Abstract
Steroid receptor coactivator 3 (SRC-3/AIB1) interacts with steroid receptors in a ligand-dependent manner to activate receptor-mediated transcription. A number of intracellular signaling pathways initiated by growth factors and hormones induce phosphorylation of SRC-3, regulating its function and contributing to its oncogenic potential. However, the range of mechanisms by which phosphorylation affects coactivator function remains largely undefined. We demonstrate here that peptidyl-prolyl isomerase 1 (Pin1), which catalyzes the isomerization of phosphorylated Ser/Thr-Pro peptide bonds to induce conformational changes of its target proteins, interacts selectively with phosphorylated SRC-3. In addition, Pin1 and SRC-3 activate nuclear-receptor-regulated transcription synergistically. Depletion of Pin1 by small interfering RNA (siRNA) reduces hormone-dependent transcription from both transfected reporters and an endogenous steroid receptor target gene. We present evidence that Pin1 modulates interactions between SRC-3 and CBP/p300. The interaction is enhanced in vitro and in vivo by Pin1 and diminished when cellular Pin1 is reduced by siRNA or in stable Pin1-depleted cell lines. Depletion of Pin1 in MCF-7 human breast cancer cells reduces the endogenous estrogen-dependent recruitment of p300 to the promoters of estrogen receptor-dependent genes. Pin1 overexpression enhanced SRC-3 cellular turnover, and depletion of Pin1 stabilized SRC-3. Our results suggest that Pin1 functions as a transcriptional coactivator of nuclear receptors by modulating SRC-3 coactivator protein-protein complex formation and ultimately by also promoting the turnover of the activated SRC-3 oncoprotein.
Steroid receptors, in response to their cognate ligands, regulate a variety of physiological processes including reproduction, development, and cellular homeostasis. They activate gene transcription by binding to hormone-responsive elements at target genes and recruiting coactivators. Steroid receptor coactivators (SRC; p160 family) are among the first cloned steroid receptor coactivators. Members of the p160 coactivator family, including SRC-1 (16, 31), SRC-2 (GRIP1/TIF2) (14, 44), and SRC-3 (AIB1/ACTR/pCIP/RAC3/TRAM-1) (2, 8, 21, 39-41), interact with ligand-bound receptors through conserved LXXLL motifs in their nuclear receptor interaction domains (10). In addition, they also contain functional activation domains that recruit proteins to modify histones and remodel chromatin. The recruited proteins include CBP/p300, which has intrinsic histone acetyltransferase activity (8), and CARM1, which has histone methyltransferase activity (7).
The SRC-3 coactivator is involved in important physiological processes, including reproductive function, cytokine signaling, cell proliferation, and somatic growth (24, 46, 52, 58). It is believed to be an oncogene (42, 58). It is amplified and overexpressed in breast and ovarian cancers (2). SRC-3 knockout mice display delayed mammary gland development, growth retardation, and impaired vasoprotection (24, 46, 52, 55). A number of extracellular signals including steroid hormones, growth factors, and cytokines can induce SRC-3 phosphorylation (48, 49). Phosphorylation of SRC-3 has been shown to be important for its interaction with CBP/p300 and nuclear receptors (12, 49) as well as for its oncogenic potential (49).
Peptidyl-prolyl isomerases are an evolutionarily conserved group of proteins that promote the cis/trans isomerization of the peptide bond preceding Pro residues (11, 36). Peptidyl-prolyl isomerase 1 (Pin1) is a unique member of one of three such protein families, the parvulins, as it specifically interacts with and isomerizes phosphorylated Ser/Thr-Pro motifs. The study of Pin1 has merged the prolyl isomerase modification with the more extensively studied protein modification of phosphorylation and implicated a role for Pin1 in cell signaling. Pin1 was originally identified as a protein that interacted with the important fungal cell cycle regulatory protein kinase NIMA in both human and Aspergillus nidulans cDNA library screens (9, 28). It is comprised of an N-terminal WW domain that is involved in protein interaction and a C-terminal prolyl isomerase domain. Interestingly, both domains recognize phosphorylated Ser/Thr-Pro motifs (32, 43). Thus, multiple such motifs within a target protein sequence, such as the 7-amino acid-(aa) repeat that comprises the C-terminal domain (CTD) of RNA polymerase II, markedly increase the affinity and efficacy of Pin1 (15, 30). The isomerization activity of Pin1 frequently results in conformational changes that can alter the function, localization, or stability of the target protein (60). In addition, Pin1-induced isomerization can result in PP2A-mediated dephosphorylation of the phosphorylated Ser/Thr-Pro motif, as PP2A is a trans-specific phosphatase (59). A number of Pin1-interacting proteins including c-Jun, NF-κB, p53, β-catenin, and c-Myc are transcription factors, and for all of these proteins, the primary role of Pin1 seems to be the regulation of stability (34, 35, 51, 54, 56). At least in the case of c-Myc, Pin1 binding facilitates PP2A-mediated dephosphorylation, which is required for ubiquitination and degradation (54).
As Pin1 is important for the function of reproductive tissues in mice (4, 5, 23), we reasoned that Pin1 could play a role in the function of sex steroid receptors such as estrogen receptor (ER) and progesterone receptor (PR) by influencing the activities of one or more of their coactivators. Our attention was directed first to SRC-3/AIB1 because it is a dominant coactivator of PR and ER in certain reproductive tissues (19, 42, 52). Recently, Wu et al. (49) identified several phosphorylation sites in SRC-3 using mass spectrometry and found that different combinations of phosphorylation sites modulate the SRC-3 transactivation function in response to different signaling pathways. Five out of the six phosphorylation sites identified in SRC-3 contain Ser/Thr-Pro motifs, raising the possibility that Pin1 could be involved in the postphosphorylation regulation of SRC-3. In addition, the activation of SRC-3 by phosphorylation appears to be coupled to its degradation. However, how transactivation by SRC-3 is coupled to its degradation has remained an enigma. Here, we demonstrate that Pin1 can function as a novel coactivator by interacting with phosphorylated SRC-3 and modulating its protein-protein interactions with other coregulators in a manner that enhances PR and ER function as well as directs the cellular turnover of SRC-3.
MATERIALS AND METHODS
Plasmids and reagents.
The Pin1 cDNA was synthesized by reverse transcriptase (RT) PCR from MCF-7 cell mRNA and was inserted into a pCM5 mammalian expression vector, pGEX (Amersham Biosciences), for glutathione S-transferase (GST) fusion protein or pMGal4 (BD Biosciences) for Gal4DBD fusion protein. The Pin1 mutant C113A or W34A was generated using a QuikChange mutagenesis kit (Stratagene). The SRC-3 fragments were inserted into pACT (Promega) for VP16-AD fusion proteins. VP16-CBP was kindly provided by Carolyn Smith. The PKA catalytic subunit c-Jun, c-Fos, and CREB expression vectors were kindly provided by Barbara Sanborn, Tse Hua Tan, and Michael Greenberg. Different transcription-element-driven luciferase vectors were from Mercury Pathway profiling luciferase systems (BD Biosciences).
The anti-SRC-3 antibody was generated as described previously (48). The anti-Pin1 antibody used in Western blot analysis was provided by Anthony Means. Anti-Pin1 antibody used in the coimmunoprecipitation experiment was from Calbiochem. Antibodies used in the chromatin immunoprecipitation assay, anti-ERα (H-184), anti-ACTR (C-20), and anti-p300 (N-15), were from Santa-Cruz Biotechnology. Other antibodies were from various sources: anti-flag, Sigma, Affinity Bioreagents; antihemagglutinin (anti-HA), Roche Molecular Biochemicals); anti-p300, Upstate.
Cell lines and transfections.
