Abstract
Fibroblast growth factor 2 (FGF-2), which is highly expressed in developing tissues and malignant cells, regulates cell growth, differentiation, and migration. Five isoforms (18 to ∼34 kDa) of FGF-2 are derived from alternative initiation codons of a single mRNA. The 18-kDa FGF-2 isoform is released from cells by a nonclassical secretory pathway and regulates gene expression by binding to cell surface receptors. This isoform also localizes to the nucleolus, raising the possibility that it may directly regulate ribosome biogenesis, a rate-limiting process in cell growth. Although several growth factors have been shown to accumulate in the nucleolus, their function and mechanism of action remain unclear. Here we show that 18-kDa FGF-2 interacts with upstream binding factor (UBF), an architectural transcription factor essential for rRNA transcription. The maximal activation of rRNA transcription in vitro by 18-kDa FGF-2 requires UBF. The 18-kDa FGF-2 localizes to rRNA genes and is necessary for the full activation of pre-rRNA synthesis in vivo. Our results demonstrate that 18-kDa FGF-2 directly regulates rRNA transcription.
Fibroblast growth factor 2 (FGF-2) belongs to a structurally related polypeptide family consisting of at least 23 members in vertebrates (25). It is widely expressed in tissues and plays a pivotal role in mediating cell proliferation, development, and migration (3). Differential initiation of translation from upstream CUG codons of a single mRNA yields four high-molecular-weight isoforms of FGF-2 with molecular masses of 22, 22.5, 24, and 34 kDa (14). They are exclusively located in the nucleus, and the nuclear localization is regulated by two nuclear localization signals (1, 29). The 18-kDa FGF-2 isoform is translated from the AUG codon. It can be secreted through a pathway independent of the endoplasmic reticulum and Golgi (16). Extracellularly, it binds and activates high-affinity cell surface receptors (13). Like other polypeptides, such as angiogenin and parathyroid hormone-related peptide, 18-kDa FGF-2 can translocate into the nucleus and nucleolus after internalization (27). Endogenous 18-kDa FGF-2 also localizes to the nucleus and nucleolus (10, 30). We recently demonstrated that a bipartite nuclear localization signal located in the C terminus of 18-kDa FGF-2 regulates its nuclear and nucleolar localization (30). The nuclear and nucleolar localization of 18-kDa FGF-2 suggests that it may directly function in the nucleus and nucleolus.
FGF-2 can stimulate rRNA transcription through intracellular or extracellular pathways (5, 41). When applied to NIH 3T3 cells, 18-kDa FGF-2 stimulates the extracellular signal-regulated protein kinase (ERK) signaling cascade and promotes RNA polymerase I (Pol I) transcription (41). Extracellular FGF-2 was shown to stimulate synthesis of total RNA 3-fold and that of rRNA 5.6-fold, and this stimulation coincided with the nucleolar translocation of extracellular FGF-2 (5). It was also proposed that FGF-2 might regulate rRNA transcription through interacting with casein kinase 2 (CK2) (4). A point mutation in FGF-2 abolished the interaction with CK2 and blocked FGF-2-dependent stimulation of CK2 phosphorylation of nucleolin (2). Phosphorylated nucleolin cannot repress rRNA transcription (2, 6). Although it has been shown that FGF-2 regulates the transcription of phosphoglycerate kinase directly in a cell-free system (24), the mechanism of action remains to be defined.
In the following study, we probed the mechanism of FGF-2-dependent stimulation of rRNA transcription. In addition to Pol I, the human rRNA transcription machinery consists of selectivity factor 1 (SL1) and the upstream binding factor (UBF) (11). SL1 is a complex containing the TATA-box binding protein (TBP) and at least three TBP-associated factors (42). UBF binds DNA and recruits SL1 to the rRNA gene promoter (11). We found that FGF-2 binds to UBF, associates with the rRNA gene promoter in vivo, and stimulates Pol I transcription in vitro and in vivo. Together, these results demonstrate that 18-kDa FGF-2 directly regulates rRNA transcription by interacting with a transcription factor in the nucleolus.
MATERIALS AND METHODS
GST pull down.
Glutathione S-transferase (GST) and GST-FGF-2 were purified as described previously (31). HeLa cell lysate was prepared by first incubating 2 × 108 cells (National Cell Culture Center) in 10 ml of buffer A (10 mM Tris-HCl [pH 7.5], 2 mM MgCl2, 1 mM EDTA, and 10% glycerol) containing 500 mM NaCl, 2% Triton X-100, 1% deoxycholic acid, and protease inhibitor cocktail (Roche) on ice for 1 h. One tablet of protease inhibitor cocktail was added per 10 ml of lysis buffer. The lysate was centrifuged at 9,000 × g for 5 min at 4°C. A 1-ml aliquot of the lysate was diluted fivefold in buffer A containing protease inhibitor cocktail. GST and GST-FGF-2 (1 nmol each) were added separately to lysates, and the mixtures were incubated for 2 h at 4°C. In competition experiments, 10 nmol of human recombinant FGF-2 (hrFGF-2; National Cancer Institute BRB Preclinical Repository) was added to the lysates incubated with GST-FGF-2. A 50% slurry (50 μl) of glutathione Sepharose 4B (Amersham Biosciences Corp.) was added to the mixtures, and they were incubated at 4°C for an additional 2 h. The resin was washed three times with phosphate-buffered saline containing 5 mM EDTA and 0.2% Triton X-100. Proteins bound to the resin were treated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, including 25 mM dithiothreitol at room temperature for 10 min, and then samples were incubated with 100 mM iodoacetamide at 37°C for 10 min. Samples containing FGF-2 were not exposed to a higher temperature because of FGF-2's lability (34). Proteins were separated on a SDS-PAGE gel and visualized using Coomassie brilliant blue. Protein bands were excised and analyzed using in-gel digest/nanospray liquid chromatography mass spectrometry (Protein Structure Laboratory, SUNY Downstate Medical Center).
