Abstract
The DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) is central to both DNA replication and repair. The ring-shaped homotrimeric PCNA encircles and slides along double-stranded DNA, acting as a “sliding clamp” that localizes proteins to DNA. We determined the behavior of green fluorescent protein-tagged human PCNA (GFP-hPCNA) in living cells to analyze its different engagements in DNA replication and repair. Photobleaching and tracking of replication foci revealed a dynamic equilibrium between two kinetic pools of PCNA, i.e., bound to replication foci and as a free mobile fraction. To simultaneously monitor PCNA action in DNA replication and repair, we locally inflicted UV-induced DNA damage. A surprisingly longer residence time of PCNA at damaged areas than at replication foci was observed. Using DNA repair mutants, we showed that the initial recruitment of PCNA to damaged sites was dependent on nucleotide excision repair. Local accumulation of PCNA at damaged regions was observed during all cell cycle stages but temporarily disappeared during early S phase. The reappearance of PCNA accumulation in discrete foci at later stages of S phase likely reflects engagements of PCNA in distinct genome maintenance processes dealing with stalled replication forks, such as translesion synthesis (TLS). Using a ubiquitination mutant of GFP-hPCNA that is unable to participate in TLS, we noticed a significantly shorter residence time in damaged areas. Our results show that changes in the position of PCNA result from de novo assembly of freely mobile replication factors in the nucleoplasmic pool and indicate different binding affinities for PCNA in DNA replication and repair.
Proper duplication, maintenance, and repair of the genome are essential for ensuring genomic stability. Defects in any of these processes contribute to the onset and progression of cancer (9), because genomic DNA is subject to damage by both environmental agents and endogenous metabolic processes. In addition, stalling of the DNA replication machinery, which occurs upon encountering damaged DNA, is a challenging problem for cells. A variety of DNA damage bypass and repair mechanisms rescue replication after encountering DNA lesions. Protein clamps on DNA have been implicated in all processes of DNA metabolism, including DNA replication and repair. The most extensively investigated protein clamp in eukaryotes is the DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA).
PCNA is a central protein in both DNA replication and repair. PCNA encircles double-stranded DNA as a trimer, forming a sliding clamp that tethers proteins such as polymerases to DNA (5). PCNA is essential not only for DNA replication but also for several forms of DNA repair, including nucleotide excision repair (NER), the major pathway by which cells remove DNA damage introduced by UV light and a variety of chemical carcinogens (10). After recognition of the lesion, the damaged strand is excised and resynthesized in a process requiring PCNA. However, some lesions, such as cyclobutyl pyrimidine dimers (CPDs), are repaired slowly by NER, and unrepaired CPDs remain in the DNA (13). During replication, mammalian cells can circumvent this damage by using a group of specialized polymerases, referred to as translesion synthesis (TLS) polymerases, which also require PCNA (16). Although TLS does not result in damage removal and can be mutagenic, damage bypass is an important component of the overall cellular response to DNA damage and makes an important contribution to cellular survival. Replication of mildly damaged DNA can be advantageous to the cell because it provides a sister chromatid that can be used as a template for repair by homologous recombination.
A central question is how eukaryotic cells accomplish the switching from a high-fidelity replicative polymerase to a specialized TLS polymerase. A recent study demonstrated that in human cells, PCNA becomes monoubiquitinated at the lysine residue at position 164 (K164) upon UV irradiation and that monoubiquitinated PCNA specifically interacts with the TLS polymerase polη (14). Therefore, it was suggested that the state of monoubiquitination of the sliding clamp, PCNA, might serve as a switch that enables or abrogates binding of PCNA to polη. To obtain insight into the mode of action of PCNA in living mammalian cells in DNA damage and repair, we generated cell lines expressing the human PCNA protein tagged with green fluorescent protein (GFP-hPCNA). Using time-lapse microscopy and photobleaching studies, we analyzed the dual involvement of PCNA in replication and repair. Furthermore, we tested the role of PCNA monoubiquitination in vivo by generating cell lines stably expressing mutant GFP-hPCNAK164R.
MATERIALS AND METHODS
Cell culture and DNA constructs.
CHO9 cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with antibiotics and 10% fetal calf serum under an atmosphere of 5% CO2. A cDNA encoding GFP-hPCNA and yellow fluorescent protein-hPCNA was constructed by digesting the PT7 vector containing the entire 1.4-kb human PCNA cDNA insert (kindly provided by Bruce Stillman, Cold Spring Harbor Laboratory, New York) with NdeI and BamHI to produce a 900-bp fragment, which was isolated. DNA ends were blunted and cloned in frame into the SmaI site of the eGFP-C1 and eYFP-C1 vectors (Clontech). The mutant construct GFP-hPCNAK164R was generated using the QuikChange site-directed mutagenesis method (Stratagene) and the primers pcna-KR (5′ AGATGCTGTTGTAATTTCCTGTGCACGTGACGGAGTGAAATTTTCTGCAAGT 3′) and pcna-KR-rev (5′ ACTTGCAGAAAATTTCACTCCGTCACGTGCACAGGAAATTACAACAGCATCT 3′). The constructs were sequenced to verify the presence of the K164R mutation. CHO9 cells stably expressing these cDNAs were generated by transfection using Lipofectamine 2000 (Invitrogen). Cells were further screened for the presence and proper expression level of nuclear fluorescence by cell sorting with a FACSVantage cell sorter (Becton Dickinson) and immunoblot analysis using a monoclonal anti-PCNA (DAKO) and anti-GFP antibodies (Roche). The generation of the CFP-hRAD51 construct has been described elsewhere (27).
