Abstract
Caspases play important roles in apoptotic cell death and in some other functions, such as cytokine maturation, inflammation, or differentiation. We show here that the 5′-flanking region of the human CASP-2 gene contains three functional response elements for sterol regulatory element binding proteins (SREBPs), proteins that mediate the transcriptional activation of genes involved in cholesterol, triacylglycerol, and fatty acid synthesis. Exposure of several human cell lines to statins, lipid-lowering drugs that drive SREBP proteolytic activation, induced the CASP-2 gene to an extent similar to that for known targets of SREBP proteins. Adenoviral vector-mediated transfer of active SREBP-2 also induced expression of the CASP-2 gene and the caspase-2 protein and increased the cholesterol and triacylglycerol cellular content. These rises in lipids were strongly impaired following small interfering RNA-mediated silencing of the CASP-2 gene. Taken together, our results identify the human CASP-2 gene as a member of the SREBP-responsive gene battery that senses lipid levels in cells and raise the possibility that caspase-2 participates in the control of cholesterol and triacylglycerol levels.
Caspases belong to a large family of serine-aspartate proteases, which are primarily involved in the proteolysis of various substrates, including essential cellular proteins and procaspases themselves (8, 12). During apoptosis induction by various stresses, such as exposure to anticancer drugs or UV irradiation, procaspases undergo sequential activation steps that ultimately lead to cell dismantling and death (46). Caspases are often classified as initiators or executioners of the apoptotic program, in correlation with the length of their N-terminal prodomain (24). Some caspases, like caspase-1, do not, however, play major roles in apoptosis but support important physiological functions, in this case maturation of pro-interleukin 1β into active interleukin 1β, or play important—although less clearly defined—roles in lymphocyte proliferation (23) and differentiation of various cell types (48) (for a review, see references 13 and 42).
One of the caspases involved in apoptosis is caspase-2 (Casp-2), whose activation can precede mitochondrial permeabilization in some proapoptotic paradigms (26, 35, 37), an effect that appears to involve processing of the zymogen but not always the associated catalytic activity (38). In addition, caspase-2 has recently been shown to participate in the inflammatory response upon recruitment in a protein complex with TRAF-2 and RIP1 and induction of NF-κB and p38 mitogen-activated protein kinase activation (25). Two main caspase-2 isoforms may be synthesized. Caspase-2L is proapoptotic (6, 50), whereas caspase-2S can oppose some of the apoptotic effects triggered by caspase-2L (7, 50). However, we have recently shown that the Casp-2S isoform encoding mRNA is subjected to nonsense-mediated decay in many human cell types (47). Therefore, most of the biological activity of caspase-2 is likely to depend on expression of the long caspase-2L isoform. We have recently reported the isolation and initial analysis of the human CASP-2 gene promoter activity (27). This promoter region allows mRNA initiation from separate domains of the two main caspase-2 transcripts. The CASP-2L promoter region is much more active than the downstream CASP-2S promoter, in good correlation with the respective levels of the transcripts found in the majority of cells and tissues (50).
A survey of putative transcription factor binding sites within the 5′-flanking region of the CASP-2 gene revealed the presence of several sites that can be recognized by sterol regulatory element binding proteins (SREBPs). These sites are found in the promoter region of genes involved in lipid homeostasis, encoding enzymes involved in the cholesterol or triacylglycerol/phospholipid synthesis pathways, such as 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase or farnesyl diphosphate synthase (FPP synthase) (11), and the fatty acid synthase (FAS) (28) or low-density lipoprotein receptor (LDLr) (51) (for a review, see references 3, 19, and 40). SREBP proteins are endoplasmic reticulum (ER)- and nuclear membrane-bound 125-kDa transcription factors with a basic helix-loop-helix leucine zipper structure, involved in the maintenance of lipid homeostasis (17, 33, 44). Three major SREBP isoforms are known in humans, mice, and hamsters: SREBP-1a and SREBP-1c are encoded by the same gene through the use of separate transcription start sites, whereas SREBP-2 is encoded by a distinct gene (22, 29). SREBP-1c, which harbors a short N-terminal acidic domain, is a much weaker transactivator than SREBP-1a and SREBP-2 (45). When sterol levels are low, i.e., in response to statins, HMG-CoA reductase inhibitors, the 125-kDa membrane-bound precursor form of SREBP is transported to the Golgi apparatus by a chaperone protein, the sterol cleavage activating protein, SCAP (2, 3, 21), which is both an escort protein and a sensor of sterol levels. Recently, two additional ER-bound proteins, Insig-1 and Insig-2, have been shown to control the transport of the SCAP-SREBP complex to the Golgi apparatus (for a review, see reference 36), where SREBPs are sequentially cleaved by the S1P and S2P proteases (3, 39), which leads to the release of the 68-kDa transcriptionally active N-terminal domain of the protein. This fragment dimerizes, migrates into the nucleus, helped by importin β (31, 32), and binds to sterol regulatory elements (SREs) or to E-box sequences to enhance transcription of target genes (3, 9). In addition to the sterol regulatory cleavage system, rapid degradation by the ubiquitin proteasome pathway (16) and a feed-forward regulation mediated by SREs present in the promoters of the SREBP-1a and SREBP-2 genes (1, 41) control SREBP activity in the nucleus. The three isoforms play partially different roles in lipid homeostasis. SREBP-1c and -1a are involved in fatty acid metabolism, and SREBP-1c appears to be a mediator of insulin/glucose signaling in lipogenesis (15), whereas SREBP-2 regulates both cholesterogenesis and lipogenesis in vitro and in vivo (34).
The identification of several putative DNA binding sites for SREBP proteins in the promoter region of the human CASP-2 gene prompted us to investigate the possible roles of these sites in the regulation of CASP-2 gene expression. Our results demonstrate that the CASP-2 gene is a member of the SREBP-responsive gene battery in human cells. Strikingly, suppression of CASP-2 gene expression strongly reduced the increase in cellular lipid levels brought about by overexpression of SREBP-2. Thus, besides its role in cell death, caspase-2 activity may be required as part of the adaptative response of human cells to variations in cholesterol and triacylglycerol levels. The implication of these findings may be twofold. First, they support and extend the notion that disturbance of lipid homeostasis may play an important role in the sensing of apoptosis stimuli. Second, they might suggest new venues of investigation to address the mechanism underlying some lipid disorders.
MATERIALS AND METHODS
Cell lines and culture.
Three human cell lines were used in this study: the HepG2 hepatoma cell line (American Type Culture Collection, Rockville, MD) as a representative liver cell line, strongly engaged in lipid synthesis, and the HeLa cervical carcinoma cell line and the HCT116 colon carcinoma cell line, representative of other cancer types (ECACC, Salisbury, United Kingdom). Cells were maintained in Eagle's minimum essential medium supplemented with 10% (vol/vol) fetal calf serum for HeLa and HCT116 cells and in Dulbecco minimum essential medium plus Ham F10 medium (1:1) in the presence of 5% (vol/vol) fetal calf serum for HepG2 cells. Both media were supplemented with 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml), and cells were cultured at 37°C in an atmosphere of 95% air and 5% CO2. All tissue culture compounds were purchased from BioWhittaker (Fontenay-sous-bois, France).
For lovastatin treatment (Sigma-Aldrich, Saint-Quentin Fallavier, France), all cell lines were preincubated in 5% lipoprotein-deficient serum (Sigma-Aldrich) during 24 to 36 h. Lovastatin was dissolved in dimethyl sulfoxide. The final concentration of dimethyl sulfoxide did not exceed 0.5% (vol/vol), a concentration that did not induce any toxicity.
Plasmid constructions.
