of which strand the mutation originated on, which can be deduced using a strain deficient in OGG1, a glycosylase that specifically initiates repair of 8-oxo-7,8-dihydroguanine (GO) lesions (70), which can base pair efficiently with adenine as well as cytosine. In an ogg1 mutant, the resulting increase in GC to TA transversions reflects specifically G-A rather than C-T mispairings, thus assigning the strand on which the original mispair occurred (51). We reasoned that if the presence of a GO lesion in the template can stimulate DNA polymerase slippage, we should observe elevated frameshifts within the 10C/10G runs of the LYS-10C alleles in ogg1 mutants. Using the same reasoning as applied to the transversions of GC to TA in ogg1 mutants, we could assign a strandedness to the underlying slippage events because the initiating GO lesions would always be on the template strand.
An ogg1Δ allele was introduced into the PMS1, pms1Δ, pms1-G882E, and pms1-H888R strains containing the LYS2F-10C allele or the LYS2R-10C allele, in which GO lesions should be present within the leading- or lagging-strand template run, respectively (see Fig. 4). In each ogg1 mutant, the rate of frameshifts in the 10G/10C run was increased at least fivefold relative to the rate in the corresponding OGG1 parent strain (Table 5). In either the PMS1 ogg1 or the pms1Δ ogg1 strain, the rate of slippage within the 10G/10C run was same regardless of whether the G run was on the leading- or the lagging-strand template. Therefore, at least in the case of the LYS2-10C alleles used here, there appears to be neither a strand-related difference in the rate of polymerase slippage within G/C runs nor a strand-specific bias in the efficiency of MMR. In addition, the very strong bias for the accumulation of +1 frameshifts was evident in the pms1-G882E ogg1 and the pms1-H888R ogg1 strains, regardless of whether the 10G run was on the leading- or lagging-strand template. We estimate that the repair efficiencies of template strand loops in these strains was >90%, while that of primer strand loops was less than 1%. These data confirm that the novel pms1 alleles reported here are specifically defective in the repair of primer strand loops generated during leading- and lagging-strand DNA replication.
pms1-G882E and pms1-H888R impact MMR and synergize with the +1-bp frameshift mutator allele, pol2-C1089Y.
Previously, a mutation in DNA polymerase ɛ, pol2-C1089Y, was reported to elevate preferentially +1-bp frameshift mutations within mononucleotide runs in yeast (33). As expected for a DNA polymerase defect, pol2-C1089Y synergized with msh2Δ for +1-bp frameshifts in mononucleotide runs (33). To address whether the pms1-G882E and pms1-H888R strains indeed reflect defects in MMR rather than some aspect of replication per se, we constructed double mutant strains containing pms1 alleles together with either msh2Δ or pol2-C1089Y. As shown in Table 6, pms1Δ, pms1-G882E, and pms1-H888R mutations displayed epistatic interactions with msh2Δ, most relevantly for the his7-2 +1-bp frameshift assay. In contrast, all three pms1 alleles showed synergistic interactions with pol2-C1089Y in both the CAN1 forward and his7-2 reversion assays. Furthermore, pol2-C1089Y did not synergize with pms1-G882E or pms1-H888R using the −1-bp frameshift specific reporter, hom3-10 (data not shown). These results strongly suggest that both pms1-G882E and pms1-H888R alleles specifically impact mismatch repair of +1-nucleotide primer-strand loops.
TABLE 6.