Cell cycle analysis.
For cell cycle experiments, cells were irradiated with 10 J/m2 of UV light (254 nm) 2 days after plating. The cells (5 × 106) were trypsinized and fixed, using 70% ice-cold ethanol, at different time points after UV irradiation (0, 8, and 16 h). After being washed with phosphate-buffered saline, the cells were resuspended in 400 μl of phosphate-buffered saline containing 0.1% Triton X-100, 8 μg propidium iodide, and 80 μg RNase. Cell suspensions were left for incubation overnight at 4°C, and the cell cycle profile was measured in the PE, TC channel on a FACScan instrument (Becton Dickinson).
Local UV irradiation and immunostaining.
Local UV irradiation and immunostaining have been described in detail previously (22).
Confocal microscopy.
Cells were treated with UV radiation and subjected to photobleaching experiments after irradiation. Confocal images of living cells expressing GFP-hPCNA were obtained using a Zeiss LSM 510 microscope equipped with a 25-mW Ar laser at 488 nm and a 40× 1.3-numerical-aperture oil immersion lens. Images of single nuclei were taken at lateral sample intervals of 100 nm. GFP fluorescence was detected using a dichroic beam splitter (HFT488), and an additional 505- to 530-nm bandpass emission filter was placed in front of the photomultiplier tube. Photobleaching experiments were performed and analyzed as described previously (6).
RESULTS AND DISCUSSION
Imaging ongoing DNA replication in living cells.
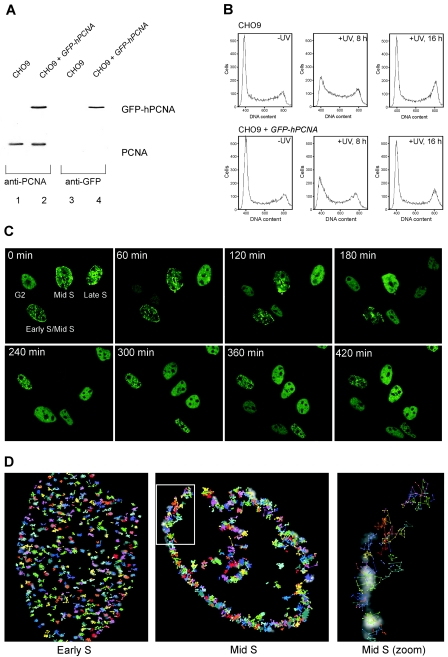
To visualize DNA replication in living cells, we generated CHO cells that stably express human PCNA tagged with GFP. Immunoblot analysis of the cells expressing GFP-hPCNA with anti-PCNA antibodies showed that the expression levels of endogenous and GFP-hPCNA were similar (Fig. 1A, left panel). Hybridization with anti-GFP antibodies revealed no GFP-hPCNA breakdown products, indicating that the fusion protein remained intact (Fig. 1A, right panel). The expression of GFP-hPCNA did not interfere with normal cell cycle progression or with cell cycle progression in response to UV irradiation (Fig. 1B). In addition, cells were tested for their UV sensitivities, and no significant difference in survival rates was detected (data not shown). We then analyzed the spatial and temporal organization of DNA replication sites. To analyze the large-scale relocalization of GFP-hPCNA, cells were followed through mitosis by time-lapse imaging (Fig. 1C; see also online movie at http://www.erasmusmc.nl/oic/essers_pcna). GFP-hPCNA displayed the specific distribution patterns of PCNA throughout the different stages of the cell cycle (17, 25). GFP-hPCNA was strictly localized in the nuclei of living cells, and the distinct focal distributions at early-, mid-, and late-S-phase replication sites were clearly identified (Fig. 1C). Cells were imaged every 10 min for 5 h. All four cells shown in Fig. 1C could be followed through mitosis while they underwent the characteristic relocalization of PCNA as they progressed through the cell cycle. From these movies, we determined the average time spans of the G1 phase (380 ± 250 min), the early S phase (266 ± 26 min), the mid-S phase (323 ± 53 min), the late S phase (141 ± 39 min), and the G2 phase (77 ± 19 min). This shows that the times needed to progress through one complete cell cycle varied greatly in individual cells, with the largest variation in the durations of the G1 phase.
FIG. 1.
