Abstract
The polypyrimidine tract binding protein (PTB) has been described as a global repressor of regulated exons. To investigate PTB functions in a physiological context, we used a combination of morpholino-mediated knockdown and transgenic overexpression strategies in Xenopus laevis embryos. We show that embryonic endoderm and skin deficient in PTB displayed a switch of the α-tropomyosin pre-mRNA 3′ end processing to the somite-specific pattern that results from the utilization of an upstream 3′-terminal exon designed exon 9A9′. Conversely, somitic targeted overexpression of PTB resulted in the repression of the somite-specific exon 9A9′ and a switch towards the nonmuscle pattern. These results validate PTB as a key physiological regulator of the 3′ end processing of the α-tropomyosin pre-mRNA. Moreover, using a minigene strategy in the Xenopus oocyte, we show that in addition to repressing the splicing of exon 9A9′, PTB regulates the cleavage/polyadenylation of this 3′-terminal exon.
Alternative splicing, the process by which multiple mRNAs can be produced from a single primary transcript, is a major mechanism in metazoans to expand the complexity of gene expression. It is now well documented that 35 to 60% of human genes possess alternative exons (3, 9, 27, 28), and a large fraction of the alternative splicing events alter the resulting protein products. Hence, alternative pre-mRNA processing is considered to be the most important source of protein diversity in vertebrates (17, 28). Resch et al. (33) proposed that alternative splicing could modulate protein interaction networks by the addition or removal of specific protein-protein interaction domains. The bioinformatic observation that ∼30% of human alternative transcripts contain premature termination codons, making them a target for nonsense-mediated decay, also suggests that alternative splicing could be part of an integrated mechanism that controls the level of gene expression (22). The consequences of alternative splicing do not only concern the protein products. Indeed, differential splicing of 3′-terminal exons associated with alternative poly(A) site selection can generate mRNAs that differ in their 3′ untranslated sequences, with consequences for stability, localization, and translation (28).
Despite the considerable efforts to dissect the mechanisms of tissue-specific alternative splicing (reviewed in reference 2), very few reports have studied the regulation of alternative splicing in a physiological context. One exception is the Drosophila model, where elegant and powerful genetic studies associated with biochemical approaches led to the dissection of the sex determination pathway and the demonstration that sex-specific factors regulate a cascade of RNA processing events (reviewed in reference 24). However, in vertebrates, the implication of tissue-specific factors in controlling alternative splicing events appears limited. Consequently, it was proposed that ubiquitously expressed splicing factors could determine a cellular code based on variations in their relative levels or activities in different tissues (37). Several RNA binding protein families that are pivotal for alternative splice site choices were identified in vertebrates by biochemical approaches. These splicing factors, which include the SR and SR-like proteins, the CELF proteins, and several hnRNP proteins, were essentially validated as regulatory splicing factors in cultured cells using minigene models. However, the role of these factors in regulating specific alternative splicing events in a physiological context is poorly documented. Loss-of-function strategies in a whole organism can provide such information, and this is an important approach to discern general from specific functions of these factors. For example, despite a strong functional redundancy of the different SR proteins in constitutive and alternative splicing regulation, it was recently demonstrated by conditional gene targeting in the mouse that the ASF/SF2 SR protein functions in specific pathways and is a key regulator of specific alternative splicing events during heart development (46).
The polypyrimidine tract binding protein (PTB) is an abundant hnRNP protein that is conserved through metazoans. In vertebrates, PTB is widely expressed, and it has been implicated in the repression of several alternative spliced exons including exon 7 of β-tropomyosin (30), exon N of GABAγ2 (1), exons IIIb and IIIc of FGF-R2 (4, 20), exon N1 of c-src (6), exon SM of α-actinin (38), exon EIIIb of fibronectin (32), exon 3 of α-tropomyosin (11), exon 9 of caspase-2 (8), exon M2 of immunoglobulin M (36), and exon 9 of CFTR (47). The physiological relevance of PTB in alternative exon repression was demonstrated by RNA interference-mediated depletion in cultured cells that leads to an increase in exon IIIb of FGF-R2 and exon EIIIb of fibronectin inclusion (42). Based on these data, it was proposed that PTB has a general function in preventing the definition of regulated exon (41). More recently, a general role for PTB in the repression of the cleavage/polyadenylation processus by competing with CstF64 binding to the downstream U or U/G region of a poly(A) site was proposed (5). In addition to these functions in pre-mRNA processing, PTB has also been implicated in cytoplasmic events such as internal initiation of translation (16), mRNA localization (7), and mRNA stability (12, 18, 40).
The well-documented general role of PTB in antagonizing exon definition does not exclude that PTB may also be involved more specifically in several pre-mRNA processing events. We have developed the Xenopus laevis embryo as an in vivo model to study the basis of tissue-specific splicing regulation. Using this model, we have previously shown that PTB is involved in the repression of the 3′-terminal exon 9A9′ of the α-tropomyosin (α-TM) pre-mRNA in nonmuscle cells. Several high-affinity PTB binding sites, whose mutation impairs PTB binding and generates exon 9A9′ derepression, were identified in the intron upstream of this exon. Using a morpholino-based translational inhibition strategy, we also conclusively demonstrated that PTB was required in vivo to repress exon 9A9′ in epidermal cells in the context of a minigene (13). The strong derepression of exon 9A9′ we observed suggested that PTB can be an important regulator of the tissue-specific 3′ end processing of the α-TM pre-mRNA. To address this question in a physiological context, we misexpressed PTB in developing embryos and studied the effects upon endogenous α-TM pre-mRNA processing. We show that morpholino-mediated depletion of PTB provokes a strong switch in nonmuscle tissues from the nonmuscle isoform to the striated muscle isoforms that result from the inclusion of exon 9A9′. Conversely, targeted overexpression of PTB in the somites causes a switch towards the nonmuscle isoform in which exon 9A9′ is skipped. Our results demonstrate that the relative level of PTB is determinant for the tissue-specific switches of α-TM isoforms and establish PTB as a key regulator in the 3′ end processing of the α-TM pre-mRNA. Exon 9A9′ being a 3′-terminal exon, its skipping in nonmuscle cells could be the result of repression of splicing or cleavage/polyadenylation, or both. Using a minigene strategy in which the 3′ splice site or the poly(A) site of exon 9A9′ has been deleted, we found that PTB can repress both processes independently. These data indicate that in addition to repressing splicing, PTB can directly regulate the cleavage/polyadenylation of a 3′-terminal exon.
MATERIALS AND METHODS
Plasmid constructs.
The α-TM minigenes pSV40.7-9B, pSV40.7-9BΔαe, and pSV40.7-9BΔ3′ss were described previously (13). The α-TM pSV40.7-9A9′ and pSV40.7-9A9′Δ3′ss constructs were derived from the pSV40.7-9B and pSV40.7-9BΔ3′ss minigenes, respectively, by generating a KpnI/SalI digestion, a T4 DNA polymerase treatment (Biolabs), and a ligation of the ends. The mutated minigenes were produced by primer-directed PCR mutagenesis and cassette substitutions as previously described (13). The pSV40.βGlobinβpA minigene was obtained by insertion of a BglII/SalI fragment derived from a PCR amplification of pVC-CMV-βGlobinβpA (5) into the pBS-SV40 vector using the following primers: 5′-GCAGATCTACTAGCAACCTCAAACAGAC-3′ and 5′-CCGTCGACTGCACTGACCTCCCACATTC-3′. The plasmids pGEM-150PY and pGEM-150pBS were described previously (13). The polymerase III (Pol III) adenovirus-associated (VA) reporter and the corresponding RNase protection probe plasmid were a gift of D. Bentley. The cardiac actin.GFP (pKS+) expression plasmid was a gift of T. J. Mohun. The cardiac actin.xPTB/V5-His plasmid was constructed as follows. The Xenopus cardiac actin promoter was amplified by PCR from plasmid 598 (29) using an upstream primer containing a BamHI restriction site (5′-GGGGATCCTGCACATTGGCTGTTTCCCA-3′) and a downstream primer containing a HindIII restriction site (5′-AAAAGCTTCCGTGCTGTGATTGAATTGG-3′). This fragment was cloned into the pcDNA3.1/V5-HisC vector (Invitrogen) digested with BglII and HindIII to remove the cytomegalovirus promoter, producing the pcDNA3.1-cardiac actin vector. The open reading frame of xPTB was amplified by PCR from the hnRNPI cDNA (7) using an upstream primer containing a BamHI restriction site (5′-GGGGATCCACCATGGAAGGAATTGTTCAAG-3′) and a downstream primer designed to remove the stop codon and containing a NotI restriction site (5′-GGGCGGCCGCAATTGTGGATTTGGAAAAG-3′). The product was cloned as a BamHI/NotI fragment into the pcDNA3.1-cardiac actin vector in frame with the carboxyl V5-His epitope tag.
