Abstract
mRNA maturation in Trypanosoma brucei depends upon trans splicing, and variations in trans-splicing efficiency could be an important step in controlling the levels of individual mRNAs. RNA splicing requires specific sequence elements, including conserved 5′ splice sites, branch points, pyrimidine-rich regions [poly(Y) tracts], 3′ splice sites (3′SS), and sometimes enhancer elements. To analyze sequence requirements for efficient trans splicing in the poly(Y) tract and around the 3′SS, we constructed a luciferase-β-galactosidase double-reporter system. By testing ∼90 sequences, we demonstrated that the optimum poly(Y) tract length is ∼25 nucleotides. Interspersing a purely uridine-containing poly(Y) tract with cytidine resulted in increased trans-splicing efficiency, whereas purines led to a large decrease. The position of the poly(Y) tract relative to the 3′SS is important, and an AC dinucleotide at positions −3 and −4 can lead to a 20-fold decrease in trans splicing. However, efficient trans splicing can be restored by inserting a second AG dinucleotide downstream, which does not function as a splice site but may aid in recruitment of the splicing machinery. These findings should assist in the development of improved algorithms for computationally identifying a 3′SS and help to discriminate noncoding open reading frames from true genes in current efforts to annotate the T. brucei genome.
In eukaryotes, a central step in generating mature mRNA from pre-mRNA is the removal of introns and the joining of the two flanking exons, a process known as cis splicing (14, 33). For successful splicing, the intron has to be correctly identified. Several sequence elements are implicated in defining the two splice sites. The 5′ end of the intron is generally defined by a GT dinucleotide, whereas the 3′ end is marked by AG. Additional characteristics of the 3′ splice site (3′SS) are a branch point sequence followed by a pyrimidine-rich [poly(Y)] tract. Enhancer regions, which may contribute to the assembly of the spliceosome or the identification of the correct 3′SS, have also been described (33). Splicing involves two trans esterification steps. During the first trans esterification, the conserved branch point adenosine forms a 2′-to-5′ phosphodiester bond with the 5′ end of the intron (13, 34). This step depends on U2 snRNP binding to the branch point sequence, which in turn requires the help of the heterodimeric auxiliary factor U2AF, consisting of 65-kDa and 35-kDa subunits. The U2AF65 subunit has been shown to bind to the poly(Y) tract, whereas U2AF35 associates with the 3′SS AG dinucleotide (20, 46, 47). During the second trans esterification, the free hydroxyl of the upstream exon attacks the phosphate of the 3′SS to join the exons and release the lariat-shaped intron (25).
The mechanism for selecting the appropriate 3′SS AG dinucleotide from other cryptic AG sites for the second trans esterification reaction has not entirely been solved. Two models have been proposed. According to the scanning model, identification of the correct 3′SS AG occurs by a linear search mechanism, assuming that the spliceosome begins scanning at the branch point and selects the first downstream AG dinucleotide (39). This model is supported by research with HeLa cells, in which splicing is blocked when hairpin loops are inserted upstream of the AG splice site, possibly preventing movement of the spliceosome (3). With a bimolecular exon ligation assay, it was also shown that the substrate with the 5′-most AG is selected among different 3′ RNA substrates and that no poly(Y) tract was necessary for the second catalytic step (1, 3). In contrast to human cells, a simple scanning model cannot explain various findings on Saccharomyces cerevisiae, where downstream AG sites can outcompete upstream AG sites if the upstream sites are located closer than 23 nucleotides (nt) to the branch point sequence (5, 31). Moreover, hairpin loops upstream of an AG splice site had an enhancing effect, leading to increased usage of that splice site instead of inhibition (8).
The second model suggests a mechanism in which the correct 3′SS AG dinucleotide is identified by its distance from the branch point. Data supporting this model stem from findings obtained with yeast and human cells that the second trans esterification step occurs most efficiently when the 3′SS AG is located 19 to 23 nt downstream of the branch point (4, 5). In addition to the branch point to AG distance, the sequence of this region itself, especially the presence or absence of a poly(Y) tract, has been shown to strongly affect splicing efficiency in a number of organisms, including humans (39), S. cerevisiae (31), and Trypanosoma brucei (15, 27).
T. brucei transcribes the majority of its genes as polycistronic units (6). To generate mature mRNAs, a 39-nt miniexon, also called the spliced leader, must be trans spliced to the primary transcript, at appropriate points, from a capped precursor of ∼140 nt (spliced leader RNA). trans splicing serves two functions: it dissects mRNAs from polycistronic primary transcripts, and the spliced leader provides the cap structure for the mRNA (6, 22). In contrast to cis splicing, trans splicing joins exons derived from two independently transcribed RNAs. trans splicing and cis splicing, however, share remarkable similarities: both require the same characteristic sequence motifs [GT at the 5′SS, an adenosine branch point, a poly(Y) tract, AG at the 3′SS, and possibly exonic enhancer motifs] (15, 16, 23, 27, 30, 32, 40), both follow the same general mechanism (two catalytic trans esterification reactions), and many of the major components of the yeast or human spliceosome are conserved in T. brucei (22). trans splicing is a prerequisite for protein expression, and changes in the poly(Y) tract, leading to a difference in trans-splicing efficiency, should be reflected in differences in protein levels. Given these characteristics, T. brucei is an excellent model to evaluate the effects of intronic sequence motifs on splicing efficiency, as one can rely on measuring protein levels derived from a carefully designed reporter system. Understanding sequence requirements for trans-splicing efficiency may help to predict splice sites and their probable efficiency. A systematic series of experimental data could also help design specific bioinformatics tools that, by identifying true splice sites, can distinguish true genes from random open reading frames (ORFs) and thereby assist in the annotation of the T. brucei genome (11).