Genetic interactions between pms1 alleles and msh2Δ, pol2-C1089Y
| Strain |
CAN1
|
his7-2
|
||
|---|---|---|---|---|
| Mutation rate (10−7)a | Increase (fold)b | Reversion rate (10−9)a | Increase (fold)b | |
| PMS1 MSH2 POL2 | 4.3 (3.4-5.2) | 1 | 5.9 (3.8-8) | 1 |
| pms1Δ | 71 (50-92) | 17 | 1,020 (990-1,050) | 172 |
| pms1-G882E | 13 (8.4-17) | 3 | 329 (277-381) | 56 |
| pms1-H888R | 21 (17-24) | 4.7 | 903 (821-984) | 153 |
| msh2Δ | 66.1 (49-83) | 15 | 1,450 (1,160-1,740) | 246 |
| msh2Δ pms1Δ | 51 (33-69) | 11.8 | 1,260 (910-1,600) | 214 |
| msh2Δ pms1-G882E | 54 (32-75) | 12.6 | 1,330 (520-2,130) | 225 |
| msh2Δ pms1-H888R | 61.7 (26-98) | 14.3 | 1,350 (1,200-1,490) | 229 |
| pol2-C1089Y | 16 (11-21) | 3.7 | 290 (30-551) | 49 |
| pms1Δ pol2-C1089Y | 890 (454-1,300) | 206 | 32,260 (7,000-57,530) | 5,468 |
| pms1-G882E pol2-C1089Y | 372 (248-495) | 86 | 26,000 (8,800-43,200) | 4,406 |
| pms1-H888R pol2-C1089Y | 356 (272-440) | 83 | 38,200 (3,160-73,240) | 6,475 |
Values in parentheses are 95% confidence intervals.
Increases are calculated relative to PMS1.
pms1-G882E and pms1-H888R efficiently repair 1-nucleotide loop mispairs during meiotic recombination.
In addition to a mitotic mutation avoidance role, MMR proteins also function during meiotic recombination. During recombination, sequence nonidentities between recombining alleles can result in mismatches in heteroduplex DNA intermediates (47, 52). Such mismatches are normally subject to mismatch correction, which can lead to gene conversion. However, failure of MMR will result in two nonidentical daughter cells when persisting heteroduplex DNA is replicated, which is termed postmeiotic segregation (PMS). Hence in wild-type cells, efficient heteroduplex repair will result in relatively high levels of gene conversion and low levels of PMS. In contrast, MMR-deficient strains will display increased levels of PMS at the expense of gene conversion events.
To determine the effect of pms1-G882E and pms1-H888R on the correction of 1-nucleotide loop mispairs in meiotic heteroduplex DNA, we used the haploid backgrounds AS4 and PD24. The resultant diploids are identical in sequence for the HIS4 locus, except for being heterozygous for his4-713, a 1-bp insertion near the C terminus of HIS4. Furthermore, these diploids show high levels of non-Mendelian (aberrant) segregation initiated by double-strand breaks in the HIS4 promoter region (11, 15). As shown in Table 7, the overall percentage of aberrant segregants, i.e., 6:2 plus 5:3, was similar in all diploid strains tested (20 to 30%). In wild type, pms1-G882E and pms1-H888R diploids, high levels of gene conversion, i.e., 6:2, and a low levels of PMS, i.e., 5:3, were observed, indicating efficient MMR of the 1-nucleotide loop mispair at his4-713 (Table 7). Given that the two HIS4 chromosomes used in the diploids strains experience double-strand breaks with identical frequencies (J. L. Arqueso and T. D. Petes, personal communication), the observed equal ratios of 6:2 versus 2:6 tetrads in the PMS1, pms1-G882E, and pms1-H888R strains suggest that there is no bias in the repair of the 1-nucleotide loop heteroduplex. As expected for MMR deficiency, pms1Δ diploids displayed significantly increased levels of PMS and reduced levels of gene conversion. (Table 7, 45% gene conversion and 55% PMS). Whereas differences between PMS levels in the pms1Δ mutant and the wild type (P < 0.0001), pms1-G882E (P = 0.0001), and pms1-H888R (P = 0.0003) are highly significant, the differences in PMS levels between wild type and pms1-G882E and pms1-H888R were not significant (data not shown). Thus, these results indicate that neither pms1-G882E nor pms1-H888R significantly alters the repair efficiency of 1-nucleotide loop mismatches in heteroduplex DNA during meiotic recombination.
TABLE 7.