Characterization of CHO9 cells expressing GFP-hPCNA. (A) Immunoblot analysis. Whole-cell extracts of CHO9 cells and CHO9 cells stably expressing GFP-hPCNA were analyzed for the presence of endogenous PCNA and GFP-hPCNA by immunoblotting using antibodies against PCNA (left panel) and GFP (right panel). (B) Cell cycle analyses. CHO9 cells and CHO9 cells expressing GFP-hPCNA were irradiated with UV light (10 J/m2). After the indicated time points, the cells were analyzed for DNA content using a fluorescence-activated cell sorter. In each histogram, the counted cells are plotted against the relative fluorescence intensities derived from the propidium iodide signal. (C) Time-lapse imaging of four cells expressing GFP-hPCNA. Cells were imaged every 10 min during a 5-h period. Shown are the images taken every 60 min. All four cells will go through cell division and follow the characteristic PCNA focal pattern. Small dots are characteristic for early S phase, staining at the periphery of the nucleus marks the mid-S phase, and big blobs mark the late S phase. The G2 cell divided first and, depending on their stages in S phase, the other cells followed. (D) Analysis of movement of PCNA replication structures in living cells done by tracking individual PCNA replication structures in single cells. In the background, the PCNA staining pattern at time point zero is shown in grayscale. Positions of the centers of replication structures were determined in each frame of a movie from the focal plane of a cell, and consecutive positions were connected by lines. Cells were examined for 30 min using time-lapse video microscopy at intervals of 30 s. The coordinates of each focus in the cell were determined at every 30-s time interval (Volocity; Improvision). Each individually colored track represents the movement of a different focus during the period captured. The last image shows the zoom image of the mid-S-phase cell.
Replication of the mammalian genome starts at thousands of origins that are activated at different times during S phase. Using tracking of individual sites of replication foci represented by PCNA, we investigated how this replication program is coordinated. We analyzed the small-scale movement of replication foci during early and mid-S phase by imaging GFP-hPCNA every 30 seconds for 30 min. Tracking of individual replication foci over time showed limited but asynchronous x and y axis shifts (Fig. 1D). This constrained motion in the microenvironment is most likely due to the Brownian motion of the chromosome fiber and has also been observed in other eukaryotes (2, 18). In these studies, the constrained diffusion of specific chromatin domains was also observed. When we analyzed the transition from early S phase to mid-S phase, we were unable to track individual foci translocating from the interior of the nucleus to the periphery. This is consistent with previously reported photobleaching experiments, which suggested that the transition from earlier to later replicons occurs by disassembly into a nucleoplasmic pool of highly mobile subcomponents and subsequent reassembly at newly activated sites (26).
Accumulation of GFP-hPCNA at sites of local UV-induced DNA damage in different stages of the cell cycle.
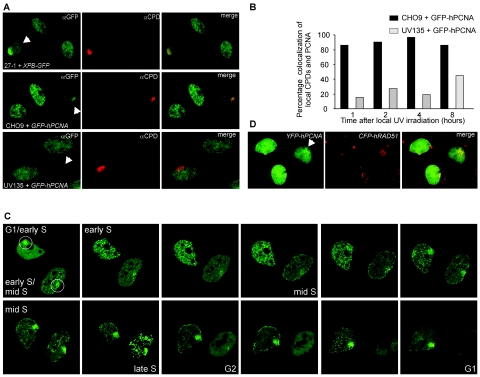
In addition to playing a pivotal role of PCNA in replication, this protein is also required for different DNA repair mechanisms, including DNA damage response towards the major UV-induced DNA lesions, i.e., CPDs and 6-4 photoproducts (1, 7, 23). To examine the dual involvement of PCNA in the repair of UV-induced DNA damage and DNA replication, we visualized both processes simultaneously in the same cells using time-lapse microscopy. To visually distinguish between repair and replication, we used a local UV irradiation technique. DNA damage was induced at restricted areas in the nucleus by covering cells with a UV-blocking membrane containing randomly distributed pores with diameters of ∼5 μm and subsequently UV irradiating them (21). Soon after UV irradiation (within ∼10 min), GFP-hPCNA accumulated in brightly fluorescent regions (7). This local accumulation could be detected in cells with diffuse GFP-hPCNA staining, indicative of G1 and G2 cells, and in cells with the different S-phase-specific PCNA patterns. The presence and position of the UV-damaged DNA was confirmed and visualized in fixed cells by immunofluorescence using CPD-specific antibodies. The involvement of NER in the repair of this local UV-induced damage was shown by indirect immunofluorescence experiments using antibodies against CPD and the NER factor TFIIH. The TFIIH component XPB concentrates in areas containing locally induced UV-induced DNA damage (Fig. 2A, top row) (21, 28). Simultaneous visualization of CPDs and PCNA by immunofluorescence revealed that CPDs and PCNA colocalized (Fig. 2A, middle row). On top of the normal PCNA replication pattern, an accumulation of PCNA at the CPDs was observed. Eighty to 90% of the cells with local CPD accumulation also showed local PCNA accumulation 1 to 8 h after local UV irradiation. The requirement of active NER for confined PCNA accumulation was confirmed in a NER-deficient cell line, UV135 (Fig. 2A, bottom row, and B). Using this cell line, which is deficient in one of the incision nucleases of NER, XPG, we found a significant reduction of the immediate accumulation of PCNA on a locally UV-damaged area.
FIG. 2.