Fragments from the promoter region of the human CASP-2 gene were inserted into the promoterless pGL3-basic vector (pGL3b; Promega, Charbonnières, France) or into the simian virus 40 (SV40) promoter-containing pGL3-promoter vector (pGL3 pm; Promega). Progressive deletion constructs of the CASP-2 gene 5′-flanking region were produced by PCR. Five-prime deletion constructs containing the CASP-2L transcription start site were inserted into pGL3b, and 3′ deletion constructs were inserted into pGL3 pm, under the control of the SV40 promoter. All plasmids were purified with the “Nucleobond ax” extraction kit (Macherey-Nagel, Hoerdt, France) and verified by sequencing (Genome Express, Meylan, France). Sequences of the primers used (Proligo, France) are available in supporting information (see Table S1 in the supplemental material). These primers also contain KpnI and XhoI or BglII restriction enzyme sites to facilitate subsequent cloning steps.
Plasmids encoding SREBP-1a and SREBP-2 transcriptionally active forms were a kind gift from T. F. Osborne (University of California, Irvine, CA). SREBP-1a and SREBP-2 expression was driven by the cytomegalovirus (CMV) promoter from the pCMV5 vector. The luciferase reporter vector for the proximal region of the LDL-receptor promoter (−171 to +57 inserted in pGL3b vector) was a kind gift from T. Huby (Inserm U551, Paris, France).
Site-directed mutagenesis.
Point mutations were introduced with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Briefly, 40 ng of parental vectors was used with the relevant oligonucleotides, and PCR was performed for 18 cycles. The mutations were detected by sequencing. PCR primer sequences are available in the supplemental material (see Table S2 in the supplemental material).
Adenoviral infections and plasmid and small interfering RNA (siRNA) transfections.
Overexpression of the mature form of SREBP-2 (nSREBP-2) or of a dominant-negative form of SREBP-2 (SREBP-2dn) (see Fig. S5 in the supplemental material) was obtained by infection with adenoviruses encoding the active transactivation domain (amino acids 1 to 475) or a truncated transactivation domain (amino acids 294 to 475) of human SREBP-2 fused to the green fluorescent protein (GFP) (Ad-S2n and Ad-S2dn, respectively). A GFP-only-containing adenovirus was used as a control (Ad-C). Cells were infected in serum-containing medium at a multiplicity of infection between 30 and 100 PFU/cell, depending on the experiment, and harvested after 28 h (HepG2) or 36 h (HeLa, HCT116).
Functional promoter activity of the 5′-flanking region of the CASP-2 gene was analyzed using pGL3 luciferase-reporter vectors. The pCMV β-galactosidase reporter vector (pCMV-βgal) was used as an internal control for normalization of transfection efficiencies. HepG2 cells were seeded into 24-well plates at the density of 200,000 cells/well 16 h before transfection by the Lipofectamine and lipoplus reagents (Invitrogen, Cergy-Pontoise, France) according to the manufacturer's instructions. One hundred nanograms of each test plasmid, 20 ng of SREBP expression vector, and 20 ng of pCMV-βgal reporter vector were used for each experiment. After 24 h, cells were harvested and analyzed for luciferase activity using a luciferase assay reagent (Promega) and a luminometer (Lumat LB 9507; EG&G Berthold) or using the β-galactosidase enzyme assay system (Promega) for β-galactosidase activity.
Sense and antisense SREBP-2 (sense-strand RNA sequence, 5′-CAACAGACGGUAAUGAUCAUU-3′) and caspase-2 (sense-strand RNA sequence, 5′-AAACAGCUGUUGUUGAGCGAA-3′) oligoribonucleotides correspond to positions 554 to 574 of the human SREBP-2 mRNA sequence (accession number U02031) and to positions 210 to 230 of the human caspase-2 mRNA sequence (accession number NM_032982). A scrambled caspase-2 siRNA was used as a control (sense-strand RNA sequence, 5′-UAUCAGCGUAAUGGCUAAGGA-3′). All oligonucleotides were purchased from Proligo. Sense and antisense oligoribonucleotides were annealed to generate the double-stranded siRNAs (SREBP-2, siS2; caspase-2, siC2; negative control, sineg) at the final concentration of 100 μM. Cells were plated in serum-containing medium (HeLa) or in serum-free medium (HCT116 and HepG2) without antibiotics the day before transfection and then transfected by adding 5 μl of oligofectamine (Invitrogen) to 5 μl of 20 μM siRNAs (final concentration, 100 nM). Cells were rinsed with medium 4 h after transfection and then maintained in culture for 2 days before analysis.
RT-PCR and quantitative real-time PCR (Q-PCR) analysis.
Total RNA was isolated with the nucleospin RNA extraction kit (Macherey-Nagel). Five hundred nanograms of RNA was reverse transcribed and then amplified in one step with the QIAGEN OneStep reverse transcriptase PCR (RT-PCR) kit (QIAGEN, Courtabeuf, France) according to the manufacturer's instructions. The identity of each PCR-generated product with its corresponding cDNA was confirmed by sequencing. Sequences of sense and antisense primers used to amplify human cDNAs (Proligo), hybridization temperatures, and number of cycles performed are available in the supplemental material (see Table S3 in the supplemental material).
For Q-PCR, five hundred nanograms of total RNA was reverse transcribed using Moloney murine leukemia virus RT (Promega) in the presence of oligo(dT) according to the manufacturer's instructions. Then, each sample was tested in duplicate and standardized to the β2-microglobulin mRNA. In brief, 0.5 μl cDNA, corresponding to about 5 ng, was used as a template with 1× buffer (Quantitect SYBR Green PCR master mix; QIAGEN) and 0.3 μM of each primer (see Table S3 in the supplemental material) in a total reaction volume of 25 μl. Analyses were performed on an ABprism 7700 detector system (Applied Biosystems, Les Ulis, France) with the sequence detector system software.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP-IT kit (Active Motif Europe, Rixensart, Belgium) according to the manufacturer's instructions. Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature to cross-link DNA with proteins, and then cells were sonicated (10 pulses of 20 s each). After incubation with protein G saturated with salmon sperm DNA, the supernatants were immunoprecipitated with 2 μg of N19 anti-SREBP-2 (SantaCruz/Tebu, Le Perrey en Yvelines, France) or 4 μg of the supplied negative control immunoglobulin G (IgG) antibodies at 4°C overnight. For PCR, 5 μl of eluted DNA was used. The primers used correspond to the S2, S3, and S5 sites of the CASP-2 gene and to the sterol regulatory element (SRE) site of the human FPP synthase gene as a positive control. PCR on material recovered for immunoprecipitation without antibody was also performed as a control. PCR products were visualized on 2% agarose gel containing ethidium bromide. Primers sequences, length of each PCR product, and number of cycles used are available in supporting information (see Table S4 in the supplemental material).
Western blotting and immunofluorescence studies.
For total extracts, subconfluent cells were harvested, washed twice in 1× phosphate-buffered saline (PBS), and lysed in boiling buffer (1% [wt/vol] sodium dodecyl sulfate, 1 mM Na3VO4, 10 mM Tris, pH 7.4) containing 0.1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 5 μg/ml leupeptin for 10 min at 4°C. Forty micrograms of proteins was boiled in Laemmli buffer for 5 min before separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were then electroblotted onto polyvinyl difluoride membranes (Bio-Rad, Ivry-sur-Seine, France), and nonspecific binding sites were blocked for 2 h at room temperature by 6% (wt/vol) fat-free milk before an overnight incubation at 4°C with specific antibodies: mouse anti-human procaspase-2 (1:1,000), rabbit anti-human procaspase-3 (1:1,000), and goat anti-human SREBP-2 (N19; 1:250). Anti-human HSC70 antibody (1:7,500; SantaCruz/Tebu) was used as a loading control. Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgGs (1:10,000; Jackson ImmunoResearch Laboratories, Villepinte, France) or rabbit anti-goat IgGs (1:5,000; Dako, Glosturp, Denmark). Blots were revealed using an Enhanced Chemioluminescence detection kit (Amersham, Les Ulis, France).