Xenopus embryos and dissection.
Xenopus laevis embryos were obtained by artificial fertilization of eggs from laboratory-reared females. Embryos were staged according to the table of Nieuwkoop and Faber (31). Embryo dissections were performed in Steinberg medium under a microscope.
Morpholino oligonucleotide injection and xPTB rescue.
Morpholino oligonucleotides (Mo) were obtained from Gene Tools, LLC. xPTB Mo and control Mo have been described previously by Hamon et al. (13). Twenty-five nanograms/blastomere was injected into both blastomeres of two-cell-stage Xenopus embryos. For xPTB rescue, 100 pg of V5-tagged xPTB mRNA described previously by Hamon et al. (13) was coinjected with xPTBMo into each blastomere of two-cell-stage embryos.
Oocyte culture and injection.
Pieces of Xenopus laevis ovary were dissected and manually treated with collagenase to isolate individual stage V or VI oocytes. Before nuclear injection, oocytes were centrifuged for 15 min at 600 × g to visualize the germinal vesicle. Injection and culture of oocytes were carried out in OR2 medium as described in reference 13. One nanogram of α-TM or β-globin βpAsite minigenes was coinjected into the nucleus with 0.1 ng of VA minigene. For titration experiments, 15 ng of in vitro-transcribed RNA was coinjected with minigenes.
Transgenesis by sperm nuclear transplantation into fertilized eggs.
Transgenic Xenopus laevis embryos were produced by using the restriction enzyme-mediated integration method (REMI) as described previously by Kroll and Amaya (19), with minor modifications. In each reaction mixture, 65 ng of cardiac actin.GFP (pKS+) and cardiac actin.xPTB/V5-His constructions linearized with SalI and SmaI, respectively, were used to generate transgenic frogs.
Western blot analysis.
Western blot analysis was carried out as described previously (13) using the following antibodies: purified xPTB antibodies (1:600), monoclonal PCNA antibodies (1:5,000), and monoclonal V5 antibodies (1:5,000). The proteins were revealed using the enhanced chemifluorescence technique (Amersham Biosciences) and quantified by PhosphorImager analysis (Amersham Biosciences).
RT-PCR analysis of the endogenous transcripts.
RNA isolated from embryos with Tri Reagent solution (MRC Inc.) was treated with RQ1 DNase (Promega). Four micrograms of RNA was then used for the reverse transcription (RT) reaction using oligo(dT) as a primer. One-fifth of the RT reaction was then used for the PCR amplification of c-src transcripts using the 32P-labeled ex3-src sense primer (5′-GGGGGAAAGACTACAGATTG-3′) and ex4-src antisense primer (5′-AGCAGACGCTCTGCTTCTC-3′) and of α-actinin transcripts using the 32P-labeled exEF1a sense primer (5′-TGCCAAAGGCATTACCCAAG-3′) and exEF2 antisense primer (5′-GTTGGGGTCAACGATGCTC-3′). Amplification products were separated on a 7% acrylamide gel and quantified by PhosphorImager analysis (Amersham Biosciences). The amplification product obtained with α-actinin primers that correspond to the double inclusion of exon NM and exon SM was sequenced to verify its identity (Genome Express).
RT-PCR assay of microinjected minigenes.
RNA isolated from oocytes was treated with Turbo DNase (Ambion). Five micrograms of RNA was then used for reverse transcription reactions using different specific primers: ex9A antisense primer (5′-CGGAATTCATTGAAGTCATGTCATTGAG-3′) for analysis of exon 8 to exon 9A α-TM splicing and ex3 antisense primer (5′-GGAGCTCCAGTTTAGTAGTTGGACTTAG-3′) for analysis of exon 2 to exon 3 β-globin splicing. One-fifth of the RT reactions was then used for the PCR amplifications which were performed using the 32P-labeled ex8 sense primer (5′-CGGGATCCGTGCTGAGTTTGCAGAGAG-3′) and ex9A antisense primer and the 32P-labeled ex2 sense primer (5′-ACGTGGATCCTGAGAACTTC-3′) and ex3 antisense primer. Amplification products were separated on a 4% acrylamide gel and quantified by PhosphorImager analysis (Amersham Biosciences).
Analysis by RNase protection assay.
The RNase protection probe (α74) used for the detection of endogenous α-TM transcripts was described previously (15). The EF1α RNA probe template was PCR amplified from plasmid pXeX (a gift of P. Krieg) using the forward (5′-CGGGATCCCAACTGATAAGCCTCTCCGT-3′) and reverse (5′-CGGAATTCACCAGTTTCCACACGACCAA-3′) primers. The MLC1f RNA probe template was PCR amplified from plasmid XMLC1f (39) using the forward (5′-CGGGATCCATGCTGAGGTCAAGAAGA-3′) and reverse (5′-CGGAATTCAAACTCAATCCTCTTGGC-3′) primers. The RNA probe template for analyzing α-TM cleaved and uncleaved transcripts was PCR amplified from the pSV40.7-9B plasmid using the forward (5′-CGGGATCCGAGCTGTATGCTCAGAAAC-3′) and reverse (5′-CGGAATTCTTGCCAGATGTCTAAACAGG-3′) primers. The RNA probe template for analyzing βglobinβpA cleaved and uncleaved transcripts was PCR amplified from the pSV40.βglobinβpA plasmid using the forward (5′-CGGGATCCTGGCTAATGCCCTGGCCCAC-3′) and reverse (5′-CGGAATTCTTGCCAGATGTCTAAACAGG-3′) primers. The RNA probe template for analyzing splicing of exon 7 to exon 8 in the α-TM reporter was PCR amplified from the XTMO5 plasmid (14) using the forward (5′-CGGGATCCAGAAGGAGGACAAATATG-3′) and reverse (5′-CGGAATTCCTGGCACTCAAGAGCAAG-3′) primers. All the PCR products generated were digested with BamHI and EcoRI and cloned into these sites of the pBS vector. The resulting plasmids were linearized with XbaI, and transcription was carried out with T7 RNA polymerase (Promega). Different amounts of total RNA were analyzed using the RNase protection assay a previously described (15). The digested products were quantified with a PhosphorImager (Amersham Biosciences).
RESULTS
A single xPTB isoform is present in Xenopus embryos.
Three alternatively spliced isoforms of PTB (PTB1, PTB2, and PTB4) have been described in mammalian cells that have distinct activities upon alternative splicing. Since the ratio of PTB isoforms varies between different cell types, it was proposed that their ratio may control some alternative splicing events (44). Western blot analysis of mammalian PTB reveals a typical doublet, the faster- and slower-migrating bands corresponding to PTB1 and PTB2/4, respectively. In Xenopus laevis, Western blot analysis using a specific antibody (13; also see Fig. 4) revealed only one band, suggesting that the situation may differ. The cloned hnRNPi cDNA we previously used encodes a protein similar to PTB4. Several data suggested that this is the major isoform: (i) it is the only cDNA present in expressed sequence tag libraries, (ii) the internal splice site within exon 9 used in mammals to produce PTB2 is absent in Xenopus, (iii) extensive PCR analysis failed to amplify cDNAs corresponding to PTB1 (data not shown), and (iv) the Xenopus PTB comigrated with PTB4 (data not shown). We concluded from these observations that a single PTB isoform corresponding to PTB4 is present in Xenopus embryos, and we designated it xPTB. This relative simplicity provides an advantageous system to study the specific role of PTB in the control of alternative splicing.