Meiotic mismatch repair of a 1-nucleotide loop heteroduplex at HIS4a
| Strain | Total no. of tetrads | No. of tetrads
|
No. of other AB tetrads | % AB tetrads | % PMS/AB tetrads | Relative increase in PMS | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4+:4− | 6+:2− | 2−:6+ | 5+:3− | 3+:5− | ||||||
| PMS1 | 198 | 158 | 16 | 22 | 1 | 1 | 0 | 20 | 5 | 1 |
| pms1Δ | 66 | 46 | 2 | 5 | 4 | 7 | 2 | 30 | 55 | 11 |
| pms1-G882E | 203 | 159 | 12 | 23 | 2 | 2 | 5 | 22 | 9 | 1.8 |
| pms1-H888R | 197 | 145 | 15 | 27 | 3 | 4 | 3 | 26 | 13 | 2.4 |
AB, aberrant.
DISCUSSION
To understand better the function of MutLα during MMR-mediated mutation avoidance, we screened for alleles of PMS1 that exhibit novel effects on mutational spectra. We identified two alleles, pms1-G882E and pms1-H888R, that greatly elevated the rate of +1-bp frameshifts in mononucleotide runs, while having relatively little effect on the rates of −1-bp frameshifts or base substitutions. The repair bias initially was observed in the hom3-10 and his7-2 frameshift reversion assays, which report −1-bp and +1-bp frameshifts in A/T runs, respectively, as well as in forward mutation spectra at CAN1 (Tables 3 and 4). The generality of the strikingly inefficient repair of +1 frameshift intermediates was confirmed using a novel assay (Fig. 4), which determined mutational rates and spectra for C/G mononucleotide runs during leading- versus lagging- strand replication (Table 5). Using this assay, we showed that the pms1-G882E and pms1-H888R alleles were predominantly defective in the repair of primer strand loops generated during both leading- and lagging-strand DNA replication. Finally, the observed efficient repair of 1-nucleotide loops in meiotic heteroduplex DNA intermediates in pms1-G882E and pms1-H888R mutants (Table 6) suggests that the repair bias for primer strand versus template strand loops is related to mismatch repair that occurs in the context of DNA replication and not meiotic recombination. Interestingly, the mutations in the pms1-G882E and pms1-H888R alleles are in close proximity, affecting the COOH-terminus of Pms1 in a previously uncharacterized but highly conserved sequence motif.
Frameshift intermediates in repeat tracts are generated during DNA synthesis when the template and primer strands dissociate transiently and then reanneal in a misaligned configuration, resulting in single-strand loops of one or more repeats. If left unrepaired, 1-nucleotide loop mispairs will give rise, in the next round of replication, to 1-bp insertions or deletions, depending on whether the extra nucleotides are in the primer or the template strand, respectively (Fig. 3). While DNA polymerase generates comparable numbers of misaligned nucleotides on the template and primer strands when replicating simple sequence repeats, the MMR system appears to be inherently more efficient at repairing template strand loops than primer strand loops (21, 64, 68). One explanation for the more efficient repair of template strand loops in wild-type strains is that the MutS complexes differentially recognize primer strand versus template strand loops. The results reported here with pms1-G882E and pms1-H888R alleles suggest, however, that this inherent inefficiency of MMR is not a MutS recognition issue but, rather, reflects a processing difference dependent on Pms1 and, likely, Mlh1.
Although mutations in PMS1 most likely affect the repair and not the generation of mutational intermediates, we asked first whether the pms1-G882E or pms1-H888R mutations impact MMR, per se, and then whether they might in some manner alter the polymerization fidelity through mononucleotide repeats. To address these questions, we performed epistasis analysis between pms1 mutations and msh2Δ, as well as the DNA polymerase ɛ allele, pol2-C19089Y, which preferentially elevates +1-bp frameshifts in monoucleotide runs. We found that the pms1Δ, pms1-G882E, and pms1-H888R alleles all displayed epistatic interactions with msh2Δ. As observed previously (33), when combined with a msh2Δ allele, pol2-C1089Y synergized with pms1Δ for + 1-bp frameshifts in mononucleotide runs (Table 6). In contrast to the epistatic relationship with msh2Δ, pms1-G884E and pms1-H888R both synergized with the pol2-C1089Y for +1-bp frameshifts in the his7-2 assay (Table 6). These results strongly suggest that pms1-G882E and pms1-H888R indeed impact mismatch repair of 1-nucleotide primer strand loops rather than affecting some aspect of the actual replication process.