Previously, the importance of a poly(Y) tract for trans splicing in T. brucei has been demonstrated by insertion of block substitution mutations (15, 27). Additionally, it has been observed that block substitution mutations in the 5′ untranslated region (UTR) of α-tubulin can affect trans splicing, demonstrating a role for 5′ UTRs in trans splicing (23). However, no extensive systematic study has been performed to determine minimal and optimal intronic or exonic sequence motifs required for efficient trans splicing.
To systematically study sequence requirements for trans splicing in T. brucei, we constructed a luciferase-β-galactosidase double-reporter system that allows a large number of sequence motifs to be evaluated with great sensitivity and reproducibility in transiently transfected cells to define their effects on splicing efficiency. We tested ∼90 constructs to explore the roles of the composition, length, and position of the poly(Y) tract and the length and composition of the spacer region separating the poly(Y) tract from the 3′SS, and we identified a single-nucleotide change in the mRNA 5′ UTR that could compensate for the low splicing efficiency observed when the 3′SS AG dinucleotide is preceded by AC.
MATERIALS AND METHODS
Cell lines and culture conditions. All experiments were performed with the procyclic (tsetse midgut) form of T. brucei strain Lister 427 cultured in SDM-79 (2) supplemented with 10% fetal calf serum and 0.25% hemin. Wild-type 427 cells were used for transient transfections of reporter constructs, and the 29.13 derivative clone, which expresses T7 RNA polymerase and the Tet repressor (45), was used for stable transfections.
Plasmid construction.
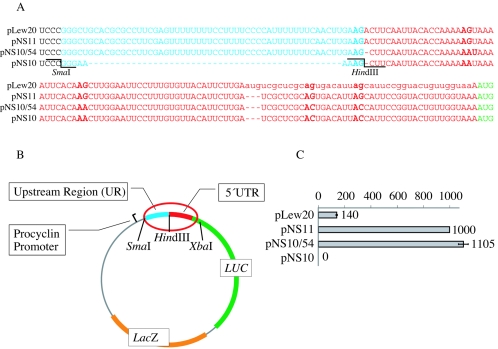
All reporter plasmids used in these experiments are derivatives of pNS10, which contains luciferase and lacZ reporter genes and restriction sites that permit the insertion of alternative upstream regions (URs) (Fig. 1A and B). pNS10 was constructed from pLew20 (44) as follows. First, pLew20 was digested with SrtI and StuI to remove the ble cassette. Next, the construct was digested with SmaI and BsmI to remove a 139-bp region between the procyclin promoter and the luciferase coding region to allow its replacement with a synthetic oligonucleotide that created sites for inserting alternative splice site motifs and eliminated all AG dinucleotides except for the wild-type splice site. The 3′ overhang generated by the BsmI digest was removed by Klenow polymerase (35). Two complementary 90-nt oligonucleotides (5′-GGGAAAAAGCTTCAATTACACCAAAAAATAAAATTCACAAACTTGGAATTCCTTTGTGTTACATTCTTGAATGTCGCTCGCAATGACATT-3′) were annealed, and the double-stranded insert was ligated into the open vector.
FIG. 1.
Design of the luciferase-β-galactosidase double-reporter system. (A) pLew20 is a construct containing a wild-type procyclin promoter, a UR (blue), and a 5′ UTR (red) preceding the luciferase ORF (LUC, green). Lowercase letters indicate a small ORF within the 5′ UTR that was carried over from the plasmid from which LUC was excised. pNS11 is a derivative of pLew20 in which the adventitious AUG was deleted from the 5′ UTR and a lacZ reporter gene was added downstream of LUC. In pNS10/54, all four cryptic AG sites were changed to AA or AC (bold) and a HindIII site was introduced at the remaining AG splice site to facilitate the insertion of different URs. pNS10 lacks the UR. (B) Organization of the pNS10/54 reporter plasmid, indicating key components and restriction sites. (C) Luciferase activity in the basic reporter plasmids. All trans-splicing efficiency values shown are measurements of relative luciferase light units normalized to β-galactosidase activity and then to the pNS11 positive control, which was set to 1,000 in this and all subsequent experiments.
To add lacZ as a second reporter, the lacZ sequence was PCR amplified from plasmid pSV-β-Galactosidase (Promega). 5′ and 3′ UTRs were PCR amplified from the ble cassette of pLew20. The three fragments, the 5′ UTR, the lacZ coding region, and the 3′ UTR, were ligated, and the product was PCR amplified. The amplified lacZ cassette was then ligated into the single-reporter construct to yield the double-reporter construct pNS5.