HeLa (American Type Culture Collection), MCF-7 (from Richard Santen), mouse embryonic fibroblast (MEF) Pin1−/− and Pin1+/+ (provided by Anthony Means), and other cell lines were maintained in Dulbecco's modified Eagle medium supplemented with 5% fetal bovine serum unless otherwise noted. T47D (CAT0) cells were maintained in 0.2 mg/ml Geneticin (Invitrogen) containing medium, and the Flp-In T-Rex 293 cells (Invitrogen) were maintained in 15-μg/ml blasticidin-100-μg/ml zeocin-containing medium. The tetracycline-inducible stable SRC-3 Flp-In T-Rex 293 cell line was generated according to instructions in the manufacturer's manual (Invitrogen) and was maintained in medium containing blasticidin and hygromycin (150 μg/ml); 0.5 μg/ml of tetracycline was used to induce the expression of flag-SRC-3. Stable Pin1-depleted HeLa cells were generated by transfection with pSuper-shPin1 (provided by Anthony Means) and selected by using 0.8 μg/μl puromycin. For assays of steroid hormone-dependent transcription, cells were maintained in charcoal-stripped fetal bovine serum (Gemini) containing phenol red-free Dulbecco's modified Eagle medium for at least 3 days before the addition of steroid hormones.
All plasmid DNAs except those transfected into MEF cells were transfected using TransIT-LT1 transfection reagent (Mirus) according to the manufacturer's protocol. Fugene6 (Roche) was used to transfect plasmid DNA into MEF cells. For measuring luciferase, steroid hormones were added to the medium 16 h after transfection. After an additional 24-h incubation, cells were lysed and cell lysates were used for a luciferase assay. The smartpool small interfering RNA (siRNA) or scrambled siRNA was obtained from Dharmacon. siRNA (4 nM) was transfected into cells by using TransIT-TKO transfection reagent (Mirus). Cells were harvested 4 days after transfection. In the case of cotransfection of siRNA and plasmid DNA experiments, cells were transfected with siRNA first; 2 days after transfection, cells were cotransfected with siRNA and plasmid DNA according to the manufacturer's protocol (Mirus), followed by the same procedure as plasmid DNA transfection alone.
GST pulldown and in vitro protein-protein interaction.
Escherichia coli-expressed glutathione S-transferase (GST)-Pin1 was bound by 10 μl glutathione Sepharose 4B (Amersham Biosciences), followed by the addition of 300 μg HeLa cell extracts or in vitro-transcribed or -translated SRC-3. After 4 h of incubation at 4°C, the beads were washed five times with wash buffer (50 mM Tris-Cl [pH 7.5], 200 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 10% glycerol, 1% NP-40, 1 mM dithiothreitol). The bound proteins were resolved in sodium dodecyl sulfate (SDS)-7% polyacrylamide gel electrophoresis (PAGE) gels and detected by Western blot analysis.
Fifty nanograms of purified baculovirus recombinant SRC-3 was immunoprecipated with anti-SRC-3 antibody in 150 μl buffer (20 mM HEPES [pH 7.6], 150 mM KCl, 0.1% NP-40, 8% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail) and then incubated with purified GST or different concentrations of GST-Pin1 (2 and 10 ng) at room temperature for 5 min. The samples were incubated at 4°C for an additional 10 min, and the excess Pin1 was removed with wash buffer. Purified baculovirus recombinant p300 was then incubated with SRC-3 at 4°C overnight, and the beads were washed extensively with wash buffer. The bound p300 was resolved in 4 to 15% SDS-PAGE and detected by Western blot analysis.
Immunoprecipitation and Western blot analysis.
Cells were washed with phosphate-buffered saline and lysed in the lysis buffer (20 mM Tris-Cl [pH 8.0], 125 mM NaCl, 0.5% NP-40, 20 mM NaF, 0.2 mM Na3VO4, 2 mM EDTA, protease inhibitor cocktail) for 30 min. After centrifugation at 13,400 × g for 15 min, the supernatant was precleared with normal immunoglobulin G (IgG) for 1 h and then incubated with the desired antibody for 2 h, followed by the addition of 20 μl protein A and G agarose beads overnight. The antibody bound complex was washed extensively with the lysis buffer. The proteins were then resolved in a 4 to 15% gradient SDS-PAGE gel and transferred to nitrocellulose membranes (Bio-Rad). The primary antibody was diluted in TBST buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% Tween 20, 5% dried nonfat milk) with overnight incubation at 4°C, followed by the appropriate secondary antibody with 1 h of incubation at room temperature. The Western blot was visualized by chemiluminescence (Pierce).
Real-time quantitative PCR and RT-PCR.
MCF-7 cell total RNA was extracted using Tri reagent (Molecular Research Center, Inc.). The pS2 mRNA and the cyclophilin mRNA (as an internal control) were quantitated by TaqMan-based reverse transcriptase PCR using the ABI Prism 7700 sequence detection system (Applied Biosystems). The primers for the pS2 mRNA were as follows: forward, 5′-GGTCGCCTTGGAGCAGA; reverse, 5′-GGGCGAAGATCACCTTGTT; probe, 5′-6-carboxyfluorescein (FAM)-TCCATGGTGGCCATTGCCTCCT-6-carboxytetramethylrhodamine (TAMRA). The primers for the cyclophilin mRNA were as follows: forward, 5′-GACAAGGTCCCAAAGACAGCAG; reverse, 5′-FCAGGAACCCTTATAACCAAATCC; probe, 5′-FAM-AAATTTTCGTGCTCTGAGC-TAMRA. The primers for the pS2 promoter in the chromatin immunoprecipitation (ChIP) assay were as follows: forward, 5′-CGTGAGCCACTGTTGTCAGG; reverse, 5′-TGGTGAGGTCATCTTGGCTG; probe, 5′-FAM-CAAGCCTTTTTCCGGCCAT-TAMRA. The primers for the cylophilin intron in the ChIP assay were as follows: forward, 5′-TGTTTAATGACATTTAGTACAAAAGGCTTC; reverse, 5′-GAACAACATTATGACTGGCAACC; probe, 5′-FAM-AGCTACCTTTCTCGTCTTG-TAMRA.
The RT-PCR analysis was performed using the Access RT-PCR kit (Promega). The primers for the flag-SRC-3 were as follows: forward, ATTACAAGGATGACGACGATAAG; reverse, CGTATCTGTCTTACTGTTTCCTTTAAAATC.
ChIP.
MCF-7 cells were transfected with Pin1 siRNA or scrambled siRNA. Four days after transfection, cells were treated with 10−8 M estradiol for 1 h and were harvested for the ChIP analysis. The procedure was essentially the same as described previously (22). The protein-bound DNA precipitated from the ChIP assay was purified with the QIAquick PCR purification kit (QIAGEN) and eluted in 50 μl of elution buffer. The DNA was quantitated using the pS2 promoter-specific primers by real-time quantitative PCR. The cyclophilin intron 3-specific primers were used as a control.
Pulse-chase metabolic labeling.
MCF-7 cells were transfected with siRNA. Four days after transfection, cells were incubated with Met- and Cys-free medium (Gibco) for 30 min and then replaced by medium containing 35S-Met/Cys and incubated for 1 h. Cells were then washed three times with phosphate-buffered saline and incubated with medium supplemented with 10 mM cold Met and Cys for 0, 4, and 8 h. Cells were harvested at the indicated time, and SRC-3 was immunoprecipitated from cell lysates by using anti-SRC-3 antibody. The precipitated SRC-3 was subjected to SDS-PAGE and detected with an autoradiograph.
Statistical analysis and quantitative analysis.
Statistical analysis was performed with a t test using simple interactive statistical analysis (SISA). Data quantitation was carried out with National Institutes of Health Image software, version 1.62.
RESULTS
Pin1 interacts selectively with phosphorylated SRC-3.