Accumulation of DNA repair proteins at sites of local UV-induced damage. (A) Upper panels: XPB and CPDs colocalize. CHO cells deficient in XPB (27-1) complemented with XPB-GFP were locally UV irradiated (locally damaged cells are indicated with the arrows). One hour after UV irradiation, DNA damage was detected with antibodies against CPDs (shown in red), and XPB-GFP accumulation was detected with antibodies against GFP (shown in green). Colocalization of XPB and CPDs is demonstrated in the merged image. Middle panels: PCNA and CPDs display colocalization. CHO9 cells expressing GFP-hPCNA were locally UV irradiated. One hour after UV irradiation, cells were fixed, DNA was denatured, and DNA damage was detected by indirect immunofluorescence using antibodies against CPDs (shown in red). GFP-hPCNA was also detected by indirect immunofluorescence using antibodies against GFP (shown in green). Shown are three GFP-hPCNA-expressing cells in different stages of the cell cycle. On top of the replication pattern of PCNA, one cell shows local accumulation of GFP-hPCNA, which colocalizes with the CPD signal (shown in yellow in the merged image). Bottom panels: No colocalization of PCNA and CPDs in a NER-deficient cell line, UV135, which is deficient in XPG. Locally irradiated UV135 cells expressing GFP-hPCNA were fixed 1 hour after irradiation and stained using antibodies against GFP (in green) and CPDs (in red). The bottom cell shows local accumulation of CPDs but no accumulation of PCNA. (B) Quantitation of the results shown in Fig. 2A, middle and bottom panels. The percentages of local CPD accumulations that also show local PCNA accumulation after local UV irradiation were determined at the indicated time points for CHO9 and UV135 cells. For all cell lines, 65 to 70 cells were examined. (C) Time-lapse imaging of GFP-hPCNA in cells treated with local UV damage shows transient accumulation of GFP-hPCNA (indicated by the circles) at sites of local UV damage in different stages of the cell cycle. Based on the PCNA replication pattern, the upper cell was at the transition from the G1 phase to the early S phase, and the lower cell was in the early S phase. In the upper cell, this local GFP-hPCNA accumulation disappeared for a short period during the early S phase but reappeared at the transition from the early S phase to the mid-S phase. In the lower cell, the local GFP-hPCNA accumulation disappeared in the G2 phase and the subsequent G1 phase in the daughter cells. Note the decrease in fluorescence signal around mitosis as a result of cells rounding up and detaching from the coverslips. (D) Colocalization of PCNA and Rad51 at site of local damage. Chinese hamster ovary cells stably expressing both YFP-hPCNA and CFP-hRAD51 were locally UV irradiated. Cells were fixed 1, 4, 8, and 16 h after UV irradiation, and local accumulation of PCNA and Rad51 was directly detected using the appropriate filter sets (Chroma) to discriminate between cyan fluorescent protein and yellow fluorescent protein. At all these different time points, we found examples of local accumulations of PCNA (shown in green) and Rad51 (shown in red) during S phase, where the focal pattern of PCNA partly colocalized with the focal pattern of Rad51 (shown in the merged image).
This information allowed us to follow the behavior of GFP-hPCNA in UV-damaged DNA repair and DNA replication. We performed time-lapse imaging of living locally irradiated CHO cells, in which UV-induced CPDs are not effectively removed by nucleotide excision repair (13), to follow the fate of local UV-damaged areas through cell cycle progression. Figure 2C (and supplementary online movie 2 at http://www.erasmusmc.nl/oic/essers_pcna) shows an example of two cells, one irradiated during G1 phase and the other during its transition from early S to mid-S phase. Both cells, showing local concentrations of PCNA at the sites of UV irradiation, were subsequently followed through their cell cycle progressions at 10-min time intervals. Surprisingly, in the upper cell, imaged from G1/early S to mid-S phase, the local PCNA accumulation was present during the entire capturing period of 11 h but temporarily disappeared in early S phase. In the lower cell, imaged from early/mid-S to G1 phase, the local GFP-hPCNA accumulation was present until late S phase and disappeared in the G2 phase. The initial local accumulation of PCNA that was observed during the G1 phase shows a homogeneous pattern indicative of the participation of PCNA in active NER, since NER factor accumulations on damaged spots show a similar distribution pattern. However, the damage-dependent accumulation temporarily disappeared during early S phase. The reappearance of PCNA accumulation in discrete foci occurred at longer time points after irradiation, which likely reflects engagements of PCNA in other genome maintenance processes dealing with lesion-stalled replication forks, such as TLS or homologous recombination. Indeed, we found that this focal distribution of PCNA at the site of the local damage during S phase partly colocalized with the focal pattern of Rad51, a key protein implicated in stalled replication fork repair through homologous recombination (Fig. 2D). Since these time-lapse experiments indicated that the behavior of PCNA molecules in the UV-damaged areas is dynamic, we analyzed the mobility of PCNA in the different stages of the cell cycle and in DNA repair.
Mobility of GFP-hPCNA in the nucleus.