Finally, two ATG sites (of which one was inadvertently introduced into the 90-nt oligonucleotide above and the other was present in a small sequence upstream of the luciferase ORF that was present in all previous luciferase-containing constructs used in this and other laboratories and created a short upstream ORF whose effect was not previously appreciated; see Results) were removed from the luciferase 5′ UTR of the double-reporter construct by PCR-based site-directed mutagenesis. To remove the first ATG site, we used a forward primer (NS35 [5′-TCTCGTCCCGGGAAAAAGCTTC-3′]) containing the sequence surrounding the SmaI site of pNS5 and a reverse primer (NS24 [5′-CCATCCTCTAGAGGATAGAATGG-3′]) containing the sequence surrounding the XbaI site. In addition to these two outside primers, two inner primers were used to introduce the desired changes in nucleotide sequence. The inner primers are complementary and contain the following sequences: NS27, 5′-GAATGTCGCTCGCAcTGACATTacCATTCCGGTACTGTTGG-3′; NS28, 5′-CCAACAGTACCGGAATGgtAATGTCAgTGCGAGCGACATTC-3′. Lowercase letters represent introduced changes. Two separate PCRs were performed with pNS5 as template and primer pairs NS23-NS28 and NS27-NS24. Next, the products from both PCRs were used as templates in a third PCR with outside primers NS23 and NS24. SmaI- and XbaI-digested amplicons were reintroduced into pNS5, replacing the original sequence between SmaI and XbaI to give pNS9. The second ATG was removed by a similar strategy with the outside primers NS35 and NS24 but with the inside primers NS36 (5′-CCTTTGTGTTACATTCTTGATCGCTCGCACTGACATTACC-3′) and NS37 (5′-GGTAATGTCAGTGCGAGCGATCAAGAATGTAACACAAAGG-3′) to generate pNS10.
To insert a UR of choice, two complementary synthetic oligonucleotides were annealed and ligated into the double-reporter construct pNS10 between the SmaI and HindIII sites.
pNS11 was generated by replacing the sequence between the SmaI and XbaI sites of pNS10 with the sequence between the SmaI and XbaI sites originally found in pLew20, which contains four cryptic AG sites. Finally, an ATG site was removed from the newly inserted sequence by PCR-based site-directed mutagenesis as described above for pNS10.
pNS10/56 and pNS10/72 to pNS10/75 all contain the T. brucei α-tubulin UR and differ only in sequences of the luciferase 5′ UTR. Changes in 5′ UTRs were generated by site-directed mutagenesis as describe above, with appropriate primers. pNS20/74 is a derivative of pNS10/74 in which we replaced the lacZ gene with a phleomycin resistance marker.
All oligonucleotides were purchased from Integrated DNA Technology and purified as recommend by the vendor. All constructs were verified by DNA sequencing.
Transient transfections and luciferase and β-galactosidase assays.
Cells (2 × 107; ∼8 × 106/ml) were transfected with 10 μg of DNA and incubated for 16 to 22 h as previously described (44). Cells were pelleted at 700 × g at 4°C for 10 min, resuspended in 1 ml of cold phosphate-buffered saline, transferred to 1.5-ml Eppendorf tubes, and centrifuged again at 8,000 × g at 4°C for 3 min. The supernatant was removed, and the cells were resuspended in 100 μl of Cell Culture Lysis Reagent (Promega). Ten microliters of the lysed cells was mixed with 45 μl of Promega luciferase assay buffer, and luciferase activity was measured immediately in a Turner TD-20e luminometer. To measure β-galactosidase activity, the remaining 90 μl of lysed cells was centrifuged at 17,900 × g at 4°C for 4 min. Thirty microliters of the cell lysate supernatant was added to a reaction mixture containing 260.4 μl of 0.1 M sodium phosphate, 6.6 μl of 10× CPRG (Roche), and 3 μl of 100× Mg2+ solution (0.1 M MgCl2, 4.5 M β-mercaptoethanol) and incubated for 8 h at 37°C (35). The reaction was terminated by adding 500 μl of 1 M Tris, and the absorbance was determined at 570 nm with a Novaspec II spectrophotometer. All transfections were done in duplicate, and the values shown are the averages of these two measurements; the error bars represent the luciferase activities in the two transfections. All transfections were repeated at least once on a different date, and all trends could be reproduced. β-Galactosidase values remained relatively constant throughout the experiments.
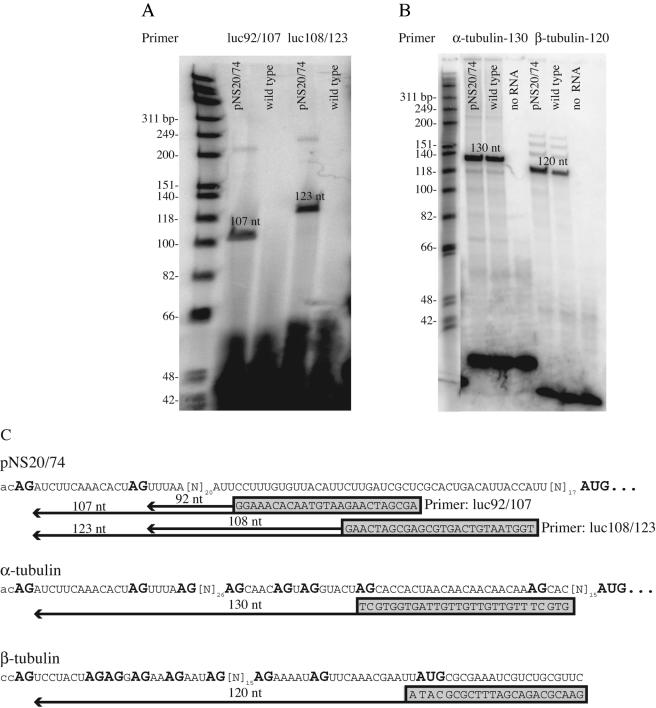
Primer extension to determine the splice site in α-tubulin 5′ UTR construct pNS20/74.