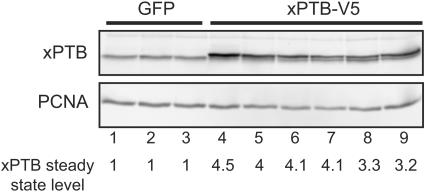
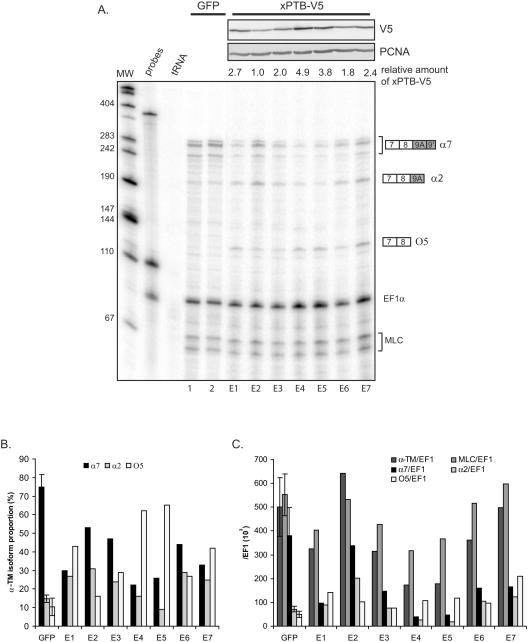
FIG. 4.
Targeted overexpression of xPTB in the somites. The somites of GFP and xPTB-transgenic embryos were dissected, and the xPTB steady-state level in each somitic fraction was analyzed by Western blot analysis using an affinity-purified xPTB antibody. PCNA was used as a loading control. Quantification of the overall xPTB signal is shown below gels with values for GFP control embryos set as 1.
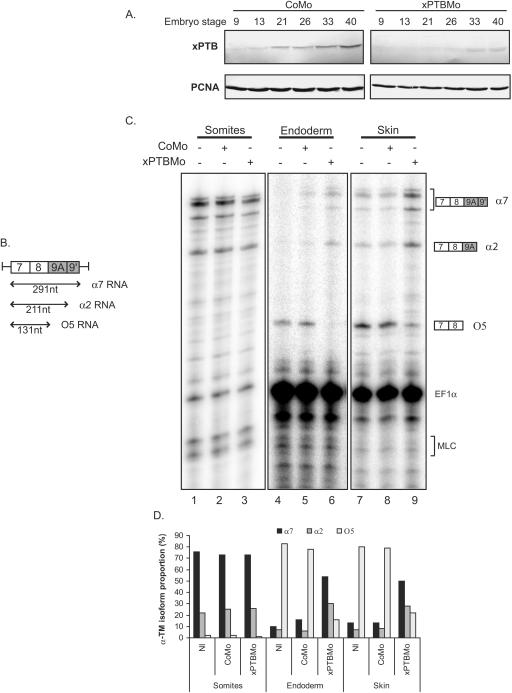
The composite internal/3′-terminal exon 9A9′ is strongly derepressed in nonmuscle tissues of xPTBMo-injected embryos.
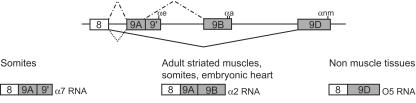
The Xenopus α-TM gene contains an alternative 3′-terminal exon (exon 9A9′) which is subjected to different splicing patterns according to the tissue environment (Fig. 1). Exon 9A9′ is skipped in nonmuscle cells to produce O5 RNA, whereas it is used as a terminal or internal exon in the somitic and adult striated muscle cells to produce α7 and α2 RNAs, respectively (14, 15). We have previously shown, using a morpholino-mediated specific knockdown of xPTB and a minigene strategy, that endogenous xPTB is required in vivo to repress the composite internal/3′-terminal exon 9A9′ in embryonic epidermal cells (13). In this study, we examined the effects of xPTB depletion upon endogenous exon 9A9′ splicing. A morpholino antisense oligomer (xPTBMo) directed against the 5′ region of xPTB mRNA or a control nonspecific morpholino oligomer (CoMo) were microinjected into both blastomeres of two-cell-stage Xenopus embryos, and the embryos were allowed to develop. The level of xPTB throughout development was analyzed by Western blot analysis (Fig. 2A). Compared to control embryos, xPTBMo strongly decreased the level of xPTB from stage 13 (neurulae) to stage 40 (early tadpole). The specificity of xPTBMo was previously demonstrated by rescue experiments with xPTB mRNA that was not a target for the morpholino (13). The xPTBMo- and CoMo-injected embryos were dissected into three parts: somites, skin, and endoderm. RNA isolated from each fraction was assayed by RNase protection using an RNA-labeled probe spanning exon 9A9′ that can distinguish between the splicing of exon 9A9′ as a 3′-terminal exon (α7 RNA) and as an internal exon (α2 RNA) and the skipping of exon 9A9′ (O5 RNA) (Fig. 2B). Two probes directed to EF1α and MLC1f/3f RNA were included in the assay as controls for RNA recovery and fraction purity, respectively. As described for Fig. 1, α7 and α2 RNAs were strongly expressed in the somites representing ∼75% and ∼20% of the α-TM transcripts, respectively, whereas O5 RNA is hardly detectable and accounts for less than 5% of the α-TM mRNA (Fig. 2C, lane 1, and D). By contrast, in the endoderm and skin fractions, O5 is the major α-TM transcript (Fig. 2C, lanes 4 and 7, and D). The faint α2 and α7 signals detected in both nonmuscle tissues are probably due to a contamination by residual somitic tissue since the somite-specific MLC1f/MLC3f RNA was also present in both fractions. No significant differences in the pattern and level of α-TM transcripts were observed in the somitic fraction of xPTBMo-injected embryos (Fig. 2C, compare lanes 1 and 3). In contrast, depletion of xPTB strongly affected the splicing pattern of endogenous α-TM transcripts in the two nonmuscle tissues. α7 and α2 RNA accumulated strongly to represent 65% and 25% of the α-TM transcripts, respectively, while O5 RNA was reduced to less than 10% of the α-TM mRNA (Fig. 2C, lanes 6 and 9, and D). The quantification of the α-TM-protected fragments normalized to EF1α transcripts indicated that the overall amount of α-TM transcripts was practically identical in the xPTBMo-injected embryos and in the noninjected embryos in the three embryonic fractions studied. These results show that decreasing xPTB in skin and endoderm switches the splicing pattern from the O5 isoform to the α2 and α7 isoforms. This indicates that xPTB depletion derepresses exon 9A9′ usage. Altogether, these results demonstrate that xPTB is required in vivo to repress exon 9A9′ as a 3′-terminal exon but also as an internal exon in the context of the endogenous pre-mRNA.
FIG. 1.
Differential processing of the 3′-terminal region of the Xenopus α-TM pre-mRNA. The 3′-terminal region of the Xenopus α-TM RNA and its alternative RNA processing pattern are schematically diagrammed. The boxes represent exons, and horizontal lines represent introns. αe, αa, and αnm represent the polyadenylation signals present within exons 9A9′, 9B, and 9D, respectively. Below the diagram, the alternative splicing products in somites, adult striated muscle, and nonmuscle tissues are indicated.
FIG. 2.
Morpholino-mediated depletion of xPTB switches the selection of the 3′-terminal exons of the α-TM pre-mRNA. A. Morpholino-mediated depletion of xPTB in developing embryos. The steady-state level of xPTB in CoMo- and xPTBMo-injected embryos throughout development was studied by Western blot using affinity-purified xPTB antibody. PCNA is used as a loading control. B. Schematic representation of the RNase protection probe used to analyze 3′ end processing of the endogenous α-TM pre-mRNA. The sizes of the predicted RNase protection fragments are indicated below the probe. C. Somites, endoderm, and skin fractions of uninjected or CoMo- or xPTBMo-injected stage 35 embryos were analyzed by RNase protection assay using the probe illustrated in panel B. EF1α and MLC1/3 probes were included in the assay as controls for RNA recovery and fraction purity, respectively. The identity or composition of each protected product is indicated at the right. Ten micrograms of the endoderm and skin RNAs and 2.5 μg of the somitic RNA were analyzed. D. Quantification of the effects of xPTB depletion upon the proportion of the different α-TM transcripts. The percentages of α2, α7, and O5 RNAs were calculated after normalization to the uridine content as a fraction of the total α-TM transcripts. Quantification was performed with a PhosphorImager. NI, noninjected embryos.