To analyze the diffusion properties of GFP-hPCNA within the different stages of the cell cycle in nuclei of living cells, fluorescence redistribution after photobleaching (FRAP) was used (12, 20, 30). In the applied FRAP variant, we irreversibly quenched the fluorescence of a small strip spanning the entire nucleus and subsequently recorded the recovery of fluorescence over a time period of 6 seconds. From these recovery curves, both the immobile fraction and the effective diffusion coefficient (Deff) of PCNA were determined (4, 11). The fluorescence recovered to 65% for cells in the G1/G2 phase of the cell cycle (since 35% of the total pool was permanently bleached), which was similar to that for free GFP in our experimental setup and indicates that the vast majority of the detected GFP-hPCNA molecules were mobile at these stages in the cell cycle (Fig. 3A) (6). However, when PCNA started forming replication foci in early S phase, about half the GFP-hPCNA molecules became (transiently) immobilized. Furthermore, a (transiently) immobile PCNA fraction was also detected in mid- and late-S-phase cells. When we analyzed the mobile fraction as shown in Fig. 3A to determine the Deff of GFP-hPCNA in the nucleoplasm during the different stages of the cell cycle, we found no significant differences in the velocities of the mobile fraction of GFP-hPCNA (Fig. 3B).
FIG. 3.
Fluorescence redistribution after photobleaching analyses of GFP-hPCNA before DNA damage induction. Cells stably expressing PCNA fused to GFP were subjected to a local bleach pulse, and the kinetics of fluorescence recovery in the bleached area was determined. (A) The fluorescence in a small strip spanning the entire nucleus was bleached with a 200-ms high-intensity laser pulse. The recovery of fluorescence in the strip was monitored at intervals of 100 ms, and the measured fluorescence intensities over time were plotted. This photobleaching protocol was applied to GFP-hPCNA-expressing cells in the different stages of the cell cycle. To determine the immobile fractions, the final measured fluorescence intensity was normalized to the prebleaching pulse fluorescence intensity. (B) Diffusion coefficient of GFP-hPCNA in different stages of the cell cycle. To determine the Deff, the fluorescence intensity immediately after bleaching and the final postbleaching pulse fluorescence intensity measured were normalized between zero and one. The Deffs in the different stages of the cell cycle were determined by fitting the experimentally obtained curves to a mathematical model describing diffusion (6). Measurements were performed in triplicate, and consistent results were obtained among different sets of experiments. Error bars indicate twice the standard errors of the mean. (C) Fluorescence loss in photobleaching and fluorescence redistribution after photobleaching of GFP-hPCNA in different stages of the cell cycle. The lower region of a cell containing replication foci was bleached by a single laser pulse. The cell was imaged at the indicated times after bleaching. FLIP was measured in the unbleached half of the cell, while FRAP was measured in the bleached half of the same cell. The same experimental protocol was applied to cells in the early, mid-, and late S and G1/G2 phases of the cell cycle. (D) Quantitation of the simultaneous FRAP and FLIP experiment on GFP-hPCNA replication foci. “n” represents the number of cells examined.
Dynamics of replication foci.
Next, we analyzed the mobility of PCNA in replication foci. Attempts to determine whether replication foci are static or dynamic structures in which PCNA shows turnover have produced conflicting conclusions (24, 26). One study reported that the DNA polymerase clamp shows no turnover at established replication sites (26), while the other reported that replication foci exhibited PCNA turnover with a relatively short half-life (∼25 s) (24). However, differences in the experimental setups of these reports may account for their conflicting conclusions. Some features of PCNA assembly and disassembly into replication foci appear not to be in dispute. Both reports agree that the transition from earlier to later replicons occurs by the dissociation of PCNA into a nucleoplasmic pool of rapidly diffusing subcomponents and reassembly at newly activated sites. The controversy arises when considering the stability of PCNA assembled into individual replication foci. Sporbert et al. bleached a small area in a mid-S-phase cell containing ∼10 replication foci and compared time overlays of the prebleaching image and the images after recovery. They concluded that the bleached foci reappear at adjacent spots. This qualitative analysis does not take into account the Brownian motion of replication foci as observed in the research presented here (Fig. 1D). Solomon et al. bleached a large area of the nucleus and measured the average recovery in the bleached area. To unambiguously determine whether PCNA is contained within established replication foci by long-lived protein interactions or in a more dynamic fashion in which PCNA proteins turn over, we combined a FRAP and a fluorescence-loss-in-photobleaching (FLIP) experiment, which was previously developed to study the dynamics of RAD52 group proteins and telomeres (6, 19). Half of a nucleus containing replication structures was bleached, and the recovery of fluorescence was subsequently monitored in the bleached part of the nucleus during a time period of 1,000 s. At the same time, the fluorescence loss in the unbleached part of the nucleus was monitored until a new equilibrium in fluorescence distribution was reached between the bleached and the unbleached regions. We performed this FLIP/FRAP analysis in the three recognizable S-phase stages: the early, mid-, and late S phases. In all three stages, the exchange of PCNA protein in the replication structures was observed (Fig. 3C). The change in fluorescence intensity over time, indicative of PCNA turnover, was quantitated. On average, the FLIP and FRAP curves converged after 570 ± 140 s, indicating that after this period, a new equilibrium between the bleached and the unbleached regions was reached (Fig. 3D). Moreover, the curves converged completely, showing an absence of a detectable stably associated PCNA fraction present in the replication foci. Note that the high mobility of the free GFP-hPCNA analyzed in Fig. 3A was not detected by this FLIP/FRAP analysis because of the time interval of 10 s, while the recovery of the free GFP-hPCNA presented in Fig. 3A had already been completed within 2 seconds.