T. brucei 29.13 cells were stably transfected with pNS20/74 as described previously (43). RNA extractions and primer extensions were performed with an RNeasy Kit (QIAGEN) and a Primer Extension Kit (Promega) by following essentially the provided protocols. Primer luc92/107 (AGCGATCAAGAATGTAACACAAAGG) anneals downstream of the 3′SS of luciferase on pNS20/74 and should yield products of 92 or 107 bp, depending on which AG is used as the splice site. Primer luc108/123 (TGGTAATGTCAGTGCGAGCGATCAAG) anneals further downstream than luc92/107 and should yield products of 108 or 123 bp. We also included primers α-tubulin130 (GTGCTTTGTTGTTGTTGTTAGTGGTGCT) and β-tubulin120 (GAACGCAGACGATTTCGCGCATA). All primers were labeled with [γ-32P]ATP (6,000 Ci/mmol).
RESULTS
Construction of a double-reporter system.
The aim of this study was to systematically characterize sequence motifs upstream of the ORF that are necessary for mRNA trans splicing in T. brucei. The UR is defined as the region between the branch point and the 5′ UTR of the mRNA (Fig. 1A and B) and contains a poly(Y) tract, a spacer region, and the 3′SS. We constructed a reporter system in which we could switch the sequence of the UR by using the T. brucei procyclin promoter to drive a luciferase (LUC) reporter gene flanked by the procyclin 5′ UTR and the aldolase 3′ UTR plus a short extraneous sequence derived from the vector from which LUC was originally cloned. We were able to use a protein assay to measure the trans-splicing efficiency of pre-mRNA by ensuring that luciferase activity would only appear if the mRNA were correctly spliced at the single available AG dinucleotide, all others having been deleted, upstream of the single essential AUG translation initiation codon. Luciferase activity can be assayed over a linear range of 5 orders of magnitude. The readout is sensitive enough to use transient transfection, which is the only practical approach to testing a large number of constructs in trypanosomes. To permit normalization for differences in transfection efficiency, we included a lacZ reporter in the plasmid as an internal control. lacZ was flanked by sites for splicing and polyadenylation and was inserted downstream of the luciferase reporter gene (Fig. 1B).
To allow the insertion of a large number of alternative URs, HindIII and SmaI restriction sites were incorporated adjacent to the 3′SS AG dinucleotide and at the upstream end of the UR, respectively. Insertion of the procyclin UR into our reporter construct led to high levels of luciferase activity (Fig. 1C, pNS10/54). Removal of an ATG site located in the 5′ UTR, which gave rise to a small ORF within the 5′ UTR, caused a sevenfold increase in luciferase activity (compare pLew20 and pNS11). All subsequent experiments were performed with constructs without an ATG site in the 5′ UTR. Elimination of four AG dinucleotides from within the luciferase 5′ UTR, leaving the construct with only a single available 3′SS, avoided the possibility that changing the length and/or composition of the UR would lead to the selection of a second cryptic splice site. This action did not decrease luciferase activity, demonstrating that those sites, in the wild-type procyclin UTR, had no essential role in regulating trans splicing (Fig. 1C, pNS10/54).
Length and composition of poly(Y) tract.
Previous reports on a wide variety of eukaryotes show the importance of a poly(Y) tract for efficient cis splicing, although S. cerevisiae can accurately remove introns that lack a poly(Y) tract, where a very conserved branch point sequence and splice site motif seem to provide the necessary recognition sites for the splicing machinery (7, 9). In T. brucei, the available data suggest a minimal requirement of ∼10 pyrimidines located between 10 and 40 nt upstream of the 3′SS (15, 27). However, neither of these parameters, nor the optimum composition of the poly(Y) tract, has been systematically and precisely investigated.
We tested poly(Y) tracts containing between 5 and 40 uridines (U) and observed a clear correlation between luciferase expression and poly(Y) tract length, in the context of two different spacer sequences between the poly(Y) tract and the 3′SS. Luciferase expression leveled out at ∼25 U (Fig. 2A and B).
FIG. 2.
Effects of increasing poly(Y) tract length on trans-splicing efficiency. Constructs containing URs with nonspecific 7-bp (A) or 17-bp (B) spacers and poly(Y) tracts of increasing length show increasing trans-splicing efficiency which plateaus at ∼25 uridines.
To investigate the effect of poly(Y) tract composition on trans splicing, we sprinkled cytidine, adenosine, or guanosine within the poly(U) tract. Interspersion of a poly(U) tract with C's, generating a true poly(Y) tract, caused a significant increase in trans splicing, but replacing a poly(U) tract with poly(C) eliminated trans splicing (Fig. 3A). Interspersion of a poly(Y) tract with a single adenosine or guanosine caused a moderate decrease in trans splicing, but two or three consecutive purines had a severe effect (Fig. 3B). These results suggest that continuity of the poly(Y) tract is important, rather than the presence of a minimum number of pyrimidines within an ∼15-nucleotide window.
FIG. 3.
Effects of poly(Y) tract composition on trans-splicing efficiency. Constructs containing a 17-bp spacer were tested for the role of C in place of U (A) and for the effect of A or G insertions into the poly(Y) tract (B). [N]17 = CAACUUAAACUUCAAUU.
Length and composition of spacer.