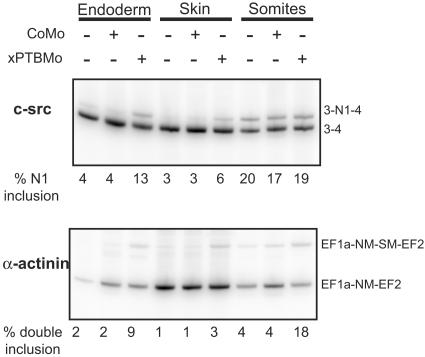
xPTB knockdown increases the inclusion of endogenous exon N1 of c-src and exon SM of α-actinin.
Since xPTB depletion dramatically changed the 3′ end maturation of the endogenous α-TM pre-mRNA, we next studied the effects of such depletion upon the splicing of several exons known to be repressed by PTB and accessible in Xenopus embryos. The splicings of exon N1 of c-src and exon SM of α-actinin were assayed by RT-PCR on the three embryonic fractions described above. In accordance with its neuron-specific splicing (21), exon N1 was strongly skipped in the endoderm and skin fractions, whereas this exon was present in 20% of the transcripts from the somitic fraction that includes the neural tube (Fig. 3, top). Depletion of xPTB resulted in a three- and twofold increase in exon N1 insertion in the endoderm and skin fractions, respectively, while no change was observed in the somitic fraction. As expected, exon SM (smooth muscle), which is specifically used in smooth muscle cells (43), was strongly skipped in the three fractions (Fig. 3, bottom). While no transcripts including only exon SM were amplified, a low but reproducible level of transcripts containing both NM (nonmuscle) and SM exons was also observed. The knockdown of xPTB resulted in a three- to fourfold increase in transcripts containing both exons NM and SM (Fig. 3, bottom).
FIG. 3.
Exon N1 of c-src and exon SM of α-actinin are partially derepressed in xPTB-depleted embryos. Endoderm, skin, and somite fractions of uninjected or CoMo- or xPTBMo-injected stage 35 embryos were analyzed by RT-PCR using primers specific for endogenous c-src and α-actinin to determine the extent of exon N1 and exon SM inclusion, respectively. The PCR products were quantified by PhosphorImager analysis. The percentage of exon N1 inclusion and NM-SM double inclusion are shown below gels.
These results confirmed, in a physiological context, the implication of xPTB in antagonizing exon N1 and exon SM definition. However, in contrast to exon 9A9′, the depletion of xPTB was not sufficient to radically switch the isoform ratio.
Somitic xPTB overexpression modifies the endogenous α-TM pre-mRNA 3′ end processing.
Since xPTB depletion resulted in a strong derepression of exon 9A9′ in nonmuscle tissues of the embryo, we tested if xPTB overexpression was sufficient to repress exon 9A9′ splicing in somitic tissues. To address this issue, we produced transgenic embryos that overexpress xPTB in the somites using the REMI method. A plasmid expressing V5-tagged xPTB and driven by the cardiac actin promoter that targets expression to the somites and the embryonic heart was constructed (actin-xPTB). A plasmid containing the green fluorescent protein (GFP) gene under the control of the cardiac actin promoter (actin-GFP) was also used to generate transgenic control embryos. GFP- and xPTB-transgenic embryos were scored for transgene expression by fluorescence and by Western blot analysis, respectively, using a V5 antibody. In both cases, ∼80% of the developing embryos were transgenic (data not shown). GFP- and xPTB-transgenic embryos presented similar small percentages of general abnormal development, and no apparent mutant phenotype was associated with these tadpoles (data not shown). Only the embryos presenting a normal development were analyzed thereafter.
With the REMI method, each transgenic embryo represents a distinct integration event; thus, some variations in the level of expression and in the phenotypic consequences are expected. Accordingly, the embryos were always analyzed individually in the following experiments. We first determined the level of xPTB overexpression in the somites. The somitic tissue from stage 39 embryos was dissected apart, and the proteins from individual fractions were analyzed by Western blot using the affinity-purified xPTB antibody (Fig. 4). As previously described (13), xPTB migrated as a single band in control GFP embryos (Fig. 4, lanes 1 to 3). In xPTB-transgenic embryos, a slower-migrating band corresponding to the V5-tagged xPTB was present (Fig. 4, lanes 4 to 9). Quantification of the overall xPTB signal and normalization with PCNA indicated that xPTB levels in xPTB-transgenic embryos were three- to fourfold higher than in control GFP siblings. Also, a decrease of the endogenous xPTB occurred in several embryos presenting a high level of the V5-tagged protein (Fig. 4, compare lanes 8 and 4, for example). This behavior is probably the consequence of a negative-feedback loop leading to nonsense-mediated decay of the xPTB mRNA as described previously for human cells (45).
We next examined the consequence of somitic xPTB overexpression upon the 3′ end processing of the endogenous α-TM pre-mRNA. For that purpose, RNA and proteins were extracted simultaneously from whole individual transgenic embryos. A whole embryo analysis reflects essentially the somitic α-TM gene expression because this gene is more strongly expressed in the somites than in the nonmuscle tissues (14). Expression of the V5-tagged xPTB was analyzed by Western blot with a V5 antibody, and RNA was assayed by RNase protection assay using the probe spanning exon 9A9′ as described in the legend of Fig. 2B. Two probes directed to EF1α RNA and MLC1f/3f were included in the assay as controls for normalization (Fig. 5A). A strong decrease of the full-length protected fragments corresponding to α7 mRNAs was observed in most xPTB-transgenic embryos compared to GFP control embryos (Fig. 5A, compare lanes E1 to E7 to lanes 1 and 2). In contrast, these same embryos presented an important rise of the partial protected fragment corresponding to O5 mRNAs. Quantification of the protected fragments revealed that the splicing pattern was strongly modified in the xPTB-transgenic embryos. The α7 RNA, which represent ∼75% of the α-TM transcripts in the GFP control embryos, was reduced in all the xPTB embryos and accounted for less than 33% in four embryos out of seven (Fig. 5B). In contrast, the O5 mRNA, which constitutes ∼10% of the α-TM transcripts in the GFP control embryos, rose in all the embryos and accounted for up to 65% in several embryos. A slight increase in the percentage of the α2 mRNA was also observed in most embryos. Western blot analysis with the anti-V5 antibody revealed a strong correlation between the level of V5-tagged xPTB and the switches of the different α-TM mRNA isoforms (Fig. 5A, top). Normalization of the α-TM-protected fragments to EF1α transcripts revealed that the changes of the α-TM isoform ratios are mainly attributable to a strong decrease (up to ninefold) in the steady-state level of α7 mRNA and a 1.5- to 2.5-fold rise in the steady-state level of O5 RNA (Fig. 5C); the steady-state level of α2 mRNA differs from one embryo to another. It is noteworthy that a decrease of the overall α-TM transcripts normalized to EF1α was observed in several embryos, indicating that the increase in O5 RNA did not compensate for the decrease in α7. A marked reduction in the steady-state level of the control MLC1f/3f mRNA was also observed in most transgenic embryos (Fig. 5C), indicating that the reduction in α-TM mRNA levels could be attributed in part to the general effect of overexpressing xPTB upon the cleavage/polyadenylation reaction (5). These data suggest that the modification of the 3′ end processing of the α-TM pre-mRNA results from a specific repression of exon 9A9′ and a general effect upon the cleavage/polyadenylation reaction. Also, they clearly demonstrate that an increase in xPTB level is sufficient to switch the somitic splicing pattern to the nonmuscle splicing pattern.
FIG. 5.