To gain further understanding into the defects of pms1-G882E and pms1-H888R, we investigated the frameshifts generated in C/G runs during leading- versus lagging-strand synthesis. In yeast, previous studies exploiting the OGG1 deficiency to assign the strand on which mutations arise suggested that mismatch repair of base/base mispairs was more efficient during lagging-strand than leading-strand replication (51). We addressed this issue for frameshifts in mononucleotide runs by a similar approach and incorporated ogg1Δ into strains containing the G/C run LYS2 reporter located near an origin of replication (Fig. 4). In these strains, the G run was present on either the leading- or lagging-strand template during replication. ogg1Δ increased frameshifts in the runs and therefore allowed the marking of the template that contained the GO lesion. In addition, ogg1Δ displayed synergistic interactions with pms1Δ, pms1-G882E, and pms1-H888R (Table 5), suggesting increased polymerase slippage due to unrepaired GO lesions. Importantly, whereas most mutations in the pms1Δ ogg1Δ strains were −1 frameshifts, more than 95% of the frameshifts in the pms1-G882E ogg1Δ and pms1-H888R ogg1Δ were +1 frameshifts. Taken together, these data indicate that both pms1 alleles primarily affect repair of primer strand loops during both leading- and lagging-strand synthesis. In turn, these findings are consistent with the current view for association between MMR and replication machinery (28, 31, 69).
Further insight into the defect of the pms1-G884E and pms1-H888R mutants emerged from examining the effects of these alleles on heteroduplex correction during meiotic recombination. Relative to a pms1Δ strain, both mutant strains efficiently repaired meiotic heteroduplexes containing 1-nucleotide loops (Table 7). This implies that there is a difference between the processing of replication- and recombination-associated mispairs in these mutants. Although there are many factors that may contribute to this difference, we suggest that the recognition of loop mispairs during meiotic recombination is likely to be independent of the DNA synthesis machinery, whereas MMR-mediated mutation avoidance is linked to replication (4, 9, 18, 22, 34, 36, 51, 69).
The pms1-G882E and pms1-H888 mutations that cause the preferential elevation of +1-bp frameshifts in mononucleotide runs map in the C-terminal 200 amino acids of Pms1 to a 13-amino-acid motif that is highly conserved in eukaryotic Pms1 homologs. Because the C-terminal region of Pms1 is required for interaction with Mlh1 (49), the novel mutator phenotypes might simply reflect abnormal MutLα heterodimer formation. However, the pms1-G882E and pms1-H888 mutations had no detectable effect on Pms1 stability or interaction with Mlh1 as determined with the two-hybrid assay. The C-terminal domain of MutL has been shown to be important for homodimerization (1, 12), interactions with MutH and UvrD (25, 26), and DNA binding (27). Furthermore, based on the recently solved crystal structure of the COOH-terminus of MutL (23), the residues affected by the pms1 alleles studied here appear to be in an exposed region of the protein and, therefore, may be important for interaction with other proteins and/or DNA during MMR. Further studies of these pms1 alleles and other mutations with specific mutator effects should shed additional light on eukaryotic MMR, possibly including the mechanism of strand discrimination.
Acknowledgments
We thank Gray Crouse, Rodney Rothstein, Marcel Wehrli, Jennifer Johnson, and Ashleigh Miller, for critical reading of the manuscript.
This work was supported by National Institutes of Health (NIH) grant 5 R0l GM45413 to R.M.L. and NIH grant GM038464 to S.J.R.; N.E. was supported by NIH fellowship F32 GM20342, and R.G. was partially supported by the Graduate Division of Biological and Biomedical Sciences at Emory University.