Mobility of GFP-hPCNA at sites of local UV damage.
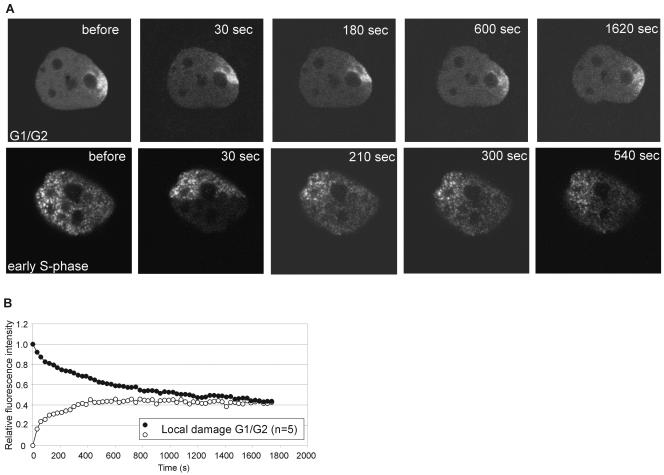
It is not known what factors signal PCNA to relocate to sites of damage or whether it remains attached once associated. Therefore, we wanted to analyze the residence time of PCNA on UV damage. After induction of local UV damage and recruitment of GFP-hPCNA, we performed a combined FLIP/FRAP experiment (Fig. 4A). As the GFP-hPCNA is diffuse and highly mobile in untreated G1 or G2 cells, we first analyzed the residence time on the site of local damage in G1/G2 cells. When we plotted the recovery and loss of fluorescence for the local damage area in these cells, we found that the curves converged after 1,200 ± 400 s, or approximately 20 min (Fig. 4B). Compared to the residence time of PCNA in replication, this suggests that the PCNA protein is engaged at least two times longer in areas undergoing repair than in replication. Next, we determined residence times in replication and repair simultaneously. We combined a FLIP/FRAP experiment on locally damaged cells that were in early S phase (Fig. 4A). After photobleaching, a new equilibrium in fluorescence in the replication area was established, while the redistribution of GFP-hPCNA in the damaged area was still ongoing. This example shows at the single-cell level that the turnover of PCNA in the damaged area is slower than in the area undergoing replication only.
FIG. 4.
Fluorescence redistribution after photobleaching analyses of GFP-hPCNA in locally UV-irradiated cells. (A) Example of FLIP/FRAP analysis in locally irradiated cells. The upper panel represents a cell in the G1 or G2 phase of the cell cycle and shows local accumulation of GFP-hPCNA in the damaged area. The lower part of the cell, including half of the site of local damage, was photobleached, and the cell was imaged every 30 s. A similar experiment (lower panel) was done on a cell that was, based on the PCNA staining pattern, in the early S phase of the cell cycle. (B) Quantitation of the simultaneous FRAP and FLIP experiments on GFP-hPCNA after local UV damage on G1/G2-phase cells. “n” represents the number of cells examined.
Kinetics of TLS-deficient PCNA.
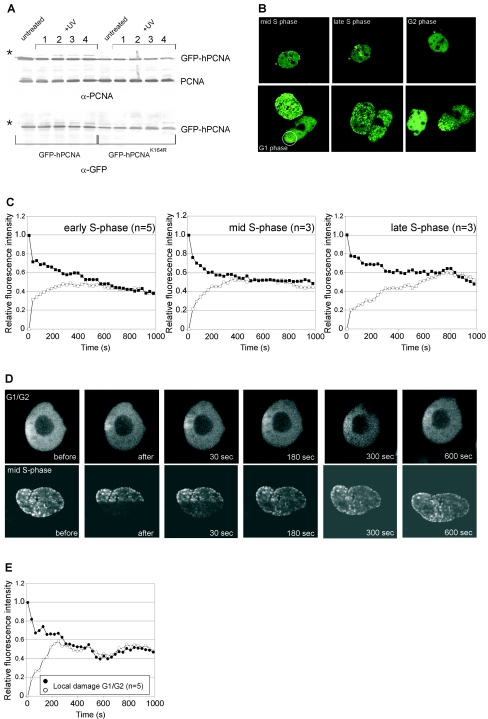
It has recently been demonstrated that in yeast cells, PCNA becomes ubiquitinated at lysine 164 following treatment with the DNA-damaging agent methyl methanesulfonate and that a yeast strain mutant in PCNA at K164 is UV sensitive (8). In addition, the monoubiquitination of PCNA appeared to be the major PCNA modification after DNA damage in mammals (14, 29). Monoubiquitinated PCNA specifically interacts with one of the TLS polymerases, polη. To test the importance of this PCNA modification in vivo, we generated cell lines stably expressing mutant GFP-hPCNAK164R. We confirmed that after UV treatment, GFP-hPCNAK164R could no longer be ubiquitinated (Fig. 5A) (24). Next, we analyzed cells expressing mutant PCNA by time-lapse microscopy (Fig. 5B) and found distribution patterns for mutant PCNA similar to those found for the wild-type GFP-hPCNA construct in DNA replication. To analyze the behavior in DNA repair, we applied local UV damage and found that the protein still relocalized to the site of local damage and, as for wild-type GFP-hPCNA, this local damage temporarily disappeared in early S phase. Solomon et al. previously reported that 4 h after treatment of cells with the chemotherapeutic agent cis-diammine-dichloroplatinum or UV light, wild-type GFP-hPCNA accumulates in numerous subnuclear foci, but mutant GFP-hPCNAK164R does not accumulate in nuclear foci upon treatment with cis-diammine-dichloroplatinum (24). It is unlikely that the DNA-damage-induced foci reported by Solomon et al. represented active NER, as the immediate repair of UV damage by NER is a pannuclear event and factors involved in NER (in contrast to factors involved in DNA double-strand break repair) have not been reported to assemble into nuclear foci after damage (3, 7, 15, 21, 22, 28). However, time-lapse experiments on locally irradiated cells reveal a diffuse accumulation of GFP-hPCNA immediately after UV damage, which changes into a focal pattern after longer periods (see