Targeted overexpression of xPTB in the somites represses the inclusion of exon 9A9′. A. Total RNAs extracted from individual GFP control and xPTB-transgenic embryos were analyzed by RNase protection using the probe described in the legend to Fig. 2B. V5-tagged xPTB expression was monitored by Western blot using a monoclonal V5 antibody (top). The relative amount of xPTB-V5 was quantified for each embryo, with the weakest value set as 1. EF1α and MLC1/3 probes were included in the assay as controls for RNA recovery and normalization. The identity or composition of each protected product is indicated at the right. B. PhosphorImager quantification of the effect of somitic xPTB overexpression on the proportion of the different α-TM transcripts. The percentage of α2, α7, and O5 RNA was calculated after normalization to the uridine content as a fraction of the total α-TM transcripts. The data for control GFP embryos are given as means ± standard deviations for eight embryos. C. PhosphorImager quantification of the effect of somitic xPTB overexpression on the α-TM and MLC1/3 transcript steady-state level. The protected fragments corresponding to the different α-TM isoforms and MLC1f/3f were normalized to the EF1α signal and uridine content. The data for control GFP embryos are given as means ± standard deviations for eight embryos.
xPTB represses the cleavage/polyadenylation of exon 9A9′ independently of splicing.
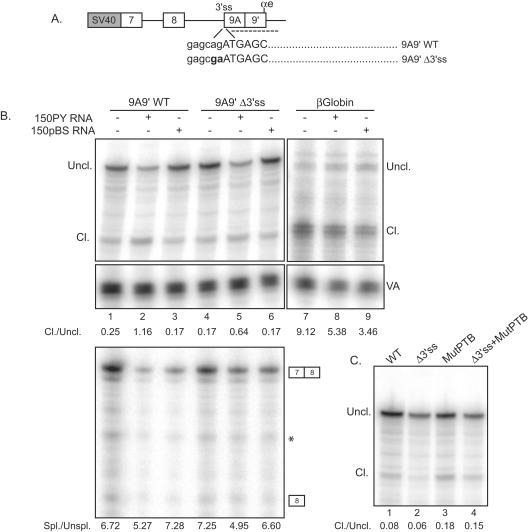
Our data demonstrate that xPTB antagonizes the definition of exon 9A9′ very efficiently compared to exon N1 of c-src and exon SM of α-actinin. Since exon 9A9′ is a 3′-terminal exon, this indicated that xPTB could act by repressing its poly(A) site. To study the cleavage/polyadenylation of exon 9A9′ independently of splicing, we took advantage of the observation that the endogenous exon 9A9′ is repressed in Xenopus oocytes and that an α-TM minigene encompassing exon 7 to exon 9B recapitulates the repression of exon 9A9′ (13). A wild-type (WT) α-TM minigene spanning exon 7 to exon 9A9′ and extending beyond the αe poly(A) site (9A9′WT) and a minigene that contains a mutation in the 3′ splice site of exon 9A9′ (9A9′Δ3′ss) were constructed (Fig. 6A). As previously reported for a minigene encompassing exon 7 to exon 9A9′ (13), RT-PCR analysis confirmed that exon 9A9′ was not spliced with this mutant (data not shown). The minigenes were injected into the germinal vesicle of oocytes with or without 150PY competitor RNA or 150pBS control RNA. The 150PY competitor RNA containing four high-affinity PTB binding sites was previously shown to bind xPTB very efficiently and to cause a complete derepression of exon 9A9′ in the oocyte in the context of a minigene. The 150pBS RNA is a 150-nt sequence from pBluescribe that does not bind xPTB (13). An RNA Pol III-VA reporter plasmid was included in all the assays to monitor the microinjection in the oocyte nucleus and RNA recovery. This gene was chosen because it is driven by a Pol III promoter, and the steady-state level of its transcripts is not affected by PTB (5). A β-globin gene construct was also used to evaluate any nonspecific effects of xPTB titration upon the 3′ end cleavage. RNase protection analysis using a probe that covers exon 9A9′ and intronic sequences downstream of the αe poly(A) site revealed that the wild-type α-TM transcripts were cleaved very inefficiently compared to the control β-globin transcripts, confirming the weakness of the αe poly(A) site (Fig. 6B, compare lanes 1 and 7). In the presence of 150PY competitor RNA, a ∼4.5-fold increase in the ratio of cleaved to uncleaved transcripts was detected for the wild-type α-TM pre-mRNAs, while the injection of the 150pBS control RNA did not change the efficiency of the 3′ end cleavage (Fig. 6B, compare lanes 1 to 3). The effects we observed are specific for the α-TM transcripts, since the 150PY competitor RNA resulted in only a slight decrease of the ratio of cleaved to uncleaved β-globin transcripts (Fig. 6B, compare lanes 7 and 8). A 3.8-fold increase in the ratio of cleaved to uncleaved transcripts was also observed in the presence of 150PY RNAs with the α-TM mutant inactive for exon 9A9′ splicing (Fig. 6B, compare lanes 4 and 5). This indicates that xPTB represses the cleavage/polyadenylation of exon 9A9′ independently of exon 9A9′ splicing. However, since the mutation of the 3′ splice site of exon 9A9′ results in the formation of a large 3′-terminal exon that spans exon 8 to exon 9A9′, it was possible that the stimulation of the 3′ end cleavage was caused by an increase in the efficiency of the splicing of exon 7 to exon 8. However, RNase protection analysis (Fig. 6B, bottom) showed that the splicing of exon 7 to exon 8 is very efficient, and quantitative comparison of the spliced to unspliced products revealed no significant change in the presence of the 150PY competitor RNAs. We previously identified four high-affinity UCUU PTB binding sites lying between the branch site and exon 9A9′ that are involved in PTB-mediated repression of exon 9A9′ (13). We therefore assayed the effects of mutating these four PTB binding sites upon the cleavage of the wild-type minigene and the splicing-deficient derivative (Fig. 6C). These PTB site mutations resulted in a ∼2-fold increase in the ratio of cleaved to uncleaved transcripts with both the wild-type reporter (Fig. 6C, compare lanes 1 and 3) and the splicing-deficient mutant (compare lanes 2 and 4), indicating that xPTB represses the cleavage/polyadenylation of exon 9A9′ in part by binding to these sites. Overall, these results demonstrate that xPTB specifically represses exon 9A9′ cleavage independently of splicing and that the four high-affinity PTB binding sites upstream of exon 9A9′ in part mediate this repression.
FIG. 6.
xPTB represses the cleavage/polyadenylation of exon 9A9′ independently of splicing. A. Schematic representation of the minigenes injected into Xenopus oocytes. The dinucleotide AG of the 3′ splice was mutated to GA to generate the 9A9′Δ3′ss mutant. The RNase protection probe used to discriminate the uncleaved and cleaved transcripts at the αe poly(A) site is represented with a broken line. B. RNase protection analysis of RNA extracted from Xenopus oocytes microinjected with the 9A9′WT and 9A9′Δ3ss constructions alone or with 150PY or 150pBS competitor RNA using the 3′ end RNase protection probe extending beyond the poly(A) site as represented in panel A (top) and a cDNA probe covering exons 7 and 8 (bottom). A representative experiment out of four is shown. A partially digested nonspecific band that appears in all six lanes is indicated with a star. A β-globin construct was included in the assay to test any general effect upon 3′ end cleavage (top). VA transcripts serve as a control for nucleus injection and loading of the RNA. Ratios of cleaved (Cl.) to uncleaved (Uncl.) or spliced (Spl.) to unspliced (Unspl.) transcripts were calculated from PhosphorImager data corrected for the uridine content of the protected fragments and are shown below each gel. C. RNA extracted from Xenopus oocytes microinjected with the 9A9′WT and 9A9′Δ3ss constructions mutated for the four PTB binding sites upstream of exon 9A9′ was analyzed by RNase protection assay as described above (B). The four PTB UCUU motifs were mutated to CCCC as described previously by Hamon et al. (13).
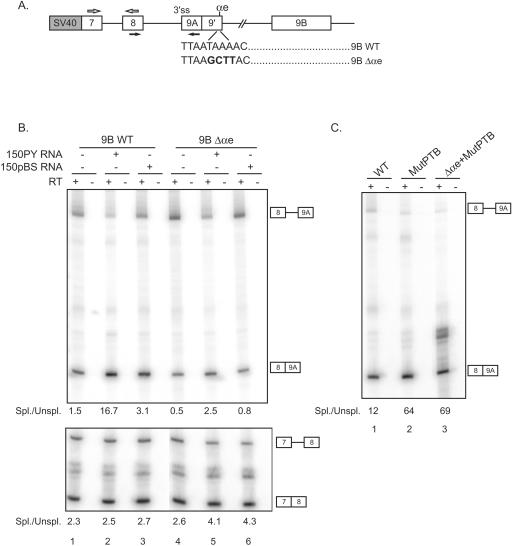
xPTB represses the splicing of exon 9A9′ independently of cleavage/polyadenylation.