It has been suggested that the position of the poly(Y) tract relative to the branch point and 3′SS may be important for efficient trans splicing (29). We therefore varied the distance between the poly(Y) tract and 3′SS in 3-nt increments while keeping all other parameters constant. Our data show a clear correlation between spacer length and trans-splicing efficiency, suggesting an optimum spacer length of ∼20 nt, which corresponds to a branch point to 3′SS distance of ∼39 nt (Fig. 4A). Subsequently, we looked for effects of spacer composition on trans-splicing efficiency (Fig. 4B). With homopolymeric spacer tracts, it was clear that a 3′SS that was preceded solely by uridines was significantly better than the heterogeneous spacer sequences included in this data set and at least as good as the natural upstream sequence derived from the highly expressed procyclin locus that was used as the positive control for all experiments in this study. The deleterious effects of a poly(A) spacer can probably be attributed to the formation of an A-U duplex region that prevents the poly(U) tract from being recognized, and the extreme inhibition of trans splicing in the poly(G) spacer construct can probably be explained by the tendency of consecutive guanosine residues to form secondary structures that could block any scanning machinery from reaching the 3′SS or prevent splicing factors from binding to the UR.
FIG. 4.
Effects of spacer length and composition on trans-splicing efficiency. Constructs containing a 10-bp poly(Y) tract were tested for the effect of variations in the length (A) and composition (B) of the spacer region. Changes of particular interest at positions −3 and −4 are underlined in panel B.
The most interesting and surprising result from this set of constructs was the 20-fold drop in luciferase activity when the 3′SS AG dinucleotide was preceded by AC rather than AU (Fig. 4B). This effect was independent of spacer length. No consensus sequence apart from the AG dinucleotide at the 3′SS has been documented in T. brucei, whereas humans and yeast appear to require a pyrimidine at the −3 position (YAG) (9). We therefore looked at the effect of the nucleotide at the −3 position in a poly(U) background [poly(U) tract and poly(U) spacer] on trans-splicing efficiency. We observed significant decreases in trans-splicing efficiency as we changed the nucleotide at the −3 position as follows: U to G, a fourfold decrease; U to A or C, a twofold decrease (Fig. 4B). We will return to this conundrum in the last section.
Native poly(Y) tracts and spacer regions.
Our results identified several trends that affected splicing efficiency when synthetic UR sequences were used. Although a few studies had compared the trans-splicing efficiencies of similar genes with different URs, we wanted to confirm that our reporter system would be influenced by differences in naturally occurring URs. We therefore tested the trans-splicing efficiency obtained with the native URs of several T. brucei genes. The T. brucei genome encodes three isoenzymes of phosphoglycerate kinase (PGK) that are coexpressed as consecutive sequences on a polycistronic pre-mRNA. This pre-mRNA gives rise to unequal amounts of PGK A, B, and C mRNAs. Very low levels of PGK A mRNA can be detected in procyclic T. brucei, compared to intermediate levels of PGK C mRNA and high levels of PGK B mRNA (10, 21). Placing the corresponding PGK UR sequences in our reporter system, we observed very high levels of luciferase activity with the UR of PGK B, intermediate levels with the UR of PGK C, and levels barely above the background with PGK A (Fig. 5), reflecting the natural mRNA levels. In a previous study, when the UR from each PGK gene was placed upstream of the luciferase ORF and the resultant constructs transiently transfected into T. brucei procyclic cells, luciferase activity levels also indicated different levels of splicing and correlated with mRNA levels (19). Enzyme activity was low when the UR from the A gene was present but indistinguishable when the B and C URs were compared. A review of these results, however, suggests that limiting reaction conditions had prevented any difference in the B and C sequences from being observed. Surprisingly, the UR of the gene encoding variant surface glycoprotein 118 (VSG118) was rather poorly spliced (the VSG represents 10% of the total cellular protein of bloodstream T. brucei) compared to efficient synthetic URs or to the UR of VSG221. Interestingly, the UR of VSG118 has an AC dinucleotide preceding the AG dinucleotide, which we had observed to result in very low levels of trans splicing.
FIG. 5.
trans-splicing efficiencies of selected native URs. The sequences upstream of the proven 3′SS for the three tandem PGK genes, two VSGs, and α- and β-tubulin are shown.
The observation that the URs of α- and β-tubulin led to drastically different levels of trans splicing (Fig. 5) was even more surprising, as tubulin exists as a heterodimer containing one α and one β subunit, thus requiring equal amounts of the two subunits. The α-tubulin UR also contains an AC dinucleotide preceding the AG at the 3′SS. One possible explanation for our observation was that the previous 3′SS assignment was incorrect (32). However, we discarded this hypothesis after sequencing of α-tubulin cDNA clones confirmed the original assignment, which was also verified by primer extension (see below). A second explanation could be that efficient trans splicing of α-tubulin genes depends upon exonic splice-enhancing elements within the α-tubulin 5′ UTR. Such splice-enhancing elements have been described in higher eukaryotes and are capable of activating weak upstream splice sites (36, 37). Splicing enhancer elements consist of short RNA sequences, which are recognized by a number of splicing factors with characteristic serine/arginine (SR)-rich domains. Two such SR proteins have been described in T. brucei, but their exact function is unknown (17, 18, 26). All our constructs contained the 5′ UTR of the procyclin gene, which might be well suited for most genes, but some URs with weak poly(Y) tracts or a suboptimal 3′SS might require additional splice-enhancing sequences in their 5′ UTR. Previously, López-Estraño and colleagues demonstrated that block substitution mutations in the α-tubulin 5′ UTR negatively affect trans splicing at the upstream 3′SS AG (23). We decided to extend the study and pinpoint the responsible UTR element.