Note: for SREBP protein detection, cells were treated with 25 μg/ml ALLN (calpain I inhibitor; Roche, Meylan, France) for the last 3 h before harvesting, in order to stabilize the short-lived nuclear forms, as described previously (16).
For immunofluorescence analysis (see Fig. S7 in the supplemental material), HeLa cells were seeded in lab-Tek II chamber slides (15,000 cells/well). Cells were fixed in 2% paraformaldehyde for 10 min at 4°C, washed three times, blocked with 50 mM NH4Cl for 10 min, and permeabilized in PBS-0.1% Triton X-100 for 4 min. Cells were then incubated with primary mouse anti-human procaspase-2 monoclonal antibody (1:75) or mouse anti-human procaspase-3 polyclonal antibody (1:100), used as a negative control (Pharmingen, Pont de Claye, France) for 30 min in PBS, 1% bovine serum albumin. After washing, cells were incubated with 488-Alexa goat anti-mouse antibody (1:1,000) for 20 min and washed three times with PBS. Immunofluorescence was analyzed using a fluorescence microscope (Nikon, Champigny, France).
Nile Red and Oil Red O staining.
Lipid level analysis was performed by using two different in situ staining methods, which both show high specificity for neutral lipids. Oil Red O and Nile Red were from Sigma-Aldrich. Oil Red O was resuspended at 0.5% (wt/vol) in 98% propane-2-ol, and Nile Red was prepared at 1 mg/ml in acetone and stored at 4°C protected from light.
HCT116 or HeLa cells were seeded in 6-well or 24-well plates, transfected with siRNAs as described above, and infected 24 h later with Ad-control or Ad-S2n adenoviruses at 30 PFU/cell during another 36 h. For Nile Red staining, cells were incubated with 20 ng/ml Nile Red during 30 min at 37°C in the dark. Fluorescence measurements were then made with an EPICS ELITE Coulter cytometer with a 488-nm wavelength excitation from an argon-ion laser at 15 mW. The optical filters were set up to capture GFP emission at 525 nm and Nile Red at 610 nm, corresponding to total lipid detection (14). Analyses were performed on a list mode of 104 events gated in GFP-positive cells and analyzed with the Win MDI software. For Oil Red O staining, cells were fixed in 4% (vol/vol) formaldehyde during 15 min, washed in 1× PBS and in 60% propane-2-ol, and then stained with freshly diluted (60% stock solution in water) and filtered Oil Red O for another 15 min. Cells were rinsed in 60% propane-2-ol, and lipid droplets were detected by light microscopy.
Determination of total cholesterol and triacylglycerol content.
Cells were seeded in 25-cm2 flasks (250,000 or 500,000 cells/flask for HeLa or HCT116, respectively), transfected with siRNAs, and infected 24 h later with Ad-control or Ad-S2n adenoviruses at 30 PFU/cell during another 36 h. Cholesterol and triacylglycerol levels were determined enzymatically with the method of Danno (4), adapted as follows. Briefly, cells were washed in 1× PBS, resuspended in tert-butanol, and centrifuged for 10 min at 10,000 rpm. The same volume of methanol-1% Triton X-100 was added to the lipid phase. Total cholesterol and triacylglycerol content was then determined by enzymatic and colorimetric methods with the “cholesterol 100” and “triglycerides 25” kits (Abx Diagnostics, Montpellier, France) and read on a Victor2 1420 multilabel counter (Perkin-Elmer, Boston, MA).
Computer analysis of transcription binding sites.
All prediction analyses for the in silico identification of putative transcription factor binding sites were performed with the MatInspector software (Genomatix), which uses matrix determination to identify DNA binding sites.
Statistical analyses.
All results are presented as means (± standard deviations) from at least triplicate determinations, unless otherwise stated. Statistical analysis was performed using the two-sided Student t test.
RESULTS
Several SREBP putative binding sites localize to the human CASP-2 gene promoter region.
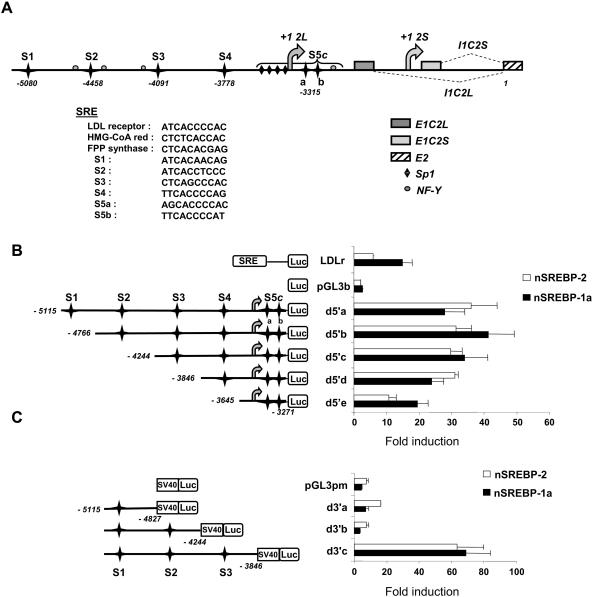
A survey of putative binding sites for transcription factors over a 5-kb region upstream from the first noncoding exon of caspase-2L revealed the presence of six putative SREBP binding sites. A schematic representation of the 5′ region of the CASP-2 gene and the location of the putative SREBP DNA binding sites is shown in Fig. 1A. All of these sites correspond to SREs, as identified in the genes coding for the LDLr, the HMG-CoA reductase, and the FPPs. Sites 2 (S2), 3 (S3), 5a (S5a), and 5b (S5b) are in close proximity with NF-Y and/or Sp1 DNA binding sites, a situation encountered in the SREBP binding regions from several responsive genes (30, 53). Most strikingly, S5a and S5b together with several Sp1 sites and one NF-Y site overlap within what we will refer to as the S5 complex (S5c). Such an organization is also found in the promoters of the mouse SREBP-1c (1), the human FAS (53), or the human HMG-CoA synthase (5) gene. These structural features prompted us to analyze further the role of the putative SREBP DNA binding sites in CASP-2 gene regulation.
FIG. 1.
5′-flanking region of the human CASP-2 gene. (A) Sequence positions are numbered relative to the major translation initiation site (position 1) located in the first coding exon (E2) common to both caspase-2 isoforms. Both transcription initiation sites (+1 2L and +1 2S) and the corresponding first introns (I1C2L and I1C2S) are indicated. Gray boxes indicate the position of the first exons, specific for each Casp-2 mRNA. The six SRE sites are represented by black stars. Putative Sp1 and NF-Y sites are also shown. S5c designates a complex composed of four Sp1 sites, two juxtaposed SREs (S5a and S5b), and one NF-Y site. The sequence of each site is indicated, together with those of sites found in the promoters of bona fide SREBP target genes. (B and C) Each CASP-2 gene promoter fragment was prepared as described in Materials and Methods. Five-prime deletions (from d5′a to d5′e) (B) were subcloned into the pGL3b vector and 3′ deletions (from d3′a to d3′c) (C) in the pGL3 pm vector. Each SRE site is identified (S1 to S5c). HepG2 cells were transfected with each reporter vector (or with LDLr, empty pGL3b, or pGL3 pm vectors as controls) and cotransfected with expression vectors for the SREBP-1a (nSREBP-1a) or SREBP-2 (nSREBP-2) mature form. Luciferase activity was measured 24 h later and normalized to β-galactosidase activity. The bars represent inductions relative to basal luciferase activity without SREBP cotransfection. The values are the means (± standard deviations) of at least three independent experiments.