In the experiments described above, the activation of the 3′ end cleavage/polyadenylation of exon 9A9′ following xPTB titration was more marked for the wild-type construct than for the splicing-inactive derivative (Fig. 6B, compare lanes 2 and 5). This observation raises the possibility that the splicing of exon 9A9′ was also repressed by xPTB. To test this possibility, a wild-type α-TM minigene encompassing exon 7 to exon 9B (9BWT) and a minigene in which the αe poly(A) site is mutated (9BΔαe) were constructed (Fig. 7A). It was previously shown that the mutation of the αe poly(A) hexanucleotide abolished the formation of α7 RNA (13). The different minigenes were injected into the germinal vesicles of oocytes with or without 150PY competitor RNA or 150pBS control RNA as described above. The efficiency of exon 9A9′ splicing was analyzed by RT-PCR using primers that hybridize in exon 8 and 9A (Fig. 7A). The splicing of exon 7 to exon 8 was also assayed to evaluate any general effect of xPTB titration upon splicing. In the presence of 150PY RNA, an ∼11-fold increase in the ratio of spliced to unspliced transcripts was observed for the α-TM wild-type construct (Fig. 7B, top, compare lanes 1 and 2), while only a twofold increase in the efficiency of splicing was noticed with the 150pBS control RNA. The effects are specific to exon 9A9′ splicing, as the efficiency of splicing of exon 7 to exon 8 was unaffected (Fig. 7B, bottom). The titration of xPTB also resulted in a fivefold increase in the ratio of spliced to unspliced transcripts for the cleavage/polyadenylation-inactive mutant 9BΔαe (Fig. 7B, top, compare lanes 4 and 5), indicating that xPTB can repress the splicing of exon 9A9′ independently of the cleavage/polyadenylation reaction. In order to determine whether xPTB represses splicing through the four high-affinity PTB binding sites present within the intron upstream of exon 9A9′, we compared the effect on splicing of their mutation on the wild-type α-TM reporter and the mutant inactive for the cleavage/polyadenylation reaction (Fig. 7C). Mutation of the four PTB binding sites resulted in a ∼5-fold increase in the ratio of spliced to unspliced transcripts with wild-type α-TM (Fig. 7C, compare lanes 1 and 2) and the mutant inactive for exon 9A9′ cleavage/polyadenylation (compare lanes 1 and 3), indicating that PTB represses the splicing of exon 9A9′ in part by binding to these sites. Together with the above-described results, these data demonstrate that xPTB represses both the splicing and the cleavage/polyadenylation of exon 9A9′ and that the four PTB binding sites lying between the branch site and the 3′ AG border are involved in the assembly of a repressor complex that targets both processes.
FIG. 7.
xPTB represses the splicing of exon 9A9′ independently of polyadenylation. A. Schematic representation of the minigenes injected into Xenopus oocytes. The hexanucleotide AATAAA of the αe poly(A) site was mutated to AAGCTT to produce the 9BΔαe mutant. The primers used to amplify the spliced and nonspliced transcripts are represented by black and white arrows. SV40, simian virus 40. B. RT-PCR analysis of RNA extracted from Xenopus oocytes microinjected with the 9BWT and 9BΔαe constructions alone or with 150PY or 150pBS competitor RNAs. A representative experiment out of three is shown. Ratios of spliced (Spl.) to unspliced (Unspl.) products were calculated from PhosphorImager data and are indicated below each gel. C. RNA extracted from Xenopus oocytes microinjected with the 9BWT and 9BΔαe constructions mutated for the four PTB binding sites upstream of exon 9A9′ (MutPTB) was analyzed by RT-PCR as described above (B). The four PTB UCUU motifs were mutated to CCCC as described previously by Hamon et al. (13).
xPTB is an essential protein in Xenopus development.
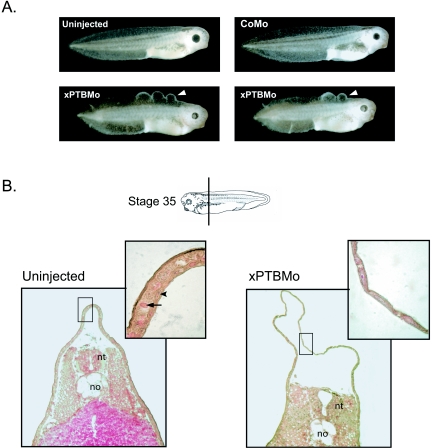
The xPTBMo-injected embryos showed a slight delay in development, but they were especially characterized by a dorsal abnormal skin morphology which resulted, from the late tailbud stage, in the development of blister-like structures back to the head (Fig. 8A, bottom). Thereafter, the majority of the embryos displayed a skin necrosis which was probably the cause of embryo death at the late tadpole stage. Histological sections of control and xPTBMo-injected embryos at stage 35 showed that xPTBMo-injected embryos had a dramatic increase of the dorsal epidermis area compared to control embryos (Fig. 8B). Higher-magnification observation revealed that a single layered epidermis was present within the blister-like structures instead of the two layers, a superficial epithelial layer and a deep sensorial one, normally observed. This suggests that the blister-like phenotype results from a defect in cell movements that leads to an intercalation of cells destined to form both layers.
FIG. 8.
The morpholino-mediated depletion of xPTB in developing embryos results in dorsal skin defects. A. Blister-like structures (white arrowheads) were induced in stage 35 embryos by the injection of 50 ng of xPTBMo; two representative embryos are shown. No skin defects are visible in uninjected embryos or CoMo-injected embryos. B. Transverse sections through uninjected or xPTBMo-injected stage 35 embryos (the plane of section is indicated by a bar) stained with hematoxylin-eosin-saffron. The epidermis area of the dorsal fin appears dramatically enlarged in xPTBMo-injected embryos. Higher magnification reveals a single-layered epidermis where blisters develop instead of two layers, a superficial epithelial layer and a deep sensorial one, in control uninjected embryos. Black arrowheads and arrows point to the nuclei and yolk platelets, respectively. no, notochord; nt, neural tube.
To confirm that the xPTBMo-induced phenotype was generated by a specific knockdown of xPTB, a rescue experiment was performed with in vitro-transcribed xPTB mRNA that has a third-base modification in the sequence spanning the translation start site. To monitor the efficiency of the rescue, the phenotype was graded according to the size and number of blister-like structures (Table 1). The rescue is efficient, with 56% of the embryos presenting a complete or partial rescued phenotype (Table 1) as opposed to 5% in the absence of injected xPTB mRNA. These results show that the effects upon development are a direct consequence of xPTB knockdown and that xPTB is an essential factor in Xenopus development.
TABLE 1.
Coinjection of sense xPTB RNA with xPTBMo results in the rescue of the mutant phenotypesa
| Embryo injection | % of embryos (no.) of phenotype:
|
||||
|---|---|---|---|---|---|
| Blistersb
|
Deformed | ||||
| − | −/+ | + | ++ | ||
| CoMo | 83 (45) | 0 (0) | 0 (0) | 0 (0) | 17 (9) |
| xPTBMo | 0 (0) | 5 (2) | 30 (13) | 56 (24) | 9 (4) |
| xPTBMo + sense xPTB RNA | 22 (18) | 34 (28) | 23 (19) | 11 (9) | 10 (8) |
Frequency of the phenotype obtained from embryos coinjected with sense xPTB RNA and xPTBMo compared to xPTBMo- and CoMo-injected embryos. The numbers of scored embryos are indicated in parentheses. The embryos were observed at stage 35, and mutant phenotypes were graded according to the size and number of blister-like structures. Embryos presenting a general abnormal development (deformed) were also scored. Three independent experiments were carried out.
−, no blister; −/+, one small blister; +, one large blister; ++, more than one large blister.