REFERENCES
- 1.Ban, C., and W. Yang. 1998. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell 95:541-552. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellacosa, A. 2001. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 8:1076-1092. [DOI] [PubMed] [Google Scholar]
- 4.Bowers, J., P. T. Tran, A. Joshi, R. M. Liskay, and E. Alani. 2001. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol. 306:957-968. [DOI] [PubMed] [Google Scholar]
- 5.Bronner, C. E., S. M. Baker, P. T. Morrison, G. Warren, L. G. Smith, M. K. Lescoe, M. Kane, C. Earabino, J. Lipford, A. Lindblom, P. Tannergard, R. J. Bollag, A. R. Godwin, D. C. Ward, M. Nordenskjold, R. Fishel, R. Kolodner, and R. M. Liskay. 1994. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature 368:258-261. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. D., A. Rathi, R. Kamath, D. I. Beardsley, Q. Zhan, J. L. Mannino, and R. Baskaran. 2003. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 33:80-84. [DOI] [PubMed] [Google Scholar]
- 7.Buermeyer, A. B., S. M. Deschenes, S. M. Baker, and R. M. Liskay. 1999. Mammalian DNA mismatch repair. Annu. Rev. Genet. 33:533-564. [DOI] [PubMed] [Google Scholar]
- 8.Chattoo, B. B., F. Sherman, T. A. Azubalis, T. A. Fjellstedt, D. Mehnert, and M. Ogur. 1979. Selection of lys2 mutants of yeast Saccharomyces cerevisiae by the utilization of α-aminoadipate. Genetics 93:51-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, A. B., F. Valle, K. Drotschmann, R. K. Gary, and T. A. Kunkel. 2000. Functional interaction of proliferating cell nuclear antigen with MSH2- MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 275:36498-36501. [DOI] [PubMed] [Google Scholar]
- 10.Dao, V., and P. Modrich. 1998. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J. Biol. Chem. 273:9202-9207. [DOI] [PubMed] [Google Scholar]
- 11.Detloff, P., M. A. White, and T. D. Petes. 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drotschmann, K., A. Aronshtam, H. J. Fritz, and M. G. Marinus. 1998. The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 26:948-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelmann, L., and W. Edelmann. 2004. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am. J. Med. Genet. C 129:91-99. [DOI] [PubMed] [Google Scholar]
- 14.Erdeniz, N., U. H. Mortensen, and R. Rothstein. 1997. Cloning-free PCR-based allele replacement methods. Genome Res. 7:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, Q., F. Xu, and T. D. Petes. 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15:1679-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishel, R., M. K. Lescoe, M. R. Rao, N. G. Copeland, N. A. Jenkins, J. Garber, M. Kane, and R. Kolodner. 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75:1027-1038. [DOI] [PubMed] [Google Scholar]
- 17.Fleig, U. N., R. D. Pridmore, and P. Philippsen. 1986. Construction of LYS2 cartridges for use in genetic manipulations of Saccharomyces cerevisiae. Gene 46:237-245. [DOI] [PubMed] [Google Scholar]
- 18.Flores-Rozas, H., D. Clark, and R. D. Kolodner. 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26:375-378. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Rozas, H., and R. D. Kolodner. 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95:12404-12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, A. L., X. Pan, and J. H. McCusker. 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15:507-511. [DOI] [PubMed] [Google Scholar]
- 21.Gragg, H., B. D. Harfe, and S. Jinks-Robertson. 2002. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:8756-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, L., Y. Hong, S. McCulloch, H. Watanabe, and G. M. Li. 1998. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26:1173-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarne, A., S. Ramon-Maiques, E. M. Wolff, R. Ghirlando, X. Hu, J. H. Miller, and W. Yang. 2004. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 23:4134-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habraken, Y., P. Sung, L. Prakash, and S. Prakash. 1997. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr. Biol. 7:790-793. [DOI] [PubMed] [Google Scholar]