Fig. 2C). This possibly represents the involvement of PCNA in active NER immediately after DNA damage and in other DNA repair processes (TLS, homologous recombination) at later time points. In the CHO9 cell line that we used, most cells are in S phase, and GFP-hPCNA therefore already shows a focal pattern in most of the untreated cells, which makes PCNA a less suitable marker for the detection of damage in the form of overall nuclear foci in our cells (27). However, regardless of the method of DNA damage detection, we find, in contrast to Solomon et al., that mutant GFP-hPCNAK164R can still efficiently relocalize to the damaged areas. As immunoprecipitation experiments showed that both wild-type and mutant GFP-hPCNA interact with the endogenous wild-type PCNA (data not shown) and as it has been demonstrated that PCNA is present in solution and functions as a trimeric ring, both wild-type and mutant GFP-hPCNA probably relocalize to damaged areas as a part of the trimeric ring.
FIG. 5.
Characterization of mutant GFP-hPCNAK164R localization and kinetics in DNA replication and repair. (A) Substitution of the lysine residue at position 164 for arginine (K164R) results in defective monoubiquitination of GFP-hPCNA. Cells expressing GFP-hPCNA or GFP-hPCNAK164R were treated with 10 J/m2 (lanes 1 and 2) or 20 J/m2 (lanes 3 and 4) UV, and cell lysates were prepared after 4 h (lanes 1 and 3) or 6 h (lanes 2 and 4). The protein blot shows total cell lysates of UV-treated and untreated cells probed with anti-PCNA antibodies (upper blot) and GFP antibodies (lower blot). Indicated are the positions of endogenous PCNA, GFP-hPCNA, and monoubiquitinated GFP-hPCNA (*). (B) Time-lapse imaging experiments on GFP-hPCNAK164R-expressing cells. Upper panels: time-lapse imaging of untreated cells expressing GFP-hPCNAK164R. An example of a cell imaged from the mid-S phase to the G2 phase of the cell cycle is shown. Lower panels: time-lapse imaging of locally irradiated cell (indicated with the circle) showing transient accumulation of GFP-hPCNAK164R at sites of UV damage. (C to E) Fluorescence redistribution after photobleaching analyses of mutant GFP-hPCNAK164R-expressing cells. (C) Quantitative FLIP/FRAP analysis of mutant GFP-hPCNAK164R replication foci in the early, mid-, and late S phases of the cell cycle. (D) Examples of FLIP/FRAP analysis in locally irradiated mutant cells. The upper panel represents a GFP-hPCNAK164R-expressing cell, which, based upon the diffuse PCNA pattern, is in the G1 or G2 phase of the cell cycle and shows local accumulation of GFP-hPCNAK164R in the damaged area. The lower part of this cell, including half of the site of local damage, was photobleached, and the cell was imaged every 30 s. A similar experiment (lower panel) was done on a cell that was, based on the peripheral PCNA staining pattern, in the mid-S phase of the cell cycle. (E) Quantitation of the simultaneous FRAP and FLIP experiment on GFP-hPCNAK164R after local UV damage in G1/G2 cells as shown in Fig. 5D, upper panel. “n” represents the number of cells examined.
Next, we analyzed the kinetics of mutant PCNA in DNA replication and repair. The turnover rate of GFP-hPCNAK164R in DNA replication foci was not significantly affected by the K164R mutation, since the FLIP/FRAP curves converged after 485 ± 218 s (Fig. 5C). In contrast, the residence time for mutant PCNA in DNA-damaged areas was much shorter than that for the wild type (Fig. 5D). The FLIP/FRAP analysis in Fig. 5E shows that the residence time for GFP-hPCNAK164R in DNA-damaged areas decreased to 300 ± 90 s, or 5 min, compared to 20 min for the wild-type GFP-hPCNA protein (Fig. 4B). Thus, although the mutant PCNA protein can still localize to the UV-damaged sites, the residence time is significantly affected. Possibly, the mutated PCNA, which is unable to load TLS polymerases, loses its affinity for stalled replication forks, and this leads to the reduced residence time. However, the mutated protein also shows reduced residence time in G1/G2 cells. As there is presumably no TLS polymerase present in these stages, the mutation is also important for PCNA dynamics at sites of local damage, independent of the TLS polymerase.