SREBP proteins transactivate the human CASP-2 gene promoter.
In order to analyze the contribution of the SREs to the regulation of CASP-2 gene expression, a set of 5′ and 3′ deletions of the CASP-2 gene promoter region was prepared and their functional activity was assayed in liver-derived HepG2 cells. The deletion constructs, as well as the proximal region of the LDLr promoter as a positive control, were then used in cotransfection assays with plasmids encoding the mature, transcriptionally active form of the SREBP-1a or SREBP-2 proteins. Both induced the activity of the 5′-deletion constructs up to 50-fold. The shorter fragment (d5′e), which contained only the S5c sites, was induced by 10- to 20-fold, whereas the largest constructs showed stronger inductions (Fig. 1B).
As shown in Fig. 1C, the smallest 3′ deletion (d3′c) was very highly responsive to either SREBP protein. By contrast, d3′b was unresponsive, and the larger d3′a deletion was only weakly responsive, mostly to SREBP-2. This 3′-deletion analysis indicated that the putative SREBP S1 and S2 binding sites played little or no role in SREBP responsiveness, whereas the S3 site had a major impact on the response.
Mutations of the putative SREBP binding sites alter CASP-2 gene promoter activity.
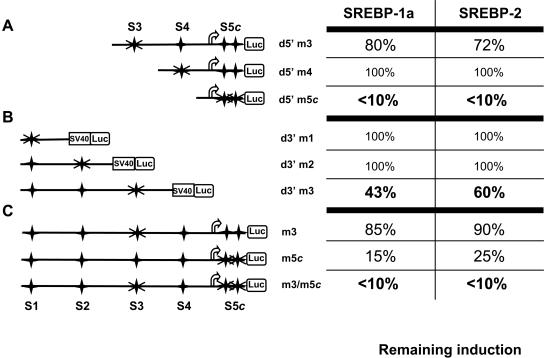
In order to analyze the functional activity of the putative SREBP binding sites, we prepared and tested mutants of each of these sites on 5′- and 3′-deletion constructs by cotransfection in HepG2 cells. As shown in Fig. 2A and B, mutation of the S4, S1, and S2 sites had no effect on SREBP transactivation by either SREBP isoform, in agreement with the results from 3′-deletion analyses (Fig. 1C). However, the transactivating potential of both SREBP isoforms was similarly influenced by mutations of the S3 and S5 sites. S3 site mutation reduced transactivation by SREBP-1a and -2 by 20 to 28% in the context of the CASP-2 gene promoter and by up to 57% in the context of the SV40 promoter. Mutation of both sites of the S5 complex abolished transactivation (Fig. 2A). To confirm the major implication of the S3 and S5c sites, we tested the effect of mutations of either site in the context of the largest CASP-2 gene promoter fragment. As shown in Fig. 2C, S3 site mutation slightly reduced transactivation by either SREBP-1a or -2, whereas mutations of S5c reduced it by up to 85%. Finally, mutations of both the S3 and S5c sites (m3/m5c) totally abolished transactivation by each SREBP. Similar results were obtained in HCT116 cells (data not shown). Taken together, these results demonstrate that most of the transcriptional activity of the CASP-2 gene observed upon overexpression of SREBP proteins may depend on the identified binding sites for these proteins, the S3 and especially the S5c sites.
FIG. 2.
S3 and S5c sites are the most responsive to SREBP proteins in HepG2 cells. The caspase-2 promoter 5′ (A) or 3′ (B) deletion constructs or the largest promoter (C) was mutated on each SRE site (as indicated by crosses) and tested by cotransfection with both SREBP-1a and SREBP-2 mature forms. Cells were then cultured for 24 h. Luciferase activity was measured and normalized to β-galactosidase activity. The table on the right shows the levels of remaining induction by either SREBP protein relative to the luciferase activity obtained for each wild-type fragment (100% indicates no change). The values were compiled from two to four independent experiments. The variation between experiments did not exceed 13%. d5′m3, S3 site mutation in context of CASP-2 gene promoter; d5′m4, S4 site mutation; d5′m5c, mutation of both sides of S5 complex; d3′m1, S1 site mutation; d3′m2, S2 site mutation; d3′m3, S3 site mutation in context of SV40 promoter; m3, S3 site mutation in context of largest CASP-2 gene promoter fragment; m3/m5c, mutations of both the S3 and S5c sites.
Cholesterol-lowering drugs increase CASP-2 gene expression.
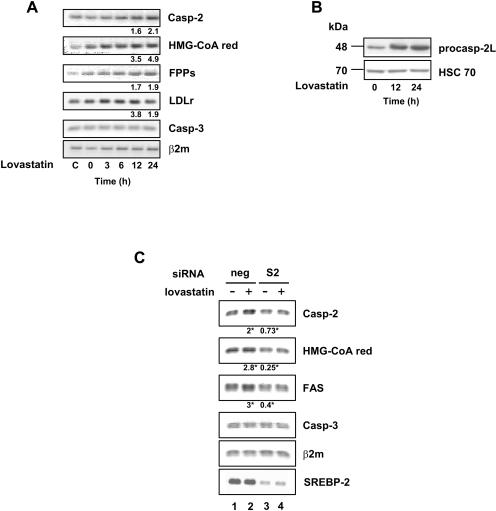
In view of the positive regulation of the CASP-2 gene promoter by SREBP proteins, we sought to determine whether the endogenous gene could be regulated via the lipid content of the cells. To this end, we looked at the effects of statins, HMG-CoA reductase inhibitors, on expression of target genes. Statins are known to trigger cholesterol depletion and to evoke a compensatory feedback mechanism that induces SREBP maturation and transactivation potential. We treated the human HepG2 hepatoma cell line, strongly engaged in lipid synthesis, with 50 μM lovastatin for 24 h to inhibit cholesterol synthesis. As shown in Fig. 3A, mRNA levels from bona fide SREBP-responsive genes, such as the HMG-CoA reductase, the FPPs, and the LDLr genes, were rapidly increased in response to lovastatin. Remarkably, the Casp-2 mRNA level was also increased to a comparable extent. As a control, the Casp-3 mRNA level remained unaffected. In addition, Western blot analysis from lovastatin-treated HepG2 cells showed a marked increase in the procaspase-2 protein level (Fig. 3B). Since statins are known to trigger SREBP-2 isoform maturation, rather than that of SREBP-1 (20, 43), we decided to focus on the putative role of SREBP-2 in the control of the endogenous CASP-2 gene. To this end, we looked at the effect of SREBP-2 gene silencing on the response of HepG2 cells to lovastatin. As shown in Fig. 3C, lovastatin treatment led to the induction of FAS, HMG-CoA red, and Casp-2 mRNAs by two- to threefold in cells transfected with control siRNA, whereas this effect was not observed in cells transfected with SREBP-2 siRNA (lane 4 versus lane 2), indicating that lovastatin-mediated induction of target genes was dependent on SREBP-2. Furthermore, SREBP-2 silencing also triggered a drop in Casp-2 (27%), HMG-CoA-Red (75%), and FAS (60%) mRNA levels compared to those of control cells (lane 3 versus lane 1), suggesting that SREBP-2 may also control basal transcription of these target genes. The Casp-3 mRNA level remained unchanged. These results strongly suggest that SREBP-2 mediates the positive effect of statins on the Casp-2 mRNA level.