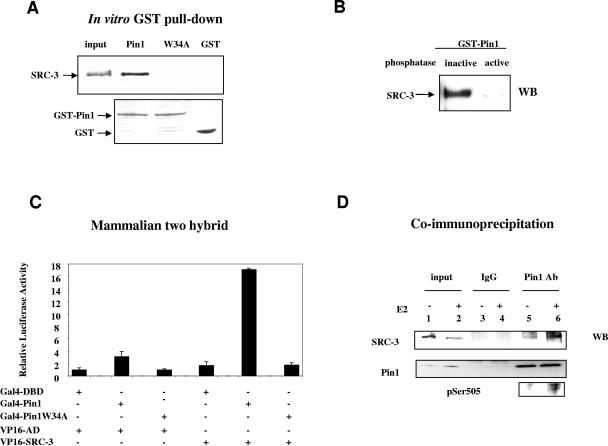
To test the potential role of Pin1 in SRC-3 function, we first tested whether Pin1 interacts with SRC-3 by using a GST pulldown assay of a HeLa cell extract. As shown in Fig. 1A, SRC-3 was detected in the Western blot following pulldown by GST-Pin1, but not by GST alone or a GST-Pin1W34A mutant which has a mutation in the WW domain and is defective in the target protein binding function (47). Similar results were also obtained when in vitro-transcribed and -translated SRC-3 was incubated with GST-Pin1 (data not shown). Given that all known Pin1 substrates are phosphoproteins and that five of the six identified phosphorylation sites of SRC-3 are Ser/Thr-Pro, we tested whether the interaction between SRC-3 and Pin1 is dependent on the phosphorylation status of SRC-3. Flag-SRC-3 was immunoprecipitated from extracts of transfected HeLa cells with anti-flag antibody and dephosphorylated by treatment with λ-phosphatase. After elution from antibody precipitates with flag peptides, the dephosphorylated SRC-3 was incubated with GST-Pin1. As shown in Fig. 1B, the interaction between SRC-3 and Pin1 was lost when SRC-3 first was treated with the active phosphatase; heat-inactivated λ-phosphatase treatment had no effect on this interaction. We conclude that Pin1 interacts primarily with phosphorylated SRC-3.
FIG. 1.
Pin1 interacts with phosphorylated SRC-3. (A) In vitro interaction of SRC-3 and Pin1. HeLa cell extracts were subjected to a GST pulldown assay and immunoblotted for SRC-3 (upper panel). The lower panel shows the Coomassie blue staining of GST-Pin1, GST-Pin1W34A, and GST. (B) Phosphorylation is required for SRC-3 to interact with Pin1. Flag-SRC-3 was immunoprecipitated from transfected HeLa cell extracts, treated with λ-phosphatase or heat-inactivated λ-phosphatase, and then eluted from the antibody. The eluted SRC-3 was used for the GST-Pin1 pulldown assay. WB, Western blot. (C and D) In vivo interaction of SRC-3 and Pin1. (C) Mammalian two-hybrid assay. HeLa cells were cotransfected with 100 ng of plasmids expressing Gal4DBD-Pin1, VP16AD-SRC-3, and the Gal4 binding reporter luciferase. (D) Coimmunoprecipitation. The nuclear extracts from MCF-7 cells treated with or without estradiol were immunoprecipitated with anti-Pin1 antibody or normal IgG and Western blotted for anti-SRC-3 (upper panel) or anti-Pin1 (middle panel). SRC-3 pSer505-phosphorylation-specific antibody indicates increased phosphorylation of SRC-3 when cells were treated with estradiol.
We next determined whether the SRC-3/Pin1 interaction also occurs in cells by using a mammalian two-hybrid assay. Pin1 or Pin1W34A was fused to a Gal4 DNA binding domain, and SRC-3 was fused to a VP16 activation domain. The cotransfection of Gal4-Pin1, but not Gal4-Pin1W34A, with VP16-SRC-3 markedly increased reporter activity (Fig. 1C), indicating that Pin1 also interacts with SRC-3 in living cells. To confirm that this interaction also occurs between endogenous SRC-3 and Pin1, coimmunoprecipitation was carried out using MCF-7 nuclear extracts (Fig. 1D). SRC-3 was detected in an anti-Pin1 antibody precipitation (lane 5) while it could not be detected in control IgG precipitations (lane 3). Furthermore, when cells were treated with estradiol, which previously was shown to stimulate SRC-3 phosphorylation (49) and was demonstrated again by Western blot anaylsis using SRC-3 pSer505-specific phosphoantibody (shown in the bottom panel), the amount of SRC-3 coimmunoprecipitated by the Pin1 antibody increased (lane 6 versus lane 5). Together, these results indicate that SRC-3 interacts with Pin1 both in vitro and in vivo and that the interaction is phosphorylation dependent.
Pin1 interacts with multiple regions of SRC-3.
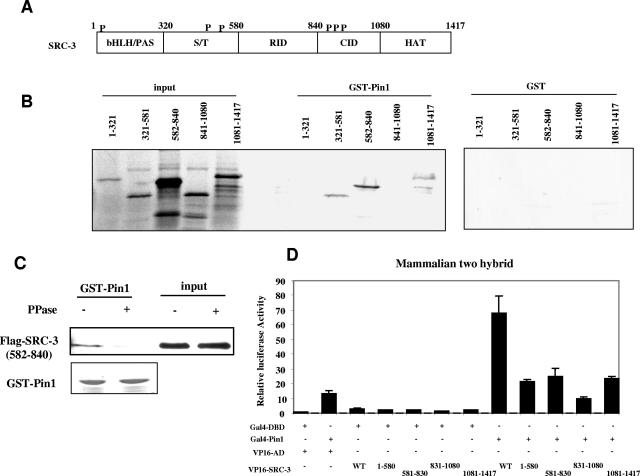
To determine potential Pin1 interaction domains in SRC-3, we used a series of in vitro-transcribed and -translated SRC-3 fragments in GST pulldown assays. SRC-3, like other members of the p160 coactivator family, contains several functional domains, as shown in Fig. 2A. It interacts with nuclear receptors through the conserved LXXLL motifs in the receptor interaction domain (aa 582 through 840). The N-terminal region contains a highly conserved bHLH/PAS domain (aa 1 through 320) and a Ser/Thr-rich region (aa 321 through 581). The C-terminal regions are the CBP/p300 interaction domain (aa 841 through 1080) and the domain that contains histone acetyltransferase (HAT) activity (aa 1081 through 1417). Three fragments of SRC-3 (aa 321 through 581, 582 through 840, and 1081 through 1417) were found to interact with GST-Pin1 but not GST (Fig. 2B). To determine whether in vitro-translated products are phosphorylated and whether the interaction we observed is dependent on phosphorylation, we immunoprecipitated in vitro-synthesized flag-SRC-3 (582 through 840) with anti-flag antibody, eluted, and then treated it with or without λ-phosphatase. The eluted SRC-3 fragment was then used for GST-Pin1 pulldown assay. As shown in Fig. 2C, no interaction was observed between Pin1 and the phosphatase-treated SRC-3 fragment, suggesting that this in vitro-translated SRC-3 fragment is phosphorylated and that its interaction with Pin1 is dependent on its phosphorylation status.
FIG. 2.
Pin1-interacting regions in SRC-3. (A) Schematic representation of SRC-3 domains. bHLH/PAS, basic helix-loop-helix/Per, ARNT, Sim homologous domain; S/T, Ser/Thr-rich region; RID, receptor interaction domain; CID, CBP/p300 interaction domain; HAT, HAT activity domain. P indicates the previously identified SRC-3 phosphorylation sites. (B) In vitro interaction of Pin1 and SRC-3 fragments. A series of 35S-labeled in vitro-transcribed and -translated SRC-3 fragments were used in the GST-Pin1 pulldown assay and were detected by autoradiograph. (C) Interaction between Pin1 and in vitro-transcribed and -translated SRC-3 fragment is dependent on phosphorylation. The procedure was similar to the one described for Fig. 1B except that flag-SRC-3 (582 through 840) was synthesized in vitro. The interaction was detected by Western blot analysis using anti-flag antibody. (D) Mammalian two-hybrid assay. The same procedure described in the legend of Fig. 1 was used. WT, wild type.
To confirm that the interaction of different SRC-3 fragments with Pin1 in vivo was similar to our in vitro results, we performed a mammalian two-hybrid assay. Consistent with the in vitro interaction results, the cotransfection of VP16-AD fused to the SRC-3 fragments and Gal4-Pin1 enhanced transcription activity, although at a lower level than full-length SRC-3; the fragment (aa 841 through 1080) that did not interact with Pin1 in the GST pulldown assay also did not interact with Pin1 in whole cells (Fig. 2D). Our results indicate that Pin1 can interact with SRC-3 at multiple sites and raise the question of whether Pin1 plays a role in regulating SRC-3 activity.