In conclusion, we found that PCNA transiently accumulates at sites of DNA damage and that PCNA has different residence times in DNA replication and repair. In addition, we found that the residence times of wild-type PCNA and of PCNA bearing the K164R mutation are similar in terms of replication but not in terms of local damage repair. This shows that one function of the monoubiquitination of PCNA is to modulate the residence time of PCNA at sites of DNA damage.
Acknowledgments
This work was supported by NWO grants 805-47-193, 912-03-012, and 917-03-012 and investment grants NWO 903-68-370 and 901-01-229 from the Netherlands Organization for Scientific Research and by EC grants HPRN-CT-2002-00240 and MRTN-CT-2003-503618.
REFERENCES
- 1.Aboussekhra, A., and R. D. Wood. 1995. Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp. Cell Res. 221:326-332. [DOI] [PubMed] [Google Scholar]
- 2.Chubb, J. R., S. Boyle, P. Perry, and W. A. Bickmore. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439-445. [DOI] [PubMed] [Google Scholar]
- 3.Dunand-Sauthier, I., M. Hohl, F. Thorel, P. Jaquier-Gubler, S. G. Clarkson, and O. D. Scharer. 2005. The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. J. Biol. Chem. 280:7030-7037. [DOI] [PubMed] [Google Scholar]
- 4.Ellenberg, J., E. D. Siggia, J. E. Moreira, C. L. Smith, J. F. Presley, H. J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellison, V., and B. Stillman. 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1:E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green, C. M., and G. Almouzni. 2003. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 22:5163-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 9.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 10.Hoogervorst, E. M., H. van Steeg, and A. de Vries. 2005. Nucleotide excision repair- and p53-deficient mouse models in cancer research. Mutat. Res. 574:3-21. [DOI] [PubMed] [Google Scholar]
- 11.Houtsmuller, A. B., S. Rademakers, A. L. Nigg, D. Hoogstraten, J. H. Hoeijmakers, and W. Vermeulen. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284:958-961. [DOI] [PubMed] [Google Scholar]
- 12.Houtsmuller, A. B., and W. Vermeulen. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 115:13-21. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, B. J., S. Toering, U. Francke, and G. Chu. 1998. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 18:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kannouche, P. L., J. Wing, and A. R. Lehmann. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14:491-500. [DOI] [PubMed] [Google Scholar]
- 15.Katsumi, S., N. Kobayashi, K. Imoto, A. Nakagawa, Y. Yamashina, T. Muramatsu, T. Shirai, S. Miyagawa, S. Sugiura, F. Hanaoka, T. Matsunaga, O. Nikaido, and T. Mori. 2001. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Investig. Dermatol. 117:1156-1161. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann, A. R. 2003. Replication of damaged DNA. Cell Cycle 2:300-302. [PubMed] [Google Scholar]
- 17.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall, W. F., A. Straight, J. F. Marko, J. Swedlow, A. Dernburg, A. Belmont, A. W. Murray, D. A. Agard, and J. W. Sedat. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7:930-939. [DOI] [PubMed] [Google Scholar]
- 19.Mattern, K. A., S. J. Swiggers, A. L. Nigg, B. Lowenberg, A. B. Houtsmuller, and J. M. Zijlmans. 2004. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 24:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 21.Mone, M. J., M. Volker, O. Nikaido, L. H. Mullenders, A. A. van Zeeland, P. J. Verschure, E. M. Manders, and R. van Driel. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2:1013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademakers, S., M. Volker, D. Hoogstraten, A. L. Nigg, M. J. Mone, A. A. Van Zeeland, J. H. Hoeijmakers, A. B. Houtsmuller, and W. Vermeulen. 2003. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 23:5755-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivji, K. K., M. K. Kenny, and R. D. Wood. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell 69:367-374. [DOI] [PubMed] [Google Scholar]
- 24.Solomon, D. A., M. C. Cardoso, and E. S. Knudsen. 2004. Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell Biol. 166:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somanathan, S., T. M. Suchyna, A. J. Siegel, and R. Berezney. 2001. Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J. Cell. Biochem. 81:56-67. [DOI] [PubMed] [Google Scholar]
- 26.Sporbert, A., A. Gahl, R. Ankerhold, H. Leonhardt, and M. C. Cardoso. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10:1355-1365. [DOI] [PubMed] [Google Scholar]
- 27.van Veelen, L. R., T. Cervelli, M. W. M. M. van de Rakt, A. F. Theil, J. Essers, and R. Kanaar. 2005. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutat. Res. 574:22-33. [DOI] [PubMed] [Google Scholar]
- 28.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe, K., S. Tateishi, M. Kawasuji, T. Tsurimoto, H. Inoue, and M. Yamaizumi. 2004. Rad18 guides pol eta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, J., and E. Stelzer. 1999. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 9:61-65. [DOI] [PubMed] [Google Scholar]