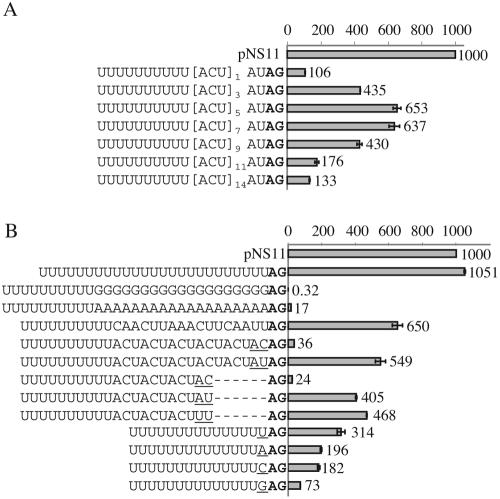
Role of the α-tubulin 5′ UTR in trans splicing.
All of the experiments described so far used constructs containing a procyclin 5′ UTR in which we replaced four cryptic AG sites with AA or AC. To test the hypothesis that the 5′ UTR contains sequence elements that, in some contexts, are important for efficient trans splicing, we first changed the 5′ UTR in our constructs in small blocks to match the 5′ UTR from α-tubulin. We observed a 60-fold increase in luciferase expression after changing nucleotides 9 to 18 nt downstream of the 3′SS to match the sequence present in endogenous α-tubulin 5′ UTRs (Fig. 6). These 10 replacement nt contained an AG dinucleotide, and a single-nucleotide change showed that high luciferase expression depended upon this AG (compare pNS10/74 and 10/75). Several other single-nucleotide substitutions in the vicinity of the AG site had no effect on trans-splicing efficiency (data not shown). We therefore needed to exclude the possibility that the second AG site was being used as the 3′SS. Because of the low luciferase mRNA levels in transiently transfected trypanosomes and our inability to distinguish the two potential alternative products by quantitative reverse transcription-PCR, we performed primer extension analyses on clones that had been stably transfected with a version of pNS10/74 (pNS20/74) in which lacZ had been replaced with a gene encoding phleomycin resistance. With two primer pairs, we demonstrated that the first AG site functioned exclusively as a splice site; no product corresponding to the second AG was observed (Fig. 7A and C). Primer extension experiments with the endogenous α-tubulin and β-tubulin mRNAs in these transfected clones confirmed the previously assigned splice sites (Fig. 7B and C). Based on these findings, we concluded that the presence of a second AG site in the 5′ UTR enhances trans splicing in constructs that contain a suboptimum sequence (AC) immediately upstream of the 3′SS.
FIG. 6.
One nucleotide in the 5′ UTR of α-tubulin is critical for splicing. The upper sequence shows the endogenous α-tubulin 5′ UTR. Sequences are shown from the −4 position relative to the 3′SS, and all constructs contain the α-tubulin UR. Only the 5′ UTR sequences differ. pNS10/56 contains the procyclin 5′ UTR lacking cryptic AG sites. In pNS10/72, 8 nt downstream of the 3′SS have been changed to match the sequence found in the endogenous α-tubulin 5′ UTR. pNS10/73 contains a 5′ UTR sequence identical to the α-tubulin 5′ UTR for 20 nt downstream of the 3′SS, except for the omission of nucleotides at positions +1 and +2. pNS10/74 is identical to pNS10/73 but with the +1 and +2 nucleotides intact. In pNS10/75, the AG within the 5′ UTR of pNS10/74 was changed to AA.
FIG. 7.
Primer extension analysis of the 3′SS used in pNS20/74 and by endogenous α- and β-tubulin. (A) Twenty micrograms of cellular RNA and primers specific for pNS20/74. The entire reaction product was loaded, and the gel was exposed to a phosphorimager screen for 60 h. (B) Ten micrograms of RNA and primers specific for α-tubulin and β-tubulin. Half the reaction product was loaded, and the gel was exposed to a phosphorimager screen for 15 h. One major product was seen in each analysis. (C) Schematic depiction of possible primer extension products. The expected lengths include the 39 nt of the miniexon which are added during the trans-splicing reaction.
DISCUSSION
Role of poly(Y) tract.
In this study, we demonstrated that trans splicing in T. brucei depends upon the length, composition, and position of a poly(Y) tract. Furthermore, our data suggest that changes in length between the branch point and splice site are acceptable as long as the poly(Y) tract and spacer region fulfill certain requirements. Our observations are consistent with recently published data indicating that T. brucei genes do not necessarily contain a single branch point and that different nucleotides may serve as branch points (24). One of the early events in spliceosome assembly is the recruitment of U2 snRNP to the branch point sequence. In mammalian cells, U2 recruitment has been confirmed to depend upon the two subunits of U2AF. The 65-kDa subunit has been shown to bind to the poly(Y) tract, while the 35-kDa subunit binds to the 3′SS AG (28, 46, 47, 49). Further evidence that poly(Y) tract variations affect the first of the two catalytic steps stems from research with HeLa cells, where no poly(Y) tract is necessary for the second step (1, 3).
No trypanosome homolog to the human U2AF65 subunit has been found, although database mining has revealed proteins with domains that could potentially bind to pyrimidine-rich RNA tracts. In contrast, the T. brucei genome contains a relatively conserved U2AF35 homolog. A similar homolog has been studied more closely in Trypanosoma cruzi, and interestingly, just like the fission yeast homolog, it is missing the C-terminal SR domain. In addition, the T. cruzi homolog is lacking Trp 134, which is highly conserved in other organisms and has been shown to allow the human U2AF35 subunit to participate in a tongue-in-groove interaction with its larger partner U2AF65 (20, 41, 42). The absence of these conserved residues appears plausible in the absence of U2AF65.