DISCUSSION
Our results highlight the importance of PTB as a regulator of tissue-specific splicing. There have been many reports of the splicing repressor activity of PTB leading to the proposal that PTB was a global repressor of regulated exons (reviewed in reference 41). Our data go beyond this proposal with the demonstration, in a physiological context, that PTB, in addition to repressing splicing, is a negative regulator of polyadenylation and a key regulator of the 3′ end processing of the α-TM pre-mRNA.
PTB in the regulation of the 3′ end processing of the α-TM pre-mRNA.
We have shown that the morpholino-mediated knockdown of xPTB resulted in a switch from the nonmuscle isoform (O5 RNA) to the somite-specific isoforms (α2 and α7 RNAs) in nonmuscle tissues. Conversely, targeted somitic overexpression of xPTB led to the synthesis of nonmuscle isoforms in the somites. These findings have important consequences for the tissue-specific regulation of the 3′ end processing of the α-TM pre-mRNA. First, they demonstrate that xPTB is the primary physiological determinant of exon 9A9′ repression in embryonic nonmuscle cells. Second, they show that embryonic endodermal and epidermal cells possess all the factors required to efficiently splice exon 9A9′ when xPTB repression is relieved, indicating that exon 9A9′ processing per se does not require tissue-specific regulatory splicing factors. Because exon 9A9′ is a 3′-terminal exon, its skipping in nonmuscle tissues could be the result of repression of splicing or cleavage/polyadenylation, or both. Our results from minigenes in which the 3′ splice site or the poly(A) site of exon 9A9′ has been deleted clearly show that xPTB can regulate both processes independently. We have previously identified an intronic element that is involved in the repression of exon 9A9′ in nonmuscle cells (13). This element harbors four high-affinity PTB binding sites, lying between the branch site and the 3′ AG border, that are essential for the repression of exon 9A9′. In this study, we show that these PTB binding sites affect both splicing and the cleavage/polyadenylation reactions. As we previously observed, the xPTB knockdown results in a stronger increase of exon 9A9′ splicing and cleavage/polyadenylation than the mutation of the four high-affinity binding sites, suggesting that the additional PTB binding sites which are present upstream and downstream of exon 9A9′ may be involved in exon 9A9′ repression. Interestingly, UCUU motifs are present within the polypyrimidine tract associated with the −274 branch point and the uridine-rich element of the αe poly(A) site. However, the functionality of these binding sites cannot be determined by a mutational approach because their mutation strongly decreases exon 9A9′ usage in muscle cells. These data led us to predict a model where the initial binding of xPTB to regulatory sites serves as a nucleus for the synergistic multimerization of xPTB leading to the sequestration of exon 9A9′ in which the 3′ splice site and the poly(A) site are inaccessible to the splicing and polyadenylation machineries. It is improbable that the tissue specificity of both the splicing and the polyadenylation of exon 9A9′ is simply controlled by the level of expression of xPTB. Rather, we favor a model in which these processes are sensitive to the amount of xPTB relative to that of other regulatory splicing factors. In support of this model, we have observed that although exon 9A9′ is repressed in both skin and endoderm, only in skin is the expression of xPTB higher (1.5-fold) than in somites (C. Le Sommer and S. Hardy, unpublished data). Accordingly, we have identified an intronic enhancer element that overlaps with the repressor sequence containing the four high-affinity PTB binding sites (C. Le Sommer et al., unpublished data). This feature suggests that activation of exon 9A9′ in somitic cells may rely on a mechanism by which regulatory splicing factors displace xPTB. The antagonistic binding of these factors could relieve xPTB repression and strengthen the recognition of the weak signals that define exon 9A9′. In contrast with our α-TM model, PTB was also reported to positively regulate the inclusion of the proximal alternative 3′-terminal exon of the calcitonin/calcium gene-related peptide by binding to a downstream intronic enhancer and activating polyadenylation by an unknown mechanism (25). While these results appear contradictory, they can be reconciled if we consider the enhancer element as a pseudoexon whose recognition by splicing factors is antagonized by PTB binding.
PTB in repressing polyadenylation.
Recently, Castelo-Branco et al. (5) reported that PTB can modulate the efficiency of polyadenylation by competing with CstF64 for binding to the downstream uridine-rich region of a poly(A) site. They proposed a model in which PTB-mediated inhibition of 3′ end processing leads to a reduction in gene expression. According to this model, weak poly(A) sites that lack a strong downstream uridine-rich region may be particularly sensitive to PTB repression. However, their observation that a fourfold overexpression of PTB had similar effects in the accumulation of mRNA produced from transfected minigenes with a weak or strong poly(A) site raises the question of whether these effects are physiologically relevant. In agreement with their data, we noticed that transgenic embryos that presented the highest increase in xPTB level (Fig. 5, embryos E4 and E5) displayed, in addition to α-TM isoform switching, ∼40% and 60% reduction in the endogenous MLC1f/3f mRNA and α-TM mRNA levels, respectively. This suggests that xPTB represses the expression of both genes by nonspecific competition with Cstf64 binding. This general effect upon gene expression is not present with the xPTB-transgenic embryos that exhibit the lowest increase in PTB (Fig. 5, embryos E6 and E7) while switching toward the repression of exon 9A9′ is still present. Moreover, the strongest rise in O5 mRNA levels was observed with these embryos, suggesting that the switch toward the nonmuscle splicing pattern is underestimated due to the nonspecific repression of the polyadenylation of the nonmuscle exon 9D. These observations indicate that an increase in the somitic xPTB level is sufficient to efficiently repress exon 9A9′ processing, although the switch toward the nonmuscle processing pattern is diminished by a general repression of polyadenylation.
xPTB is an essential protein in Xenopus.
Using a morpholino-based translational inhibition of xPTB, we have shown that xPTB is an essential protein in Xenopus. Its depletion resulted first in a specific phenotype with epidermal blister-like structures and subsequently in lethality. This is in contrast with the double-stranded RNA interference results obtained in Caenorhabditis elegans, in which no obvious phenotype and a complete viability of the worm were observed (23). In Drosophila, several mutations in the dmPTB locus are homozygous lethal (10). A mutation in the dmPTB locus that causes male sterility was also characterized. This mutation disrupts the expression of an abundant dmPTB transcript which is expressed only in the male germ line and affects spermatid differentiation (34). The skin blister-like phenotype we observed probably results from a defect in epidermis morphogenesis, as histological studies revealed that the typical organization of the epiderm with an epithelial layer and a sensorial layer was altered to produce a monostratified epithelium. This suggests that an intercalation of the cells of both layers occurred to give a single layer with an increased area. The reason why xPTB knockdown causes the epidermis to organize in a single layer is not clear. We and others (see above for references) have shown that PTB can control the processing of pre-mRNAs transcribed from a number of genes. Therefore, the blister-like phenotype observed could result from multiple partial changes in splicing or instead in a dramatic change in the splicing pattern of a specific transcript. This issue could be addressed by designing a microarray analysis to assess how alternative splicing events are altered by the knockdown of xPTB.
Dual function for PTB?
We have shown that a change in the xPTB steady-state level is sufficient to switch the splicing pattern of α-TM exon 9A9′. This result is in contrast with previous reports in which overexpression or RNA interference-mediated depletion of PTB (42) produced only partial effects on the use of a particular splicing pattern. Accordingly, xPTB-depleted embryos displayed only limited effects on the splicing pattern of exon N1 of c-src and exon SM of α-actinin. While these effects confirm the implication of PTB in the regulation of these exons, they indicate that additional factors acting through a PTB-independent pathway are involved in their repression or alternatively that the relief of PTB repression is not sufficient to cause an efficient splicing because tissue-specific activator factors are required. In agreement with these observations, in vitro studies showed that hnRNPA1 can also repress exon N1 splicing by a different mechanism than PTB (35). Moreover, efficient splicing of exon N1 requires the assembly of an activator complex that comprises the KSRP factor that is expressed at a higher level in neuronal cells (26). From these results, we suggest a classification of the alternative exons regulated by PTB into two classes. In the first class, represented by exon N1 of c-src and exon SM of α-actinin, PTB is part of a negative regulatory complex that comprises other regulatory proteins. The presence of multiple partners may increase the combinatorial controls of these exons and authorize the inclusion of the alternative exon at different levels depending on the cell type. In the second class, represented by exon 9A9′ of α-TM, PTB is the major actor in the tissue-specific repression. The important effect of xPTB upon exon 9A9′ processing could be due to the notable property that exon 9A9′ is a 3′-proximal terminal exon whose poly(A) site usage is regulated by PTB. Cleavage at this proximal poly(A) site following PTB derepression excludes any competition with the distal 3′-terminal exon and leads, therefore, to a strong inclusion of exon 9A9′. In addition, the use of exon 9A9′ in nonmuscle and muscle tissues responds to a binary on/off switch, and there is no need to control the level of its inclusion. According to this classification and in view of the importance of xPTB in repressing the processing of the 3′-terminal exon 9A9′, we suggest that PTB could be a major factor in preventing the definition of regulated proximal 3′-terminal exons.