Two of the interacting fragments (aa 321 through 581 and aa 582 through 840) contain previously identified phosphorylation sites. However, the third fragment (aa 1081 through 1417), which does not contain a previously identified phosphorylation site, still interacts with Pin1, implying that additional phosporylation sites may be present in that fragment. Indeed GST-Pin1 pulldown experiments comparing wild-type SRC-3 with the mutant in which all six Ser/Thr residues are changed to Ala show that GST-Pin1 still has some ability to interact with the mutant (data not shown). The fragment from aa 841 through 1080 does not bind to Pin1, although it contains previously identified phosphorylation sites. This is probably due to the lower affinity or inaccessibility of Pin1 to these sites. These results suggest that additional phoshorylation sites in SRC-3 may influence the interaction with Pin1.
Pin1 synergizes with SRC-3 to coactivate steroid receptor transcriptional activity.
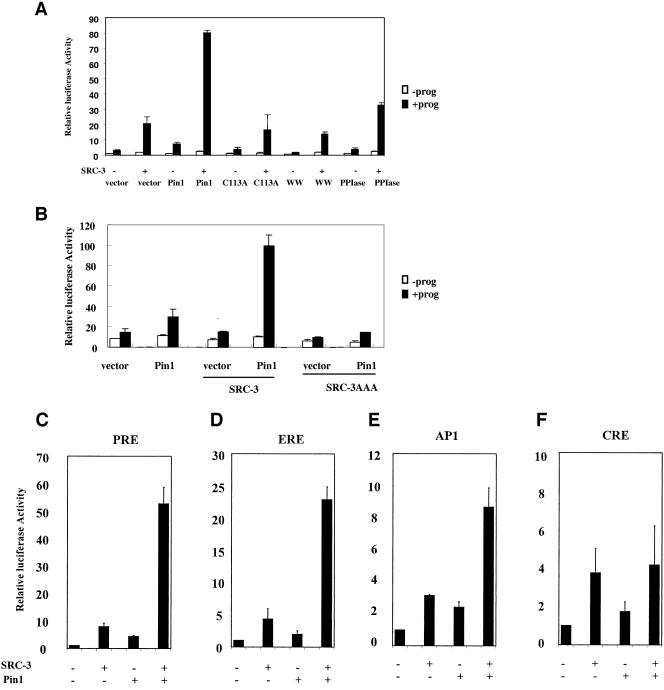
To determine if the interaction between Pin1 and SRC-3 plays a functional role, we examined the effects of Pin1 on SRC-3-activated PR activity. A PR expression vector and progesterone response element (PRE)-driven luciferase reporter were cotransfected into HeLa cells. Transfection of SRC-3 enhanced luciferase activity in the presence of 10 nM progesterone (Fig. 3A). Transfection of Pin1 also stimulated PR target gene transcription. However, cotransfection of both SRC-3 and Pin1 synergistically increased PR-dependent luciferase activity (Fig. 3A and C). In contrast, the Pin1C113A mutant, which is impaired in PPIase catalytic activity (47, 59), did not produce any activation of PR-mediated transcription. Similarly, neither of the two separate domains of Pin1 (WW or PPIase domain) could activate transcription synergistically with SRC-3. These experiments show that a fully intact and enzymatically active Pin1 is required for SRC-3-dependent synergistic activation of the PR target gene reporter.
FIG. 3.
Pin1 and SRC-3 activate steroid receptor transactivation synergistically. (A) HeLa cells were cotransfected with vectors expressing progesterone receptor B (PRB), SRC-3, Pin1 or Pin1 mutants, and PRE-luciferase reporter in the absence (−prog) or presence (+prog) of 10−8 M progesterone. (B) HeLa cells were cotransfected with vectors expressing PRB, SRC-3, or SRC-3 mutant with mutations in LXXLL motifs (SRC-3AAA), Pin1, and PRE-driven reporter in the absence or presence of 10−8 M progesterone. (C through F) HeLa cells were transfected with expression vectors containing SRC-3, Pin1, various transcription factors, and the corresponding reporters. (C) PRE-luciferase and the coexpression of PRB in the presence of 10−8 M progesterone. (D) Estrogen response element-luciferase and the coexpression of ERα in the presence of 10−7 M estradiol. (E) AP-1-luciferase and the coexpression of c-Jun and c-Fos. (F) CRE-luciferase, stimulated by the cotransfection of the vector expressing the PKA catalytic subunit.
To determine whether the effect of Pin1 on PR transcriptional activity is mediated through SRC-3, we cotransfected Pin1 with an SRC-3 mutant containing three LXXLL motifs mutated to LXXAA (SRC-3AAA), which cannot interact with PR. As shown in Fig. 3B, Pin1 did not activate transcription when SRC-3AAA mutant was cotransfected. This suggests that SRC-3 mediates the effect of Pin1 on PR activity.
Although originally identified as a nuclear receptor coactivator, SRC-3 was reported to coactivate several other transcription factors including CREB, AP-1, E2F, serum response factor, p53, and STAT (3, 13, 17, 18, 20, 27). To test whether Pin1 and SRC-3 can synergize to activate other transcription factors, we examined estrogen, AP-1, and cyclic AMP response element (CRE) responses. As shown in Fig. 3C through F, Pin1 or SRC-3 alone moderately enhanced transcription of each of the four reporter genes tested. When Pin1 and SRC-3 were cotransfected, only the estrogen response element and PRE reporter genes were synergistically activated. Additive or weak synergy of the AP-1 response was observed for SRC-3 and AP-1. Whereas either Pin1 or SRC-3 caused small increases in CRE-mediated transcription, no additivity of these responses was observed upon cotransfection of SRC-3 and Pin1. Finally, transcription of simian virus 40 promoter-driven renilla luciferase was not altered when cells were cotransfected with Pin1 and SRC-3 (data not shown), thus serving as a negative internal control.
Down-regulation of cellular Pin1 impairs steroid receptor target gene transcription.
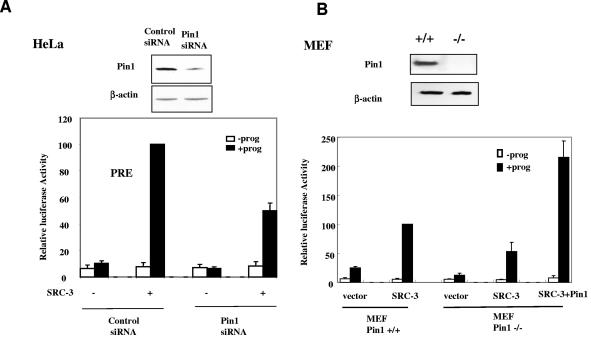
Since overexpression of Pin1 significantly enhanced SRC-3-activated steroid receptor-mediated transcription, we next wished to examine whether a decrease in endogenous Pin1 decreased such transcription reactions. To examine this question, we first transfected HeLa cells with an siRNA against Pin1 or a scrambled control siRNA. Two days after transfection, PR, SRC-3, and PRE-driven luciferase reporter were cotransfected and progesterone-stimulated luciferase activity was measured. As shown in the upper panel of Fig. 4A, the Pin1 siRNA reduced the Pin1 protein level in HeLa cells and decreased the SRC-3-activated PR activity by 50% compared to control siRNA (Fig. 4A, lower panel). Similarly, SRC-3-mediated PR transactivation was reduced by 50% in MEF Pin1−/− cells compared to that in MEF Pin1+/+ cells. Importantly, reexpression of exogenous Pin1 was able to increase transcription (Fig. 4B, lower panel).
FIG. 4.