FIG. 3.
Lovastatin induces caspase 2 mRNA and protein. HepG2 cells were incubated for 36 h in medium containing 5% lipoprotein-deficient serum before treatment with 50 μM lovastatin during the indicated times (C refers to cells with normal serum). (A) Total RNA was extracted, and expression levels were analyzed by RT-PCR and quantified for the 12-h and 24-h time points by Q-PCR. Beta-2 microglobulin mRNA (β2m) was used as a loading control. The numbers show the n-fold induction from one typical experiment among two with identical results. The variation between experiments did not exceed 10%. (B) Expression of procaspase-2L was examined by Western blotting. (C) Prior to 24-h lovastatin treatment, HepG2 cells were transfected with control siRNA (neg) or SREBP-2 siRNA (S2). Total RNA was used for RT-PCR and for Q-PCR. Numbers indicate induction factors relative to control cells (lane 1) and represent the means from three independent experiments (*, statistical significance was reached in each case; Student's t test, P < 0.05).
SREBP-2 controls endogenous CASP-2 gene expression.
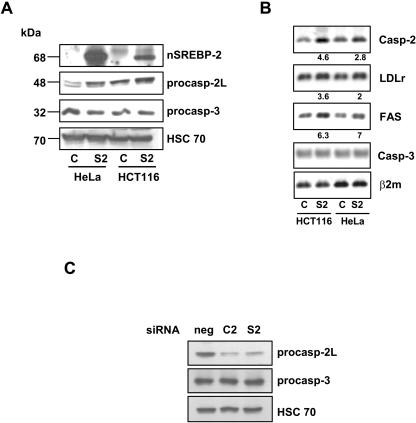
In order to confirm the role of SREBP-2 in the control of CASP-2 gene expression, we looked at the effect of SREBP-2 mature form overexpression. HCT116 and HeLa cells were infected by an adenovirus encoding the transactivation domain of SREBP-2 fused to GFP (S2) or with a GFP-only-containing virus as a control. Both nSREBP-2 and procaspase-2L, but not procaspase-3, protein levels were increased in S2-infected cells (Fig. 4A). As shown in Fig. 4B, overexpression of nSREBP-2 allowed a strong increase of FAS mRNA and a 2- to 4.6-fold increase of LDLr and Casp-2 mRNAs, with no effect on the caspase-3 mRNA level. Furthermore, the silencing of SREBP-2 by a siRNA also triggered a drop in the procaspase- 2 protein level (Fig. 4C). Finally, infection of HCT116 cells with a dominant-negative form of SREBP-2 (S2dn) led to a slight but consistent decrease of the Casp-2, LDLr, and FAS mRNA levels (see Fig. S5 in the supplemental material). These results strongly indicate that SREBP-2 controls the basal expression of the CASP-2 gene.
FIG. 4.
nSREBP-2 induces the CASP-2 gene. HCT116 and HeLa cells were infected by 100 PFU/cell of Ad-control (C) or Ad-nS2 (S2) adenoviruses. The average efficiency of infection (GFP-positive cells) was close to 75%. (A) The levels of nSREBP-2 and procaspase-2 and -3 protein species were analyzed by Western blotting following infection by each adenovirus. HSC70 was used as a loading control. One representative experiment from two independent experiments with identical results is shown. (B) Total RNA was used for RT-PCR and for Q-PCR (numbers indicate factors of induction by Ad-nS2 versus those by Ad-control). The results from one representative experiment among two with similar results is shown. The variation between experiments did not exceed 7.5%. (C) HCT116 cells were transfected with control siRNA (sineg [neg]), siC2 (C2), or siS2 (S2), and total protein extracts were used in Western blotting experiments to analyze expression of procaspases-2 and -3 and HSC70 as a loading control.
SREBP-2 binds to the S3 and S5 sites of the CASP-2 gene.
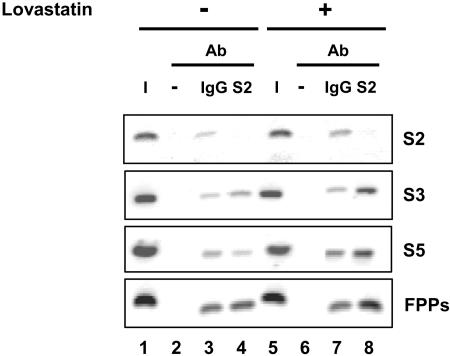
In order to obtain direct evidence for the in vivo binding of the SREBP-2 protein to its cognate response elements from the endogenous CASP-2 gene, we used ChIP assays with HepG2 chromatin extracts. As shown in Fig. 5 (lane 4), in HepG2 naive cells, SREBP-2 bound the S3 and S5 sites, albeit less strongly than to the FPP synthase response element. Such a difference was not observed in another cell line much less prone to lipid synthesis (HCT116 cells) (see Fig. S5, lane 4, in the supplemental material). Following lovastatin treatment, however, binding of SREBP-2 on the S3 and especially the S5 sites was strongly increased (S3 and S5, lane 8 versus lane 4), similarly to the FPP site, but not on the S2 site used as a negative control. The “no antibody” condition did not give any signal. Similar results after lovastatin treatment were obtained in HCT116 cells (see Fig. S6, lane 8 versus lane 4, in the supplemental material).
FIG. 5.
SREBP-2 binds to the S3 and S5 sites of the caspase-2 promoter. Chromatin was prepared from untreated HepG2 cells (−) or cells treated with 50 μM lovastatin (+) for 36 h. Cells were incubated with ALLN for the last 3 h before harvesting to stabilize the nuclear SREBP-2 form. Each cross-linked chromatin extract was incubated with anti-SREBP-2 antibody (S2), with negative control IgG (IgG), or without antibody (−). DNA extracted from each immunoprecipitate was analyzed by PCR using the relevant primers (see Materials and Methods; also see Table S4 in the supplemental material) to test the binding of SREBP-2 to the S2, S3, and S5 sites from the CASP-2 gene and on the SRE-binding sequences of the FPP synthase gene as a positive control. The input chromatin (I) corresponds to a 1:100 dilution of total DNA extracted prior to immunoprecipitation. One representative experiment of two independent experiments with similar results is shown. Optimization of PCR amplification parameters (PCR cycle numbers and input DNA amount) was performed in pilot experiments.
Inhibition of CASP-2 gene expression reduces cholesterol and triacylglycerol levels.