Poly(Y) tract lengths shorter than 10 pyrimidines may interfere with the binding of such poly(Y)-binding proteins, thereby affecting initial assembly of the spliceosome machinery. This hypothesis is in agreement with our data showing that shorter poly(Y) tracts are only well tolerated as long as they are not positioned too close to the 3′SS. Our results suggest the absence of a threshold poly(Y) tract length for optimum binding. Incremental increases in poly(Y) tract length led to a continuous increase in trans-splicing efficiency, which ultimately reaches a plateau.
Identification of the 3′SS.
In humans, there is a very strong preference for a pyrimidine preceding the AG at the 3′SS and a strong preference against G (9). In T. brucei, there is a small but statistically significant underrepresentation of G in the −3 position, but the other nucleotides are found with equal frequency (Shuba Gopal, personal communication).
However, with the procyclin 5′ UTR, we observed very low splicing efficiency when the −3 and −4 positions were occupied by A and C, respectively. These results suggest, as did earlier work by López-Estraño et al. (23), that certain URs might require distinct splice enhancer motifs in the 5′ UTR for efficient trans splicing. In HeLa cells, a very heterogeneous population of splice enhancer sequences has been described (12, 37); in contrast, we were able to show that a single downstream AG site was required for efficient trans splicing at the upstream AG site. This result is consistent with earlier observations that show that an AG dinucleotide, newly introduced upstream of the original 3′SS AG dinucleotide, can function as a new 3′SS as long as the downstream AG is present (48). These findings can be explained by the requirement of U2AF35 binding to the 3′SS splice during early stages of spliceosome assembly.
Our data can be explained assuming, first, that the first AG site functions as a splice site and is recognized by a scanning mechanism that identifies the first AG downstream of the branch point and poly(Y) tract; second, that the first AG site does not function as a U2AF35 binding site, possibly due to unfavorable residues at positions −3 and −4; and third, that U2AF35 does not have to bind at the 3′SS AG itself as long as it can bind to an AG close by, allowing interactions with a putative poly(Y) tract binding protein, helping the latter to bind to the poly(Y) tract.
The strong effect of the 5′ UTR on trans-splicing efficiency observed in our studies, compared to a more moderate effect observed by López-Estraño et al. (23), probably stems from the fact that we previously deleted all other cryptic AG sites, whereas López-Estraño et al. only deleted one AG site per block substitution mutation.
Data from studies with HeLa cells suggest a scanning model for the recognition of the 3′SS AG prior to the second catalytic step (38, 39). This model is supported by data showing that sequences that can form stable secondary structures lead to a dramatic decrease in splicing efficiency, potentially by blocking factors scanning along the RNA from the branch point to the splice site (3). We observed little splicing of constructs containing poly(A) or poly(G) spacers, which would probably form secondary structures, with the upstream poly(U) tract or otherwise.
The data obtained in this study should aid in the development of algorithms to identify splice sites computationally in T. brucei and in other members of the order Kinetoplastida. There may be differences within this broad family, however, as the poly(Y) tracts of Leishmania major are predominantly C, whereas those of T. brucei are predominantly U (Shuba Gopal, personal communication).
Predicted trans-splicing efficiency is in agreement with levels of mRNA.
In this study, we exploited the advantage of enzymatic assays (luciferase and β-galactosidase) to study trans-splicing efficiency. However, many factors besides trans-splicing efficiency might influence enzyme levels in a cellular context (RNA stability or protein degradation, for example). We are confident, however, that our assays accurately reflect the amount of trans-spliced RNA, since only exonic sequence elements should influence RNA stability, and no changes in the 5′ UTR or coding region were introduced when analyzing the UR. In our analysis of the 5′ UTR, we did change parts of the mRNA sequence, but it seems unlikely that those changes, as little as a single nucleotide change generating an additional AG site, affected RNA stability, since the deletion of four cryptic AG sites from the procyclin 5′ UTR had no effect on luciferase activity.
Previous work showed that the PGK A, B, and C mRNA levels differ, although they are transcribed as a polycistronic unit (10). These results led the authors to suggest that the differences in mRNA concentration must be controlled posttranscriptionally, by differences in trans-splicing efficiency. We were able to confirm that the different URs for PGK A, B, and C indeed resulted in very different levels of trans-splicing efficiency. These results strongly suggest that trans splicing can play an important role in posttranscriptional gene control.
Acknowledgments
This work was supported by NIH grant AI21729 and by Rockefeller University Institutional Funds.
We thank our colleagues, especially Magda Konarska, Shuba Gopal, and Terry Gaasterland, and members of the Cross lab, especially Christian Janzen and Bibo Li, for encouragement and useful discussions.