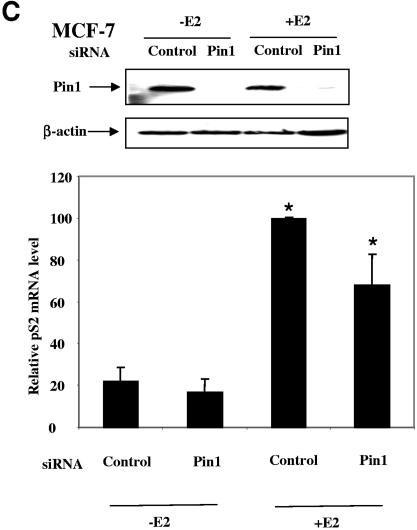
Depletion of Pin1 impairs the steroid-receptor-targeted transcription. (A) Knockdown of Pin1 reduces transient-transfected PR target gene transcription. HeLa cells were transfected with Pin1 siRNA or scrambled control siRNA and progesterone receptor B (PRB), SRC-3 expression vectors, and PRE-luciferase in the absence (−prog) or presence (+prog) of 10−8 M progesterone. Cell lysates were subjected to Western blot analysis for Pin1 and β-actin antibodies (upper panel), and the reporter luciferase activities were measured (lower panel). (B) PR target gene transcription is reduced in MEF Pin1−/− cells. MEF Pin1−/− and Pin1+/+ cells were transfected with PRE-luciferase, PRB, SRC-3, and Pin1 expression or empty vectors in the absence or presence of 10−8 M progesterone. (C) Knockdown of Pin1decreases endogenous ER target gene transcription. MCF-7 cells were transfected with Pin1 siRNA and stimulated with 10−8 M estradiol. Cell lysates were used in Western blot analysis (upper panel). Total RNA extracted from cells was subjected to real-time quantitative RT-PCR using pS2 mRNA-specific primers (lower panel). The pS2 mRNA level was normalized against the cyclophilin mRNA level (n = 4; *, P < 0.05).
We next tested whether Pin1 depletion affects the expression of an endogenous steroid receptor target gene. For this purpose, we used an MCF-7 cell line to examine estrogen-receptor-regulated pS2 transcription. As shown in the upper panel of Fig. 4C, Pin1 siRNA reduced Pin1 expression in MCF-7 cells. Consistent with the result observed in transfected HeLa cells, the endogenous estrogen-dependent pS2 mRNA level decreased 30% when Pin1 was knocked down, as revealed by real-time quantitative PCR (Fig. 4C, lower panel) (P < 0.05).
Thus, in HeLa, MEF, and MCF-7 cells, a decrease in Pin1 results in a reduction in PR- and ER-dependent transcription.
Pin1 activity enhances the interaction between SRC-3 and CBP/p300.
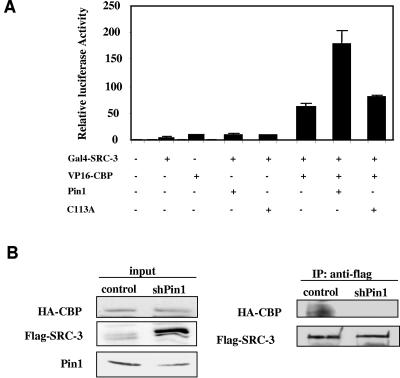
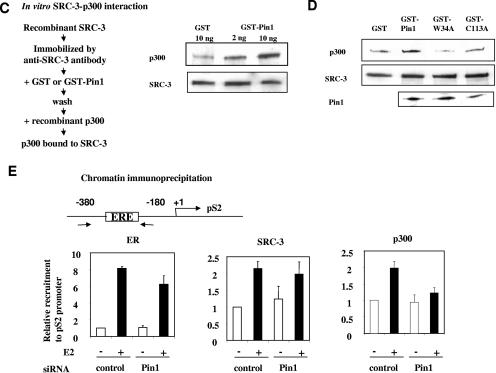
SRC-3 exerts its function as a nuclear receptor coactivator primarily through recruiting additional coregulator proteins such as CBP/p300 to the promoter-bound receptor-coactivator complex. We previously showed that mutations of the Ser/Thr-Pro SRC-3 phosphorylation sites impair the ability of SRC-3 to form complexes with coactivators such as CBP (49). Since Pin1 binds specifically to phosphorylated SRC-3, we wished to determine whether Pin1 affects the interaction between SRC-3 and CBP/p300. Mammalian two-hybrid assays were carried out using Gal4-SRC-3 and VP16-CBP (Fig. 5A). The cotransfection of Gal4-SRC-3 and VP16-CBP significantly increased reporter transcription, indicating a cellular interaction between SRC-3 and CBP. Transcription was further augmented by cotransfection of Pin1, but not the catalytically inactive Pin1 C113A mutant. Although there are several possible explanations for this result, one likely possibility is that Pin1 functions to facilitate the cellular interaction of SRC-3 and CBP/p300 to enhance the transcriptional activity. To confirm this idea, a coimmunoprecipitation experiment was carried out in HeLa cells stably knocked down with Pin1 by cotransfecting with flag-SRC-3 and HA-CBP. As shown in Fig. 5B, the amount of HA-CBP coimmunoprecipitated by anti-flag antibody is markedly reduced in cells stably knocked down with Pin1 compared to that in control cells. Interestingly, we found that the flag-SRC-3 protein level in the input increased when Pin1 was knocked down. To determine whether Pin1 directly affects the complex formation, SRC-3 and p300 interactions were assessed in vitro using recombinant proteins purified from baculovirus-infected insect cells. SRC-3 was first immunoprecipitated with a specific antibody. Different concentrations of Pin1 were then incubated with bead-bound SRC-3, and the excess Pin1 was then washed away before the addition of p300. After a further incubation period, the SRC-3/p300 complex was coimmunoprecipitated with SRC-3 antibody and detected by Western blot analysis (Fig. 5C). The amount of precipitated p300 associated with SRC-3 was increased as a function of Pin1 concentration. However, mutant Pin1 proteins with altered WW or PPIase domains (Fig. 5D) did not promote the interaction between SRC-3 and p300. These data confirm that Pin1 facilitates the interaction between SRC-3 and CBP/p300.
FIG. 5.
Pin1 enhances the interaction of SRC-3 and CBP/p300. (A) Pin1 increases the functional interaction between SRC-3 and CBP in a mammalian two-hybrid assay. (B) HeLa cells with stably integrated scrambled control or shRNA against Pin1 were transfected with HA-CBP and Flag-SRC-3. Cell lysates were immunoprecipitated with anti-flag antibody and then subjected to Western blot analysis with antibody against HA or flag. (C and D) Pin1 increases in vitro interaction between SRC-3 and p300. Recombinant SRC-3 purified from baculovirus was immunoprecipitated with SRC-3 antibody, incubated with increasing concentrations of Pin1 (C) or Pin1 mutants (D), and followed by the addition of purified recombinant p300 as described in Materials and Methods. The SRC-3-associated p300 was detected by Western blot analysis. (E) Depletion of Pin1 reduces the recruitment of p300 to pS2 promoter in response to estrogen stimulation. MCF-7 cells treated with ethanol or 10−8 M estradiol were subjected to ChIP analysis using ERα (P < 0.05), SRC-3 (P = 0.436), and p300 (P < 0.001) antibodies (n = 5). The protein-bound pS2 promoter DNA was quantitated using real-time quantitative PCR (lower panel). The arrows (upper panel) indicate the pS2-promoter-specific primers used in the PCR.
To further substantiate this finding, we next examined whether Pin1 affects the recruitment of p300 to an endogenous pS2 promoter in MCF-7 cells. Chromatin immunoprecipitation assays were carried out in MCF-7 cells transfected with control siRNA or Pin1 siRNA. The recruitments of ER, SRC-3, and p300 to the pS2 promoter upon E2 stimulation were detected by real-time quantitative PCR (Fig. 5E). The results in Fig. 5E show that knocking down Pin1 did not significantly affect the recruitment of ERα and SRC-3 to the pS2 promoter. However, the recruitment of p300 to the promoter was significantly reduced (P < 0.001) when Pin1 was knocked down. These results provide further evidence that Pin1 facilitates the recruitment of CBP/p300 to a steroid-receptor-targeted promoter.
Pin1 modulates the cellular levels of SRC-3 protein.