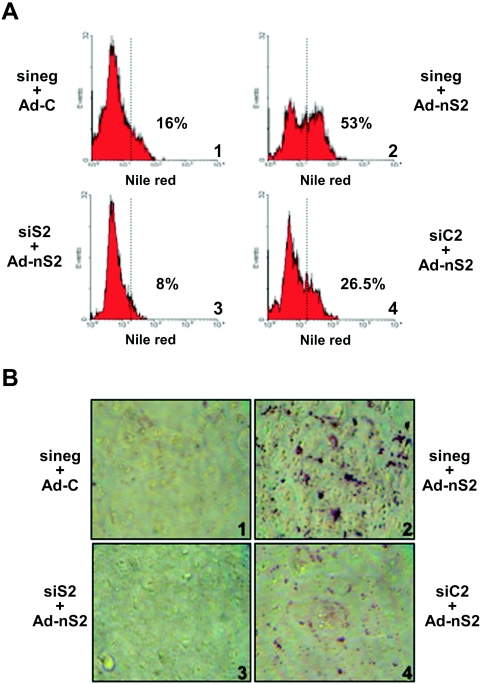
In view of the positive response of the CASP-2 gene to SREBP-2 and to lipid-lowering drugs, we looked at the effect of CASP-2 gene silencing on lipid levels in HCT116 cells. As shown in Fig. S7A and B in the supplemental material, transfected double-stranded Casp-2 siRNAs (26) led to a strong decrease of caspase-2, but not caspase-3, immunostaining and levels in protein extracts. To determine the impact of SREBP overexpression on lipid levels in the cell, we first determined, by flow cytometry analysis, the lipid content of Nile Red-stained HCT116 cells infected with the SREBP-2 mature form-encoding adenovirus. As shown in Fig. 6A, SREBP-2 overexpression resulted in a large increase in lipid levels (panel 2). As expected, a siRNA against SREBP-2 fully prevented the lipid increase and even suppressed basal levels from 16% to 8% positive cells (panel 3 versus panel 1). Strikingly, a siRNA against Casp-2 reduced the SREBP-2-mediated increase in lipid levels by 50% (from 53% to 26.5%; panel 4 versus panel 2). To confirm these results, we used Oil Red O staining, which labels all neutral lipids (Fig. 6B). This analysis showed a strong color increase in SREBP-2-transduced HCT116 cells (panel 2 versus panel 1), an effect which was fully prevented in the presence of a SREBP-2 siRNA (panel 3) and partially, but significantly, decreased in the presence of Casp-2 siRNA (panel 4). Finally, the SREBP-2-induced increase in cholesterol (2.3-fold) and triacylglycerol (5-fold) cellular levels was almost fully prevented by SREBP-2 siRNA and was reduced by 30% to 50% with Casp-2 siRNA (Table 1). Western blot analysis confirmed that SREBP-2 siRNA was indeed able to reduce SREBP-2 protein levels (see Fig. S7C in the supplemental material). All of these observations were also made with HeLa cells (data not shown). Taken together, these results strongly suggest that caspase 2 might participate in SREBP-2-driven lipid synthesis and/or accumulation in human cells.
FIG. 6.
Casp-2 silencing reduces cellular lipid content. HCT116 cells were transfected with specific siRNAs (siS2 or siC2) or with a control siRNA (sineg) and infected 24 h later by Ad-C or Ad-nS2 adenoviruses (30 PFU/cell) for 36 h. (A) Total lipids of HCT116 cells were stained by Nile Red and detected by flow cytometry. The dotted vertical line sets the fluorescence limits obtained in absence of Nile Red (the percentage of positive cells is indicated in each case). The results from one representative experiment of three independent experiments with similar results are shown. The variation between experiments did not exceed 5%. (B) Neutral lipids were labeled by Oil Red O staining. The results from one representative experiment of three independent experiments with similar results are shown.
TABLE 1.
Casp-2 silencing reduces cholesterol and triacylglycerol levelsa
| Treatment | Total cholesterol (mmol/106 cells) | i.f.e | Triacylglycerols (μmol/106 cells) | i.f.e |
|---|---|---|---|---|
| sineg + Ad-C | 0.76 ± 0.17 | 52 ± 26 | ||
| sineg + Ad-nS2 | 1.72 ± 0.63b | 2.26 | 259 ± 120b | 4.98 |
| siS2 + Ad-nS2 | 0.89 ± 0.16c | 1.17 | 44 ± 9c | 0.84 |
| siC2 + Ad-nS2 | 1.18 ± 0.35c | 1.55 | 125 ± 34d | 2.41 |
HeLa cells were transfected with specific siRNAs (siS2 or siC2) or with a control siRNA (sineg) and infected 24 h later by Ad-C or Ad-nS2 adenoviruses (30 PFU/cell) for 36 h before measuring total cholesterol and triacylglycerol contents by colorimetric assays as described in Materials and Methods. The values are the means (± standard deviations) for five independent experiments.
P < 0.01; significance level compared to result for sineg + Ad-C (Student's t test).
P < 0.05; significance level compared to result for sineg + Ad-nS2 (Student's t test).
P < 0.01; significance level compared to result for sineg + Ad-nS2 (Student's t test).
i.f., induction factor versus control (sineg + Ad-C).
DISCUSSION
The results from the present study allowed us to identify, and to functionally characterize, the transactivation potential of SREBP proteins for the CASP-2 gene promoter region. We identified six potential SRE binding sites, including one SRE-containing complex similar to that found in the promoter of bona fide SREBP-responsive genes (1, 5, 53). Transient transfection and mutational analysis demonstrated that the CASP-2 gene promoter is fully responsive to the SREBP-1a and -2 proteins and that the S3 and S5c sites are responsible for most of the activating potential of SREBPs. The evidence that SREBP proteins also regulate the endogenous CASP-2 gene is severalfold. First, treatment of HepG2 liver cells, which synthesize large amounts of lipids, with the cholesterol-lowering drug lovastatin, known to induce maturation of the SREBP-2 isoform, led to induction of the CASP-2 gene to a similar extent to that observed for several SREBP-responsive genes. Remarkably, the same effects were found in other cell lines, including HEK 293, HeLa, and HCT116, as well as with another statin, mevastatin, in all cell lines (data not shown). Second, ChIP analysis demonstrated SREBP-2 binding to the S3 and S5c sites, an effect that was strongly increased in response to statins. Third, the use of an adenoviral vector encoding the mature form of SREBP-2 allowed stimulation of CASP-2 gene expression to the same extent as, or even more than, that of reference genes. Fourth, siRNA-mediated silencing of the SREBP-2 gene decreased expression of the CASP-2 gene, together with that of other SREBP-2-dependent genes. Therefore, CASP-2 is a true SREBP-2 target gene.
In view of these findings, we wished to address the possibility that the controlled expression of the CASP-2 gene could be linked to a novel function of caspase-2, i.e., the control of lipid levels in cells. As expected, siRNA-mediated silencing of SREBP-2 abrogated the rise in lipids triggered by SREBP-2 overexpression. Much strikingly, however, this rise in cellular lipid levels was also strongly dampened by Casp-2 mRNA silencing. This observation strongly suggests that caspase-2 may participate in SREBP-2-driven lipid synthesis and/or accumulation in human cells.
How caspase-2 is connected to lipid biosynthesis pathways in human cells is unclear at this stage. Overexpression of procaspase-2L did not trigger any rise in lipids, nor did it lead to an improved response to SREBP-2 overexpression, which suggests an additional requirement of some limiting factor for caspase-2 activity, a situation that may not be symmetrical with caspase-2 silencing (data not shown). Indeed, caspase-2 function has been shown to depend on its recruitment within macromolecular protein complexes, such as the so-called PIDDosome, in which protein partner stoichiometry might be tightly controlled (49). In addition, use of the VDVAD-fmk permeant caspase-2 inhibitor did not block the increased lipid levels induced by SREBP-2 in any of the cell lines (data not shown), which indicates that caspase-2 enzymatic activity could be dispensable. It was in fact recently shown that caspase-2 activity might not be required to fulfill its role as an apoptotic initiator caspase (10, 38) or its role in inflammation (25). If indeed caspase-2 catalytic activity is dispensable for enhancing the activation of lipid synthesis by SREBP-2, presumably caspase-2 function may be largely related to its ability to interact with other cellular macromolecules, either proteins or lipids (10). Alternatively, since the activity of caspase inhibitors in live cells may not be fully quantitative, their lack of effects in some experimental paradigms might not be a fully reliable indicator of caspase activity and therefore of caspase function.