- 25.Hall, M. C., J. R. Jordan, and S. W. Matson. 1998. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 17:1535-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, M. C., and S. W. Matson. 1999. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 274:1306-1312. [DOI] [PubMed] [Google Scholar]
- 27.Hall, M. C., P. V. Shcherbakova, J. M. Fortune, C. H. Borchers, J. M. Dial, K. B. Tomer, and T. A. Kunkel. 2003. DNA binding by yeast Mlh1 and Pms1: implications for DNA mismatch repair. Nucleic Acids Res. 31:2025-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 29.Harfe, B. D., and S. Jinks-Robertson. 2000. Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics 156:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harfe, B. D., B. K. Minesinger, and S. Jinks-Robertson. 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10:145-148. [DOI] [PubMed] [Google Scholar]
- 31.Jiricny, J. 1998. Eukaryotic mismatch repair: an update. Mutat. Res. 409:107-121. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, R. E., G. K. Kovvali, L. Prakash, and S. Prakash. 1996. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J. Biol. Chem. 271:7285-7288. [DOI] [PubMed] [Google Scholar]
- 33.Kirchner, J. M., H. Tran, and M. A. Resnick. 2000. A DNA polymerase epsilon mutant that specifically causes +1 frameshift mutations within homonucleotide runs in yeast. Genetics 155:1623-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleczkowska, H. E., G. Marra, T. Lettieri, and J. Jiricny. 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15:724-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 36.Lau, P. J., H. Flores-Rozas, and R. D. Kolodner. 2002. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 22:6669-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea, D., and C. Coulson. 1948. The distribution of the number of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 38.Leach, F. S., N. C. Nicolaides, N. Papadopoulos, B. Liu, J. Jen, R. Parsons, P. Peltomaki, P. Sistonen, L. A. Aaltonen, M. Nystrom-Lahti, X.-Y. Guan, J. Zhang, P. S. Meltzer, J.-W. Yu, F.-T. Kao, D. J. Chen, K. M. Cerosaletti, R. E. K. Fournier, S. Todd, T. Lewis, R. J. Leach, S. L. Naylor, J. Weissenbach, J.-P. Mecklin, H. Jarvinen, G. M. Petersen, S. R. Hamilton, J. Green, J. Jass, P. Watson, H. T. Lynch, J. M. Trent, A. de la Chapelle, K. W. Kinzler, and B. Vogelstein. 1993. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215-1225. [DOI] [PubMed] [Google Scholar]
- 39.Liang, Q., and T. Richardson. 1992. A simple and rapid method for screening transformant yeast colonies using PCR. BioTechniques 13:730-732, 735. [PubMed] [Google Scholar]
- 40.Ma, H., S. Kunes, P. J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201-216. [DOI] [PubMed] [Google Scholar]
- 41.Marsischky, G. T., N. Filosi, M. F. Kane, and R. Kolodner. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407-420. [DOI] [PubMed] [Google Scholar]
- 42.Marti, T. M., C. Kunz, and O. Fleck. 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol. 191:28-41. [DOI] [PubMed] [Google Scholar]
- 43.Mechanic, L. E., B. A. Frankel, and S. W. Matson. 2000. Escherichia coli MutL loads DNA helicase II onto DNA. J. Biol. Chem. 275:38337-38346. [DOI] [PubMed] [Google Scholar]
- 44.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 45.New, L., K. Liu, and G. F. Crouse. 1993. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol. Gen. Genet. 239:97-108. [DOI] [PubMed] [Google Scholar]
- 46.Newlon, C. S., L. R. Lipchitz, I. Collins, A. Deshpande, R. J. Devenish, R. P. Green, H. L. Klein, T. G. Palzkill, R. B. Ren, S. Synn, et al. 1991. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics 129:343-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolas, A., and T. D. Petes. 1994. Polarity of meiotic gene conversion in fungi: contrasting views. Experientia 50:242-252. [DOI] [PubMed] [Google Scholar]
- 48.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101:228-245. [DOI] [PubMed] [Google Scholar]