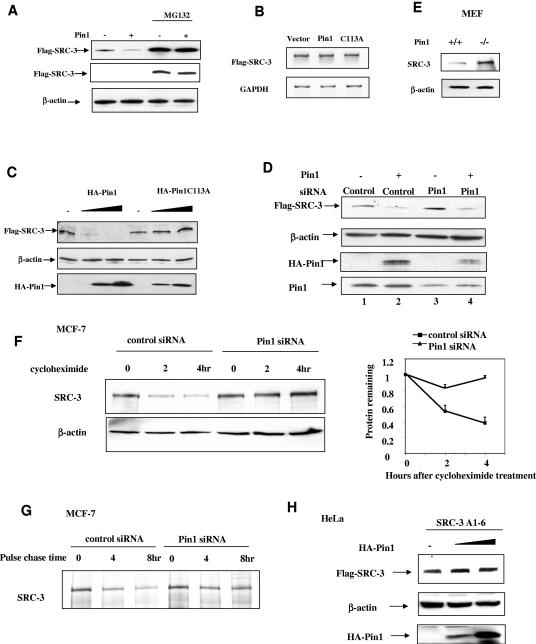
Prolyl isomerization catalyzed by Pin1 has been shown to alter the stability of target proteins such as NF-κB (35), p53 (50, 56, 57), and c-Myc (54). Since we found that the flag-SRC-3 protein level increased in Pin1 knocked-down cells (Fig. 5B), we next addressed whether the concentration of Pin1 could also modulate the stability of SRC-3 in HeLa cells. A Pin1 expression vector (or an empty expression vector) was cotransfected with flag-SRC-3 into HeLa cells. The level of the flag-SRC-3 protein was detected by Western blot analysis using a flag-specific antibody. As shown in Fig. 6A, the amount of flag-SRC-3 was reduced when cells were cotransfected with Pin1. Conversely, treatment of HeLa cells with the proteasome inhibitor MG132 resulted in a substantial increase in flag-SRC-3, and this level was unaffected by overexpression of Pin1 (as shown by shorter exposure of the Western blot). As overexpression of Pin1 did not increase the amount of flag-SRC-3 mRNA (Fig. 6B), the data suggest that Pin1 affects SRC-3 protein stability rather than altering expression of the SRC-3 gene. To determine whether the effect of Pin1 on SRC-3 protein level is cell type specific and to substantiate that Pin1 PPIase activity is involved in regulating SRC-3 level, we transfected different concentrations of Pin1 or Pin1C113A into a 293 cell line stably expressing an inducible flag-SRC-3 gene. As illustrated in Fig. 6C, increasing the concentration of Pin1 decreased the level of flag-SRC-3 whereas the Pin1C113A mutant had no effect on flag-SRC-3 protein levels. Thus, the effect of Pin1 on SRC-3 level requires the PPIase activity and is not cell specific.
FIG. 6.
Pin1 alters the steady-state SRC-3 protein level. (A) Western blot analysis of transfected flag-SRC-3 in HeLa cells using flag antibody with the cotransfection of Pin1 or empty vector in the absence or presence of 10 μM MG132 for 24 h. The middle panel shows the shorter exposure of the chemiluminescence for the upper panel. (B) RT-PCR analysis on flag-SRC-3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA from transfected HeLa cells with the cotransfection of empty vector, Pin1, or Pin1 C113A. (C) PPIase activity of Pin1 is required for the destabilization of SRC-3. Western blot analysis of flag-SRC-3 from inducible flag-SRC-3-stable Flp-In T-Rex 293 cells with the cotransfection of increasing concentrations of HA-Pin1 or HA-Pin1 C113A expression plasmids was performed using flag, β-actin, and HA antibodies. (D) Overexpression of Pin1 rescues the effect of Pin1 siRNA on SRC-3 protein level. HeLa cells were transfected with scrambled control and Pin1 siRNA, followed by the transfection of Pin1 (lanes 2 and 4) or empty vector (lanes 1 and 3). Cell lysates were subjected to Western blot analysis using flag, HA, and Pin1 antibodies. (E) MEF Pin1−/− cells have higher SRC-3 protein levels than Pin1+/+ cells. MEF cell lysates were subjected to Western blot analysis using SRC-3 antibody. (F and G) Depletion of Pin1 increases the half-life of SRC-3. (F) MCF-7 cells were transfected with Pin1 siRNA, stimulated with estradiol, and treated with 200 μg/ml cycloheximide for 0, 2, and 4 h. Cell lysates were subjected to Western blot analysis using SRC-3 antibody (left panel). The quantitation of SRC-3 protein levels normalized against β-actin during the course of cycloheximide treatment is shown in the right panel (n = 4). (G) MCF-7 cells were treated with control or Pin1 siRNA, incubated with [35S]Met/Cys containing medium for 1 h, and then incubated with cold Met/Cys containing medium for 0, 4, and 8 h. SRC-3 was immunoprecipitated and then subjected to SDS-PAGE and autoradiography. (H) Pin1 does not affect SRC-3A1-6 phosphomutant protein level. HeLa cells were transfected with flag-SRC-3A1-6 and increasing concentrations of Pin1. Cell lysates were subjected to Western blot analysis.
We also addressed the relationship between Pin1 and flag-SRC-3 levels by using siRNA to reduce the amount of Pin1 in HeLa cells (Fig. 6D). In contrast to the effect caused by the overexpression of Pin1, administration of Pin1 siRNA increased the level of flag-SRC-3 compared to the addition of a scrambled control siRNA (Fig. 6D, lane 3 versus lane 1). The increase in SRC-3 mediated by Pin1 siRNA was due specifically to the decrease in the Pin1 protein level since cotransfection of Pin1 reverted the Pin1 siRNA effect and resulted in a decreased level of flag-SRC-3 (Fig. 6D, lanes 4 and 2 versus lane 1). Similarly, an increased SRC-3 protein level was found in MEF Pin1−/− cells compared to that in Pin1+/+ cells (Fig. 6E).
To determine if the regulation of SRC-3 protein level by Pin1 was due to a change in SRC-3 stability, we directly examined the half-life of endogenous SRC-3 in MCF-7 cells treated with either scrambled control siRNA or Pin1 siRNA. The cells were treated with estradiol overnight prior to the addition of cycloheximide to inhibit cellular protein synthesis. Figure 6F shows that the SRC-3 protein levels gradually decreased during the time course of cycloheximide treatment when control siRNA was transfected into cells, and this rate of SRC-3 turnover is consistent with that previously reported (26). In cells transfected with Pin1 siRNA, however, the SRC-3 protein was more stable (Fig. 6F). To further substantiate our cycloheximide treatment results, we also did pulse-chase experiments to examine SRC-3 protein stability using [35S]Met/Cys to label SRC-3 for 1 h and then to chase it with cold Met/Cys in MCF-7 cells treated with either control or Pin1 siRNA. Our results were similar to and confirmatory of the cycloheximide treatment results (Fig. 6G). To test whether the ability of Pin1 to alter SRC-3 turnover requires phosphorylation of SRC-3, we examined the effect of the overexpression of Pin1 on an SRC-3 mutant in which all six previously identified phosphorylation sites are mutated to Ala (SRC-3A1-6) (49). Flag-SRC-3A1-6 was cotransfected with increasing concentrations of Pin1 into HeLa cells. Although this mutant can still interact with Pin1, its level is unaffected by increasing levels of Pin1 (Fig. 6H). These data support our contention that Pin1 modulates SRC-3 turnover in a manner that requires the phosphorylation of SRC-3 and in fact may be required for its intracellular proteolytic degradation.
DISCUSSION
Protein phosphorylation and dephosphorylation are important mechanisms regulating the activities of nuclear receptors and cofactors in response to extracellular signals. While phosphorylation of CBP by ERK1 increases its HAT activity (1), phosphorylation of ARA55 by Pyk2 decreases its activity by reducing its interaction with androgen receptor (45). Although it is not entirely clear as to how phosphorylation regulates coactivator activities, it is suspected that phosphorylation leads to conformational changes that provide surface binding sites for other proteins. Pin1 has been found to regulate the postphosphorylation events of a number of its target proteins by catalyzing the cis and trans isomerization of phosphorylated Ser/Thr-Pro bonds. Given that SRC-3 contains several phosphorylated Ser/Thr-Pro motifs (49) and SRC family coactivators are central to the recruitment of multiprotein coactivator complexes to promoters, we tested whether Pin1 is involved in the regulation of SRC-3 function.