REFERENCES
- 1.Anderson, K., and M. J. Moore. 1997. Bimolecular exon ligation by the human spliceosome. Science 276:1712-1716. [DOI] [PubMed] [Google Scholar]
- 2.Brun, R., and M. Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 3.Chen, S., K. Anderson, and M. J. Moore. 2000. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc. Natl. Acad. Sci. USA 97:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiara, M. D., L. Palandjian, R. Feld Kramer, and R. Reed. 1997. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 16:4746-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K., and R. Reed. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 21:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csank, C., F. M. Taylor, and D. W. Martindale. 1990. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 18:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshler, J. O., and J. J. Rossi. 1991. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 5:1252-1263. [DOI] [PubMed] [Google Scholar]
- 9.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-559. In R. F. Gesteland, T. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Gibson, W. C., B. W. Swinkels, and P. Borst. 1988. Post-transcriptional control of the differential expression of phosphoglycerate kinase genes in Trypanosoma brucei. J. Mol. Biol. 201:315-325. [DOI] [PubMed] [Google Scholar]
- 11.Gopal, S., G. A. M. Cross, and T. Gaasterland. 2003. An organism-specific method to rank predicted coding regions in Trypanosoma brucei. Nucleic Acids Res. 31:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graveley, B. R., K. J. Hertel, and T. Maniatis. 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7:806-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guth, S., T. O. Tange, E. Kellenberger, and J. Valcarcel. 2001. Dual function for U2AF35 in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 21:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J., and L. H. T. van der Ploeg. 1991. Requirement of a polypyrimidine tract for trans-splicing in trypanosomes: splice acceptor site. EMBO J. 10:3877-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummel, H. S., R. D. Gillespie, and J. Swindle. 2000. Mutational analysis of 3′ splice site selection during trans-splicing. J. Biol. Chem. 275:35522-35531. [DOI] [PubMed] [Google Scholar]
- 17.Ismaili, N., D. Perez-Morga, P. Walsh, M. Cadogan, A. Pays, P. Tebabi, and E. Pays. 2000. Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70 kDa proteins. Mol. Biochem. Parasitol. 106:109-120. [DOI] [PubMed] [Google Scholar]
- 18.Ismaili, N., D. Perez-Morga, P. Walsh, A. Mayeda, A. Pays, P. Tebabi, A. R. Krainer, and E. Pays. 1999. Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol. Biochem. Parasitol. 102:103-115. [DOI] [PubMed] [Google Scholar]
- 19.Kapotas, N., and V. Bellofatto. 1993. Differential response to RNA trans-splicing signals within the phosphoglycerate kinase gene cluster in Trypanosoma brucei. Nucleic Acids Res. 21:4067-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielkopf, C. L., N. A. Rodionova, M. R. Green, and S. K. Burley. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106:595-605. [DOI] [PubMed] [Google Scholar]
- 21.Le Blancq, S. M., B. W. Swinkels, W. C. Gibson, and P. Borst. 1988. Evidence for gene conversion between the phosphoglycerate kinase genes of Trypanosoma brucei. J. Mol. Biol. 200:439-447. [DOI] [PubMed] [Google Scholar]
- 22.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Estraño, C., C. Tschudi, and E. Ullu. 1998. Exonic sequences in the 5′ untranslated region of α-tubulin mRNA modulate trans splicing in Trypanosoma brucei. Mol. Cell. Biol. 18:4620-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucke, S., K. Jurchott, L. H. Hung, and A. Bindereif. 2005. mRNA splicing in Trypanosoma brucei: branch-point mapping reveals differences from the canonical U2 snRNA-mediated recognition. Mol. Biochem. Parasitol. 142:248-251. [DOI] [PubMed] [Google Scholar]
- 25.Madhani, H. D., and C. Guthrie. 1994. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28:1-26. [DOI] [PubMed] [Google Scholar]
- 26.Manger, I. D., and J. C. Boothroyd. 1998. Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol. Biochem. Parasitol. 97:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 28.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 29.Metzenberg, S., and N. Agabian. 1996. Human and fungal 3′ splice sites are used by Trypanosoma brucei for trans splicing. Mol. Biochem. Parasitol. 83:11-23. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, W. J., K. P. Watkins, and N. Agabian. 1986. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell 47:517-525. [DOI] [PubMed] [Google Scholar]
- 31.Patterson, B., and C. Guthrie. 1991. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell 64:181-187. [DOI] [PubMed] [Google Scholar]
- 32.Patzelt, E., K. L. Perry, and N. Agabian. 1989. Mapping of branch sites in trans-spliced pre-mRNAs of Trypanosoma brucei. Mol. Cell. Biol. 9:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 34.Ruskin, B., A. R. Krainer, T. Maniatis, and M. R. Green. 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38:317-331. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schaal, T. D., and T. Maniatis. 1999. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaal, T. D., and T. Maniatis. 1999. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19:1705-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, C. W., T. T. Chu, and B. Nadal-Ginard. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 13:4939-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. W., E. B. Porro, J. G. Patton, and B. Nadal-Ginard. 1989. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Sutton, R. E., and J. C. Boothroyd. 1988. Trypanosome trans-splicing utilizes 2′-5′ branches and a corresponding debranching activity. EMBO J. 7:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vazquez, M., C. Atorrasagasti, N. Bercovich, R. Volcovich, and M. J. Levin. 2003. Unique features of the Trypanosoma cruzi U2AF35 splicing factor. Mol. Biochem. Parasitol. 128:77-81. [DOI] [PubMed] [Google Scholar]
- 42.Wentz-Hunter, K., and J. Potashkin. 1996. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 24:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirtz, E., C. Hartmann, and C. Clayton. 1994. Gene expression mediated by bacteriophage T3 and T7 RNA polymerases in transgenic trypanosomes. Nucleic Acids Res. 22:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirtz, E., M. Hoek, and G. A. M. Cross. 1998. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 26:4626-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 46.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 47.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355:609-614. [DOI] [PubMed] [Google Scholar]
- 48.Zhuang, Y., and A. M. Weiner. 1990. The conserved dinucleotide AG of the 3′ splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene 90:263-269. [DOI] [PubMed] [Google Scholar]
- 49.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]