- 49.Pang, Q., T. A. Prolla, and R. M. Liskay. 1997. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol. 17:4465-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadopoulos, N., N. C. Nicolaides, Y.-F. Wei, S. M. Ruben, K. C. Carter, C. A. Rosen, W. A. Haseltine, R. D. Fleischmann, C. M. Fraser, M. D. Adams, J. C. Venter, S. R. Hamilton, G. M. Petersen, P. Watson, H. T. Lynch, P. Peltomaki, J.-P. Mecklin, A. de la Chapelle, K. W. Kinzler, and B. Vogelstein. 1994. Mutation of a mutL homolog in hereditary colon cancer. Science 263:1625-1629. [DOI] [PubMed] [Google Scholar]
- 51.Pavlov, Y. I., I. M. Mian, and T. A. Kunkel. 2003. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 13:744-748. [DOI] [PubMed] [Google Scholar]
- 52.Petes, T. D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2:360-369. [DOI] [PubMed] [Google Scholar]
- 53.Prolla, T. A., D. M. Christie, and R. M. Liskay. 1994. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 14:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prolla, T. A., Q. Pang, E. Alani, R. D. Kolodner, and R. M. Liskay. 1994. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265:1091-1093. [DOI] [PubMed] [Google Scholar]
- 55.Scherer, S., and R. W. Davis. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76:4951-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]
- 57.Shcherbakova, P. V., and T. A. Kunkel. 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 19:3177-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 59.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17:2851-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stapleton, A., and T. D. Petes. 1991. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stojic, L., R. Brun, and J. Jiricny. 2004. Mismatch repair and DNA damage signalling. DNA Repair 3:1091-1101. [DOI] [PubMed] [Google Scholar]
- 63.Storici, F., L. K. Lewis, and M. A. Resnick. 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19:773-776. [DOI] [PubMed] [Google Scholar]
- 64.Strand, M., M. C. Earley, G. F. Crouse, and T. D. Petes. 1995. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:10418-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 66.Tran, H. T., J. D. Keen, M. Kricker, M. A. Resnick, and D. A. Gordenin. 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran, P. T., and R. M. Liskay. 2000. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLα. Mol. Cell. Biol. 20:6390-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Twerdi, C. D., J. C. Boyer, and R. A. Farber. 1999. Relative rates of insertion and deletion mutations in a microsatellite sequence in cultured cells. Proc. Natl. Acad. Sci. USA 96:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 70.van der Kemp, P. A., D. Thomas, R. Barbey, R. de Oliveira, and S. Boiteux. 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93:5197-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Borstel, R. C., E. A. Savage, Q. Wang, U. G. Hennig, R. G. Ritzel, G. S. Lee, M. D. Hamilton, M. A. Chrenek, R. W. Tomaszewski, J. A. Higgins, C. J. Tenove, L. Liviero, P. J. Hastings, C. T. Korch, and C. M. Steinberg. 1998. Topical reversion at the HIS1 locus of Saccharomyces cerevisiae. A tale of three mutants. Genetics 148:1647-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, T. F., N. Kleckner, and N. Hunter. 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96:13914-13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welz-Voegele, C., J. E. Stone, P. T. Tran, H. M. Kearney, R. M. Liskay, T. D. Petes, and S. Jinks-Robertson. 2002. Alleles of the yeast PMS1 mismatch-repair gene that differentially affect recombination- and replication-related processes. Genetics 162:1131-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamaguchi, M., V. Dao, and P. Modrich. 1998. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J. Biol. Chem. 273:9197-9201. [DOI] [PubMed] [Google Scholar]
- 75.Zhao, X., B. Georgieva, A. Chabes, V. Domkin, J. H. Ippel, J. Schleucher, S. Wijmenga, L. Thelander, and R. Rothstein. 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 20:9076-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu, J., C. S. Newlon, and J. A. Huberman. 1992. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]