We provide evidence here that Pin1 is a novel secondary coactivator for steroid receptors. It synergizes with SRC-3 in the coactivation of steroid receptors (Fig. 3), and depletion of Pin1 reduces hormone-dependent steroid receptor activity (Fig. 4). Pin1 knockout mice were reported to have severe defects in full mammary gland expansion during pregnancy (23), which, if correct, is similar to the PR knockout mice phenotype (29). Pin1 binds SRC-3 in a phosphorylation-dependent manner, and we found that phosphorylation of SRC-3, along with the cellular concentration of Pin1, is important for SRC-3 turnover. Indeed, the half-life of SRC-3 protein was significantly increased when Pin1 levels were reduced and the altered turnover is dependent on both SRC-3 phosphorylation and the prolyl isomerase activity of Pin1 (Fig. 6).
We also provide evidence that Pin1 promotes the formation of SRC-3 and CBP/p300 coactivator complexes as Pin1 enhances the functional interaction between SRC-3 and CBP/p300 in mammalian two-hybrid assays while knocking down Pin1 levels decreases this interaction (Fig. 5A and B). In addition, Pin1 also increases SRC-3 and CBP/p300 interaction in an in vitro-purified system (Fig. 5C), supporting the idea that Pin1 may have a direct effect on SRC-3. We also observed decreased recruitment of p300 to the estrogen-receptor-targeted promoter in the presence of Pin1 siRNA in a ChIP assay (Fig. 5E), explaining in part the down-regulation of pS2 transcription by Pin1 depletion. To exclude the possibility that reduced p300 recruitment to the pS2 promoter is due to reduced p300 protein levels in the presence of Pin1 siRNA, we also measured the p300 protein level in both Pin1 knocked-down MCF-7 cells and MEF Pin1−/− cells. We found that p300 level, similar to SRC-3 level, actually was increased in both cells (data not shown). Since p300 forms a complex with SRC-3, this suggests that Pin1 is able to regulate the complex formation and likely the turnover. It also indicates that the reduced p300 recruitment is caused not by decreased p300 protein level but probably by a decreased interaction between SRC-3 and p300.
We found that Pin1 interacts with several regions in SRC-3, using both in vitro GST pulldown and in vivo mammalian two-hybrid analyses (Fig. 2), and the interaction appears to be phosphospecific because phosphatase treatment of SRC-3 results in the loss of Pin1 binding (Fig. 1B). We conclude from our studies that Pin1 may interact with multiple phosphorylated Ser/Thr-Pro motifs in SRC-3. This idea has precedent as Pin1 interacts with the 7-amino-acid repeats that constitute the CTD of RNA polymerase II. In this case, phosphorylation of Ser 2 and Ser 5 of a single motif provides the highest Pin1 binding avidity and each repeat is capable of binding Pin1 (53). Although not formally proven, it is believed that Pin1 binding alters the conformation of the CTD. The mitotic phosphatase cdc25c also contains a serine/proline-rich region whose conformation is changed upon Pin1 binding (38) and, depending on the kinase that phosphorylates cdc25c, Pin1 can alter the activity of cdc25c in different ways (38). c-Myc is also a Pin1 binding protein, and Pin1 binds to a form of c-Myc that is doubly phosphorylated on Thr 58 and Ser 62 (54). In this case, Pin1 binding increases the activity of c-Myc as a transcription factor but also facilitates the dephosphorylation of Ser 62 by PP2A, which is required for the ubiquitination and degradation of c-Myc. Thus, Pin1 apparently positively influences the activity and turnover of c-Myc.
Our data reveal that Pin1 also has multiple effects on SRC-3. On one hand, Pin1 promotes the interaction between SRC-3 and CBP/p300 and activates SRC-3-mediated steroid receptor transactivation. On the other hand, it enhances the turnover of SRC-3. Given the multiple sites of interaction between Pin1 and SRC-3, we suggest that Pin1 could regulate different aspects of SRC-3 functions by interacting with different sites. However, based on our data, we cannot exclude the possibility that Pin1 affects SRC-3 protein stability indirectly.
The question arises as to how the effects of Pin1 on both SRC-3 turnover and its interaction with p300 contribute to overall transcriptional activation. While promotion of the SRC-3 and CBP/p300 interaction no doubt will enhance transcription, ample evidence indicates that protein turnover may also directly contribute to transcriptional activation. Protein turnover has been shown to be tightly coupled to its transcriptional potential (25, 33). Cyclic turnover of ER was shown to be inherently linked to transcription (25, 33, 37). During the course of transcriptional regulation, coactivators are ubiquitinylated and subjected to turnover (25, 33, 37). Proteasome subunits have been found to be associated with actively transcribed chromatin. Similarly, retinoic acid induces increased p300 HAT activity while it destabilizes the protein (6). Given that Pin1 has multiple binding sites in SRC-3, it may regulate different aspects of SRC-3 function simultaneously. Transcription is a transient and dynamic process, and transcriptional products are accumulative while the SRC-3 protein level is an end point readout. We suggest that Pin1 binds to phosphorylated SRC-3 and induces a conformational change, protein-protein interactions, and transcriptional activation. Subsequently, Pin1 also stimulates SRC-3 turnover and facilitates the cyclic recruitment of nascent phosphorylated SRC-3 to the promoter. A generally similar process has been reported for c-Myc (54). We cannot rule out an alternative explanation that the stimulation of SRC-3 and p300 interaction is so much more potent that the activity is sufficient to overcome any short-term decrease in SRC-3 protein levels.
It is clear that phosphorylation of SRC-3 regulates its transcriptional coactivation function (12, 49). We showed recently that different cellular signals can induce different combinations of phosphorylation sites in SRC-3, which in turn differentially regulate the function of SRC-3 in coactivating transcription factors downstream of diverse signaling pathways (49). Evidence suggests that phosphorylated SRC-3 needs to be either dephosphorylated or degraded so that SRC-3 can properly function when new signals arise. We have observed that SRC-3 phosphomutants can be neither activated nor degraded and that activation of this oncogene is closely coupled with its degradation. Importantly, we observed herein that Pin1 is ineffective in regulating the turnover of phosphomutant SRC-3. Thus, it is entirely logical that Pin1 binds to the phosphorylated form of SRC-3, enhancing its function initially, and then continues to function in promoting its subsequent degradation. Based on this, we proposed a model on how Pin1 regulates SRC-3 function (Fig. 7). By this mechanism, potent coactivators such as SRC-3 can quickly respond to changes in environmental signals but activated phosphorylated SRC-3/AIB1 does not survive sufficiently long in cells to promote the unwanted enhancement of oncogenic functions. The role of Pin1 in this series of reactions emphasizes the critical requirements for precise conformational interactions among coactivators in NR-mediated transcriptional regulation. Pin1 serves as an important secondary coactivator for steroid receptors that functions in the postphosphorylation regulation of primary coactivators to achieve these requisite functional conformations.
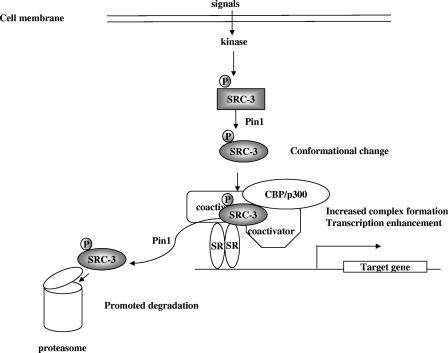
FIG. 7.
Model for Pin1 regulation on SRC-3 function. Extracellular signals induce the phosphorylation of SRC-3. Pin1 serves as a secondary coactivator by binding to the phosphorylated SRC-3 and inducing its conformational change. This conformational change increases the interaction between SRC-3 and CBP/p300, thereby enhancing steroid receptor (SR)-mediated transcription. Ultimately, Pin1 promotes the degradation of the phosphorylated SRC-3.
Acknowledgments
We thank Barbara Sanborn, Tse Hua Tan, Michael Greenberg, Carolyn Smith, and Hongwu Chen for kindly providing the PKA catalytic subunit, c-Jun, c-Fos, CREB, VP16-CBP, and SRC-3AAA expression vectors.