Acknowledgments
We thank Nick Proudfoot for the β-globin construct used to derive pSV40βglobinβpA and David Bentley for the VA reporter plasmid. We thank Chris Smith for critically reading the manuscript. We also thank Beverley Osborne for helpful suggestions and his continual support.
This work was supported by grants to S.H. from the Association Française contre les Myopathies.
REFERENCES
- 1.Ashiya, M., and P. J. Grabowski. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3:996-1015. [PMC free article] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-mRNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Brett, D., J. Hanke, G. Lehmann, S. Haase, S. Delbruck, S. Krueger, J. Reich, and P. Bork. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474:83-86. [DOI] [PubMed] [Google Scholar]
- 4.Carstens, R. P., E. J. Wagner, and M. A. Garcia-Blanco. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelo-Branco, P., A. Furger, M. Wollerton, C. Smith, A. Moreira, and N. Proudfoot. 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 24:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, R. C., and D. L. Black. 1997. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 17:4667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cote, C. A., D. Gautreau, J. M. Denegre, T. L. Kress, N. A. Terry, and K. L. Mowry. 1999. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell 4:431-437. [DOI] [PubMed] [Google Scholar]
- 8.Cote, J., S. Dupuis, and J. Y. Wu. 2001. Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J. Biol. Chem. 276:8535-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croft, L., S. Schandorff, F. Clark, K. Burrage, P. Arctander, and J. S. Mattick. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24:340-341. [DOI] [PubMed] [Google Scholar]
- 10.Dansereau, D. A., M. D. Lunke, A. Finkielsztein, M. A. Russel, and W. J. Brook. 2002. hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development 129:5553-5566. [DOI] [PubMed] [Google Scholar]
- 11.Gooding, C., G. C. Roberts, and C. W. Smith. 1998. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA 4:85-100. [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton, B. J., A. Genin, R. Q. Cron, and W. F. Rigby. 2003. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol. Cell. Biol. 23:510-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamon, S., C. Le Sommer, A. Mereau, M. R. Allo, and S. Hardy. 2004. Polypyrimidine tract-binding protein is involved in vivo in repression of a composite internal/3′-terminal exon of the Xenopus α-tropomyosin pre-mRNA. J. Biol. Chem. 279:22166-22175. [DOI] [PubMed] [Google Scholar]
- 14.Hardy, S., M. Y. Fiszman, H. B. Osborne, and P. Thiebaud. 1991. Characterization of muscle and non muscle Xenopus laevis tropomyosin transcribed from the same gene. Eur. J. Biochem. 202:431-440. [DOI] [PubMed] [Google Scholar]
- 15.Hardy, S., S. Hamon, B. Cooper, T. Mohun, and P. Thiébaud. 1999. Two skeletal alpha-tropomyosin transcripts with distinct 3′UTR have different temporal and spatial patterns of expression in the striated muscle lineages of Xenopus laevis. Mech. Dev. 87:199-202. [DOI] [PubMed] [Google Scholar]
- 16.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kan, Z., E. C. Rouchka, W. R. Gish, and D. J. States. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoch, K. P., H. Bergert, B. Borgonovo, H. D. Saeger, A. Altkruger, P. Verkade, and M. Solimena. 2004. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 6:207-214. [DOI] [PubMed] [Google Scholar]
- 19.Kroll, K. L., and E. Amaya. 1996. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122:3173-3183. [DOI] [PubMed] [Google Scholar]
- 20.Le Guiner, C., A. Plet, D. Galiana, M. C. Gesnel, F. Del Gatto-Konczak, and R. Breathnach. 2001. Polypyrimidine tract-binding protein represses splicing of a fibroblast growth factor receptor-2 gene alternative exon through exon sequences. J. Biol. Chem. 276:43677-43687. [DOI] [PubMed] [Google Scholar]
- 21.Levy, J. B., T. Dorai, L. H. Wang, and J. S. Brugge. 1987. The structurally distinct form of pp60c-src detected in neuronal cells is encoded by a unique c-src mRNA. Mol. Cell. Biol. 7:4142-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, B. P., R. E. Green, and S. E. Brenner. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 100:189-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longman, D., I. L. Johnstone, and J. F. Caceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 25.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 27.Mironov, A. A., J. W. Fickett, and M. S. Gelfand. 1999. Frequent alternative splicing of human genes. Genome Res. 9:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modrek, B., A. Resch, C. Grasso, and C. Lee. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29:2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohun, T. J., N. Garrett, and J. B. Gurdon. 1986. Upstream sequences required for tissue-specific activation of the cardiac actin gene in Xenopus laevis embryos. EMBO J. 5:3185-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulligan, G. J., W. Guo, S. Wormsley, and D. M. Helfman. 1992. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem. 267:25480-25487. [PubMed] [Google Scholar]
- 31.Nieuwkoop, P., and J. Faber. 1956. Normal table of Xenopus laevis. Daudin, North-Holland, Amsterdam, The Netherlands.
- 32.Norton, P. A. 1994. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 22:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resch, A., Y. Xing, B. Modrek, M. Gorlick, R. Riley, and C. Lee. 2004. Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome Res. 3:76-83. [DOI] [PubMed] [Google Scholar]
- 34.Robida, M. D., and R. Singh. 2003. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J. 22:2924-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rooke, N., V. Markovtsov, E. Cagavi, and D. L. Black. 2003. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen, H., J. L. Kan, C. Ghigna, G. Biamonti, and M. R. Green. 2004. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA 10:787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 38.Southby, J., C. Gooding, and C. W. Smith. 1999. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. Mol. Cell. Biol. 19:2699-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thézé, N., S. Hardy, R. Wilson, M. R. Allo, T. Mohun, and P. Thiebaud. 1995. The MLC1f/3f is an early marker of somitic muscle differentiation in Xenopus laevis embryo. Dev. Biol. 171:352-362. [DOI] [PubMed] [Google Scholar]
- 40.Tillmar, L., C. Carlsson, and N. Welsh. 2002. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J. Biol. Chem. 277:1099-1106. [DOI] [PubMed] [Google Scholar]
- 41.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288.11313454 [Google Scholar]
- 42.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 43.Waites, G. T., I. R. Graham, P. Jackson, D. B. Millake, B. Patel, A. D. Blanchard, P. A. Weller, I. C. Eperon, and D. R. Critchley. 1992. Mutually exclusive splicing of calcium-binding domain exons in chick α-actinin. J. Biol. Chem. 267:6223-6271. [PubMed] [Google Scholar]
- 44.Wollerton, M. C., C. Gooding, F. Robinson, E. C. Brown, R. J. Jackson, and C. W. Smith. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wollerton, M. C., C. Gooding, E. J. Wagner, M. A. Garcia-Blanco, and C. W. Smith. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13:91-100. [DOI] [PubMed] [Google Scholar]
- 46.Xu, X., D. Yang, J. H. Ding, W. Wang, P. H. Chu, N. D. Dalton, H. Y. Wang, J. R. Bermingham, Jr., Z. Ye, F. Liu, M. G. Rosenfeld, J. L. Manley, J. Ross, Jr., J. Chen, R. P. Xiao, H. Cheng, and X. D. Fu. 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59-72. [DOI] [PubMed] [Google Scholar]
- 47.Zuccato, E., E. Buratti, C. Stuani, F. E. Baralle, and F. Pagani. 2004. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 279:16980-16988. [DOI] [PubMed] [Google Scholar]