As an initial attempt to try to understand how caspase-2 may be able to exert some control on lipid synthesis and/or accumulation, we looked at expression of SREBP-2 target genes following siRNA-mediated CASP-2 gene silencing. In all cases surveyed, i.e., for the HMG-CoA reductase, the LDLr, and the FAS genes, a slight but consistent 17 to 30% reduction in the stimulation brought about by SREBP-2 overexpression was observed (see Fig. S8 in the supplemental material). Such a pleiotropic effect might be best explained by an activity of caspase-2 on a regulatory step common to expression of all these genes. One immediate possibility might be that physiological levels of caspase-2 would be able to interfere with SREBP-2 function. For instance, enzymatically active caspase-2 could degrade SREBP-2 into a cleavage product that would drive better transcriptional activation. However, upon use of recombinant active caspase-2, we obtained no evidence that caspase-2 could cleave SREBP-2, whereas caspase-3 was able to do so (see Fig. S9 in the supplemental material), as previously reported (52). Alternatively, caspase-2 could degrade inhibitory transcription factors, thus activating cholesterol and/or triacylglycerol synthesis. One candidate protein is the ATF6 transcription factor, an ER stress-inducible protein that undergoes proteolysis by the same S1P and S2P proteases as SREBPs (54) and that has been shown recently to antagonize SREBP-2 activity upon direct interaction. Indeed, the nuclear form of ATF6 is able to form a complex with the SRE-bound nSREBP-2, which leads to a strong impairment of SREBP-2 transcriptional activity (55). A schematic model that summarizes our observations is presented in Fig. 7.
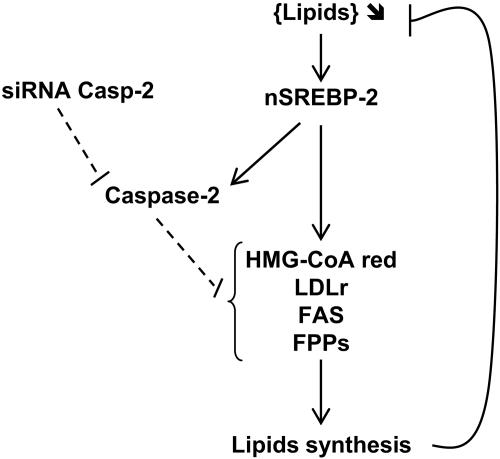
FIG. 7.
Hypothetical model for implication of caspase-2 in lipid biosynthesis pathway. Under depletion of lipids, the nuclear active form of SREBP-2 (nSREBP-2) is released to activate target genes involved in cholesterol and/or triacylglycerol synthesis pathways in order to restore lipid contents in cells. CASP-2 is also induced by nSREBP-2. Since Casp-2 siRNA reduces lipidogenic gene expression induced by nSREBP-2 as well as lipid synthesis, we propose that caspase-2 is required, although not sufficient by itself, to account for roughly 50% of the SREBP-mediated effects on cellular lipid anabolism.
In order to analyze CASP-2 gene regulation in live animals, we turned to mice. However, we failed to observe any effect of cholesterol and statin treatments on Casp-2 mRNA level in mouse liver (L. Corcos, A. Roth, and U. Meyer, unpublished observations). In addition, microarray analysis of liver RNA from SREBP-overexpressing transgenic mice or from mice deficient in the SREBP activation pathway did not detect any effect of SREBPs on Casp-2 mRNA (18). In fact, the 5′-flanking region of the mouse Casp-2 gene is strikingly different from the human sequence, with many fewer putative SREBP binding sites and a distinct organization. Because of such fundamental differences in the structure of the CASP-2 gene between humans and mice, mice might just not be appropriate for evaluating the influence of caspase-2 on cholesterol and triacylglycerol levels. A contribution of caspase-2 to lipid anabolism might thus be better searched for directly in humans.
In conclusion, in addition to its role in cell death and to the recently identified role in inflammation (25), caspase-2 appears to be a positive regulator of cholesterol and triacylglycerol homeostasis in human cells, which adds to the growing list of nonapoptotic functions of caspases (13). It is tempting to speculate that any condition that would dampen sterol levels could activate SREBP binding sites from the CASP-2 gene, which would then be turned on to provide either a direct trigger or an amplification loop aimed at restoring normal cholesterol or triacylglycerol levels. Depending on the activity of SREBP proteins, caspase-2 could thus act as a second-order sensor for satisfying the lipid demand.
Supplementary Material
Acknowledgments
We thank A. Roth and U. Meyer for the in vivo mice experiment; T. Huby for his gift of the LDLr promoter construct; T. Osborne for his gift of SREBP expression plasmids; A. Bettaieb for his encouragements and suggestions; P. Grimaldi, G. Lizard, and L. Lagrost for their critical reading and suggestions; and F. Ménétrier for his help with lipid staining.
E. Logette was a recipient of a fellowship from the Ministry of Research and Education. S. Solier was appointed by the University Hospital, Dijon. This work was supported by the Ligue Nationale Française contre le Cancer, the Conseil Général de Bourgogne, and the INSERM.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amemiya-Kudo, M., H. Shimano, T. Yoshikawa, N. Yahagi, A. H. Hasty, H. Okazaki, Y. Tamura, F. Shionoiri, Y. Iizuka, K. Ohashi, J. Osuga, K. Harada, T. Gotoda, R. Sato, S. Kimura, S. Ishibashi, and N. Yamada. 2000. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 275:31078-31085. [DOI] [PubMed] [Google Scholar]
- 2.Brown, A. J., L. Sun, J. D. Feramisco, M. S. Brown, and J. L. Goldstein. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10:237-245. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 4.Danno, H., Y. Jincho, S. Budiyanto, Y. Furukawa, and S. Kimura. 1992. A simple enzymatic quantitative analysis of triglycerides in tissues. J. Nutr. Sci. Vitaminol. (Tokyo) 38:517-521. [DOI] [PubMed] [Google Scholar]
- 5.Dooley, K. A., S. Millinder, and T. F. Osborne. 1998. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 273:1349-1356. [DOI] [PubMed] [Google Scholar]
- 6.Droin, N., F. Bichat, C. Rebe, A. Wotawa, O. Sordet, A. Hammann, R. Bertrand, and E. Solary. 2001. Involvement of caspase-2 long isoform in Fas-mediated cell death of human leukemic cells. Blood 97:1835-1844. [DOI] [PubMed] [Google Scholar]
- 7.Droin, N., C. Rebe, F. Bichat, A. Hammann, R. Bertrand, and E. Solary. 2001. Modulation of apoptosis by procaspase-2 short isoform: selective inhibition of chromatin condensation, apoptotic body formation and phosphatidylserine externalization. Oncogene 20:260-269. [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, P. A., D. Tabor, H. R. Kast, and A. Venkateswaran. 2000. Regulation of gene expression by SREBP and SCAP. Biochim. Biophys. Acta 1529:103-113. [DOI] [PubMed] [Google Scholar]
- 10.Enoksson, M., J. D. Robertson, V. Gogvadze, P. Bu, A. Kropotov, B. Zhivotovsky, and S. Orrenius. 2004. Caspase-2 permeabilizes the outer mitochondrial membrane and disrupts the binding of cytochrome c to anionic phospholipids. J. Biol. Chem. 279:49575-49578. [DOI] [PubMed] [Google Scholar]
- 11.Ericsson, J., S. M. Jackson, B. C. Lee, and P. A. Edwards. 1996. Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc. Natl. Acad. Sci. USA 93:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, U., R. U. Janicke, and K. Schulze-Osthoff. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10:76-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido, C., and G. Kroemer. 2004. Life's smile, death's grin: vital functions of apoptosis-executing proteins. Curr. Opin. Cell Biol. 16:639-646. [DOI] [PubMed] [Google Scholar]