This work is supported by NIH grants (HD08818 and HD07857) (to B.W.O.), NIDDK NURSA program (to B.W.O.), CA grant (CA082845) (to A.R.M.), and a postdoctoral fellowship from the Department of Defense Breast Cancer Research Program (W81XWH-04-1-0552) (to P.Y.).
REFERENCES
- 1.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. [DOI] [PubMed] [Google Scholar]
- 2.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 3.Arimura, A., M. van Peer, A. J. Schroder, and P. B. Rothman. 2004. The transcriptional co-activator p/CIP (NCoA-3) is up-regulated by STAT6 and serves as a positive regulator of transcriptional activation by STAT6. J. Biol. Chem. 279:31105-31112. [DOI] [PubMed] [Google Scholar]
- 4.Atchison, F. W., B. Capel, and A. R. Means. 2003. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development 130:3579-3586. [DOI] [PubMed] [Google Scholar]
- 5.Atchison, F. W., and A. R. Means. 2003. Spermatogonial depletion in adult Pin1-deficient mice. Biol. Reprod. 69:1989-1997. [DOI] [PubMed] [Google Scholar]
- 6.Brouillard, F., and C. E. Cremisi. 2003. Concomitant increase of histone acetyltransferase activity and degradation of p300 during retinoic acid-induced differentiation of F9 cells. J. Biol. Chem. 278:39509-39516. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 9.Crenshaw, D. G., J. Yang, A. R. Means, and S. Kornbluth. 1998. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 17:1315-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolinski, K., S. Muir, M. Cardenas, and J. Heitman. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Font de Mora, J., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giraud, S., F. Bienvenu, S. Avril, H. Gascan, D. M. Heery, and O. Coqueret. 2002. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J. Biol. Chem. 277:8004-8011. [DOI] [PubMed] [Google Scholar]
- 14.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, D. M., K. Saxena, M. Vogtherr, P. Bernado, M. Pons, and K. M. Fiebig. 2003. Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J. Biol. Chem. 278:26174-26182. [DOI] [PubMed] [Google Scholar]
- 16.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. J., J. H. Kim, and J. W. Lee. 1998. Steroid receptor coactivator-1 interacts with serum response factor and coactivates serum response element-mediated transactivations. J. Biol. Chem. 273:28564-28567. [DOI] [PubMed] [Google Scholar]
- 18.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 19.Kuang, S. Q., L. Liao, H. Zhang, A. V. Lee, B. W. O'Malley, and J. Xu. 2004. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 64:1875-1885. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. K., H. J. Kim, J. W. Kim, and J. W. Lee. 1999. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol. Endocrinol. 13:1924-1933. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou, Y. C., A. Ryo, H. K. Huang, P. J. Lu, R. Bronson, F. Fujimori, T. Uchida, T. Hunter, and K. P. Lu. 2002. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA 99:1335-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.List, H. J., K. J. Lauritsen, R. Reiter, C. Powers, A. Wellstein, and A. T. Riegel. 2001. Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. J. Biol. Chem. 276:23763-23768. [DOI] [PubMed] [Google Scholar]
- 25.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 26.Lonard, D. M., S. Y. Tsai, and B. W. O'Malley. 2004. Selective estrogen receptor modulators 4-hydroxytamoxifen and raloxifene impact the stability and function of SRC-1 and SRC-3 coactivator proteins. Mol. Cell. Biol. 24:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louie, M. C., J. X. Zou, A. Rabinovich, and H. W. Chen. 2004. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol. Cell. Biol. 24:5157-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, K. P., S. D. Hanes, and T. Hunter. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544-547. [DOI] [PubMed] [Google Scholar]
- 29.Lydon, J. P., F. J. DeMayo, C. R. Funk, S. K. Mani, A. R. Hughes, C. A. Montgomery, Jr., G. Shyamala, O. M. Conneely, and B. W. O'Malley. 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9:2266-2278. [DOI] [PubMed] [Google Scholar]
- 30.Myers, J. K., D. P. Morris, A. L. Greenleaf, and T. G. Oas. 2001. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry 40:8479-8486. [DOI] [PubMed] [Google Scholar]
- 31.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 32.Ranganathan, R., K. P. Lu, T. Hunter, and J. P. Noel. 1997. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89:875-886. [DOI] [PubMed] [Google Scholar]
- 33.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 34.Ryo, A., M. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3:793-801. [DOI] [PubMed] [Google Scholar]
- 35.Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y. C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K. P. Lu. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12:1413-1426. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, F. X. 2001. Prolyl isomerases. Adv. Protein Chem. 59:243-282. [DOI] [PubMed] [Google Scholar]
- 37.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol 3:15-23. [DOI] [PubMed] [Google Scholar]
- 38.Stukenberg, P. T., and M. W. Kirschner. 2001. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell 7:1071-1083. [DOI] [PubMed] [Google Scholar]
- 39.Suen, C. S., T. J. Berrodin, R. Mastroeni, B. J. Cheskis, C. R. Lyttle, and D. E. Frail. 1998. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J. Biol. Chem. 273:27645-27653. [DOI] [PubMed] [Google Scholar]
- 40.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 41.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 42.Torres-Arzayus, M. I., J. F. De Mora, J. Yuan, F. Vazquez, R. Bronson, M. Rue, W. R. Sellers, and M. Brown. 2004. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 6:263-274. [DOI] [PubMed] [Google Scholar]
- 43.Verdecia, M. A., M. E. Bowman, K. P. Lu, T. Hunter, and J. P. Noel. 2000. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 7:639-643. [DOI] [PubMed] [Google Scholar]
- 44.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X., Y. Yang, X. Guo, E. R. Sampson, C. L. Hsu, M. Y. Tsai, S. Yeh, G. Wu, Y. Guo, and C. Chang. 2002. Suppression of androgen receptor transactivation by Pyk2 via interaction and phosphorylation of the ARA55 coregulator. J. Biol. Chem. 277:15426-15431. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Z., D. W. Rose, O. Hermanson, F. Liu, T. Herman, W. Wu, D. Szeto, A. Gleiberman, A. Krones, K. Pratt, R. Rosenfeld, C. K. Glass, and M. G. Rosenfeld. 2000. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl. Acad. Sci. USA 97:13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler, K. E., K. I. Swenson, S. Kornbluth, and A. R. Means. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287:1644-1647. [DOI] [PubMed] [Google Scholar]
- 48.Wu, R.-C., J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2002. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by IκB kinase. Mol. Cell. Biol. 22:3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol. Cell 15:937-949. [DOI] [PubMed] [Google Scholar]
- 50.Wulf, G. M., Y. C. Liou, A. Ryo, S. W. Lee, and K. P. Lu. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277:47976-47979. [DOI] [PubMed] [Google Scholar]
- 51.Wulf, G. M., A. Ryo, G. G. Wulf, S. W. Lee, T. Niu, V. Petkova, and K. P. Lu. 2001. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20:3459-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, Y. X., Y. Hirose, X. Z. Zhou, K. P. Lu, and J. L. Manley. 2003. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 17:2765-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell. Biol. 6:308-318. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, Y., L. Liao, D. A. Tulis, and J. Xu. 2002. Steroid receptor coactivator-3 is required for inhibition of neointima formation by estrogen. Circulation 105:2653-2659. [DOI] [PubMed] [Google Scholar]
- 56.Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, F. Avolio, S. Volinia, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419:853-857. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, H., H. You, X. Z. Zhou, S. A. Murray, T. Uchida, G. Wulf, L. Gu, X. Tang, K. P. Lu, and Z. X. Xiao. 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419:849-853. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, G., Y. Hashimoto, I. Kwak, S. Y. Tsai, and M.-J. Tsai. 2003. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol. Cell. Biol. 23:7742-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, X. Z., O. Kops, A. Werner, P. J. Lu, M. Shen, G. Stoller, G. Kullertz, M. Stark, G. Fischer, and K. P. Lu. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 6:873-883. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, X. Z., P. J. Lu, G. Wulf, and K. P. Lu. 1999. Phosphorylation-dependent prolyl isomerization: a novel signaling regulatory mechanism. Cell. Mol. Life Sci. 56:788-806. [DOI] [PMC free article] [PubMed] [Google Scholar]