- 14.Greenspan, P., E. P. Mayer, and S. D. Fowler. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasty, A. H., H. Shimano, N. Yahagi, M. Amemiya-Kudo, S. Perrey, T. Yoshikawa, J. Osuga, H. Okazaki, Y. Tamura, Y. Iizuka, F. Shionoiri, K. Ohashi, K. Harada, T. Gotoda, R. Nagai, S. Ishibashi, and N. Yamada. 2000. Sterol regulatory element-binding protein-1 is regulated by glucose at the transcriptional level. J. Biol. Chem. 275:31069-31077. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, Y., M. Yoshida, M. Shimizu, and R. Sato. 2001. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 276:36431-36437. [DOI] [PubMed] [Google Scholar]
- 17.Horton, J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 109:1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton, J. D., N. A. Shah, J. A. Warrington, N. N. Anderson, S. W. Park, M. S. Brown, and J. L. Goldstein. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 100:12027-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton, J. D., and I. Shimomura. 1999. Sterol regulatory element-binding proteins: activators of cholesterol and fatty acid biosynthesis. Curr. Opin. Lipidol. 10:143-150. [DOI] [PubMed] [Google Scholar]
- 20.Horton, J. D., I. Shimomura, M. S. Brown, R. E. Hammer, J. L. Goldstein, and H. Shimano. 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 101:2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua, X., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87:415-426. [DOI] [PubMed] [Google Scholar]
- 22.Hua, X., J. Wu, J. L. Goldstein, M. S. Brown, and H. H. Hobbs. 1995. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics 25:667-673. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy, N. J., T. Kataoka, J. Tschopp, and R. C. Budd. 1999. Caspase activation is required for T cell proliferation. J. Exp. Med. 190:1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., and P. A. Colussi. 1999. Prodomains-adaptors-oligomerization: the pursuit of caspase activation in apoptosis. Trends Biochem. Sci. 24:1-4. [DOI] [PubMed] [Google Scholar]
- 25.Lamkanfi, M., K. D'Hondt, L. Vande Walle, M. van Gurp, G. Denecker, J. Demeulemeester, M. Kalai, W. Declercq, X. Saelens, and P. Vandenabeele. 2005. A novel caspase-2 complex containing TRAF2 and RIP1. J. Biol. Chem. 280:6923-6932. [DOI] [PubMed] [Google Scholar]
- 26.Lassus, P., X. Opitz-Araya, and Y. Lazebnik. 2002. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297:1352-1354. [DOI] [PubMed] [Google Scholar]
- 27.Logette, E., A. Wotawa, S. Solier, L. Desoche, E. Solary, and L. Corcos. 2003. The human caspase-2 gene: alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene 22:935-946. [DOI] [PubMed] [Google Scholar]
- 28.Magana, M. M., and T. F. Osborne. 1996. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J. Biol. Chem. 271:32689-32694. [DOI] [PubMed] [Google Scholar]
- 29.Miserez, A. R., G. Cao, L. C. Probst, and H. H. Hobbs. 1997. Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2). Genomics 40:31-40. [DOI] [PubMed] [Google Scholar]
- 30.Nagai, M., J. Sakakibara, Y. Nakamura, F. Gejyo, and T. Ono. 2002. SREBP-2 and NF-Y are involved in the transcriptional regulation of squalene epoxidase. Biochem. Biophys. Res. Commun. 295:74-80. [DOI] [PubMed] [Google Scholar]
- 31.Nagoshi, E., N. Imamoto, R. Sato, and Y. Yoneda. 1999. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol. Biol. Cell 10:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagoshi, E., and Y. Yoneda. 2001. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin beta. Mol. Cell. Biol. 21:2779-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborne, T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275:32379-32382. [DOI] [PubMed] [Google Scholar]
- 34.Pai, J. T., O. Guryev, M. S. Brown, and J. L. Goldstein. 1998. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J. Biol. Chem. 273:26138-26148. [DOI] [PubMed] [Google Scholar]
- 35.Paroni, G., C. Henderson, C. Schneider, and C. Brancolini. 2002. Caspase-2 can trigger cytochrome C release and apoptosis from the nucleus. J. Biol. Chem. 277:15147-15161. [DOI] [PubMed] [Google Scholar]
- 36.Rawson, R. B. 2003. The SREBP pathway—-insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4:631-640. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, J. D., M. Enoksson, M. Suomela, B. Zhivotovsky, and S. Orrenius. 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277:29803-29809. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, J. D., V. Gogvadze, A. Kropotov, H. Vakifahmetoglu, B. Zhivotovsky, and S. Orrenius. 2004. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity. EMBO Rep. 5:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai, J., R. B. Rawson, P. J. Espenshade, D. Cheng, A. C. Seegmiller, J. L. Goldstein, and M. S. Brown. 1998. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2:505-514. [DOI] [PubMed] [Google Scholar]
- 40.Sakakura, Y., H. Shimano, H. Sone, A. Takahashi, N. Inoue, H. Toyoshima, S. Suzuki, and N. Yamada. 2001. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem. Biophys. Res. Commun. 286:176-183. [DOI] [PubMed] [Google Scholar]
- 41.Sato, R., J. Inoue, Y. Kawabe, T. Kodama, T. Takano, and M. Maeda. 1996. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271:26461-26464. [DOI] [PubMed] [Google Scholar]
- 42.Schwerk, C., and K. Schulze-Osthoff. 2003. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem. Pharmacol. 66:1453-1458. [DOI] [PubMed] [Google Scholar]
- 43.Sheng, Z., H. Otani, M. S. Brown, and J. L. Goldstein. 1995. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. USA 92:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimano, H. 2002. Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism. Vitam. Horm. 65:167-194. [DOI] [PubMed] [Google Scholar]
- 45.Shimano, H., J. D. Horton, I. Shimomura, R. E. Hammer, M. S. Brown, and J. L. Goldstein. 1997. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Investig. 99:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slee, E. A., C. Adrain, and S. J. Martin. 1999. Serial killers: ordering caspase activation events in apoptosis. Cell Death Differ. 6:1067-1074. [DOI] [PubMed] [Google Scholar]
- 47.Solier, S., E. Logette, L. Desoche, E. Solary, and L. Corcos. 2005. Nonsense-mediated mRNA decay among human caspases: the caspase-2S putative protein is encoded by an extremely short-lived mRNA. Cell Death Differ. 12:687-689. [DOI] [PubMed] [Google Scholar]
- 48.Sordet, O., C. Rebe, S. Plenchette, Y. Zermati, O. Hermine, W. Vainchenker, C. Garrido, E. Solary, and L. Dubrez-Daloz. 2002. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood 100:4446-4453. [DOI] [PubMed] [Google Scholar]
- 49.Tinel, A., and J. Tschopp. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304:843-846. [DOI] [PubMed] [Google Scholar]
- 50.Wang, L., M. Miura, L. Bergeron, H. Zhu, and J. Yuan. 1994. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 78:739-750. [DOI] [PubMed] [Google Scholar]
- 51.Wang, X., M. R. Briggs, X. Hua, C. Yokoyama, J. L. Goldstein, and M. S. Brown. 1993. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J. Biol. Chem. 268:14497-14504. [PubMed] [Google Scholar]
- 52.Wang, X., N. G. Zelenski, J. Yang, J. Sakai, M. S. Brown, and J. L. Goldstein. 1996. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 15:1012-1020. [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong, S., S. S. Chirala, and S. J. Wakil. 2000. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc. Natl. Acad. Sci. USA 97:3948-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 55.Zeng, L., M. Lu, K. Mori, S. Luo, A. S. Lee, Y. Zhu, and J. Y. Shyy. 2004. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 23:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.