To detect the direct interaction of FGF-2 and UBF, 50 μg of purified GST or GST-FGF-2 and 2 μg of purified FLAG-UBF were mixed together in 100 μl of buffer A and incubated at 4°C for 2 h. A 50% slurry (20 μl) of glutathione Sepharose 4B was added, and the resulting mixtures were incubated for 2 h at 4°C. The resin was washed three times with phosphate-buffered saline containing 5 mM EDTA and 0.2% Triton X-100. Bound proteins were eluted using 25 mM glutathione in 50 mM Tris-HCl (pH 8.0). FLAG-UBF was detected on Western blots using a 1:1,000 dilution of mouse monoclonal FLAG antibody (Sigma), and GST-FGF-2 was detected by using a 1:1,000 dilution of rabbit polyclonal FGF-2 antibody (Santa Cruz Biotechnology, Inc.).
Immunofluorescence microscopy.
Immunofluorescence microscopy was carried out as described previously (30). Briefly, HeLa (8 × 104) cells were transfected with 1 μg of plasmid encoding either GFP, GFP-FGF-2, or GFP-UBF (8) (gift from S. Huang, Northwestern University Medical School) using 1 μl of Lipofectamine (Invitrogen). Endogenous FGF-2 and UBF were detected by using a 1:50 dilution of mouse monoclonal FGF-2 antibody (Oncogene) or a 1:10 dilution of mouse monoclonal UBF antibody (Santa Cruz Biotechnology, Inc.), respectively. Fluorescence was detected by using a Radiance 2000 confocal microscope (Bio-Rad). In some experiments, cells were treated with actinomycin D (0.08 μg/ml) (Sigma).
Coimmunoprecipitation.
COS-7 cells (7 × 105) were cotransfected with 1 μg of p3XFLAG-CMV-14 or pFGF-2-3XFLAG-CMV-14 and 5 μg of either pEGFP-N1 or pEGFP-C1-UBF (8) using 6 μl of Lipofectamine. After 24 h, cells were incubated in buffer B (50 mM Tris-HCl [pH 7.9], 12.5 mM MgCl2, 1 mM EDTA, and 10% glycerol) containing 300 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, and protease inhibitor cocktail on ice for 1 h. Cell lysate was centrifuged at 9,000 × g for 10 min. The supernatant was diluted fivefold in buffer B containing protease inhibitor cocktail and then incubated at 4°C overnight. A total of 2 μg of mouse monoclonal FLAG antibody (Sigma) or 0.5 μl of rabbit polyclonal green fluorescent protein (GFP) antibody (Abcam, Inc.) was added. The mixture was incubated at 4°C for 2 h. Protein A Sepharose beads (30 μl of 50% slurry) were added. The mixture was incubated at 4°C for an additional 2 h. The beads were washed three times in buffer B containing 0.2% Triton X-100. Proteins were treated as described above and detected by using Western blotting as described previously (31). FLAG fusion proteins were detected by using either a 1:1,000 dilution of the mouse monoclonal FLAG antibody or a 1:1,000 dilution of the mouse monoclonal FGF-2 antibody (Oncogene). GFP fusion proteins were detected by using either a 1:2,000 dilution of mouse monoclonal GFP antibody (Clontech) or a 1:100 dilution of mouse monoclonal UBF antibody (Santa Cruz Biotechnology, Inc.).
Analysis of ERK signaling pathway.
HEK293 cells (2 × 105) were plated in 3 ml of FreeStyle 293 expression medium and grown overnight. Cells were transfected with 1 μg of pN1, pN1-FGF-2, or pN1-FGF-2-K128G (30) using 1 μl of Lipofectamine. After 24 h, cells were incubated in buffer B containing 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, and protease inhibitor cocktail on ice for 1 h. Cell lysate was collected by centrifugation (10 min at 9,000 × g). Proteins in medium were precipitated using 10% trichloroacetic acid. Proteins were then detected by using Western blotting as previously described (31). FGF-2 was detected by using a mouse monoclonal FGF-2 antibody. Actin was detected by using a mouse monoclonal actin antibody (Oncogene). Phosphorylated ERK was detected by using a rabbit phospho-ERK1/ERK2 (T202/Y204) antibody (R & D Systems).
Analysis of pre-rRNA synthesis.
HEK293 cells (2 × 105) were plated in 3 ml of FreeStyle 293 expression medium and grown overnight. Cells were cotransfected with 1 μg pf pN1, pN1-FGF-2, or pN1-FGF-2-K128G (30) and 2 μg of either pcDNA3 or pcDNA3-FLAG-UBF (23). After 24 h, total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instruction. The concentration of total RNA was determined by the absorbance at 260 nm. Total RNA (0.5 μg) was loaded onto a 1% agarose gel. 18S and 28S RNA were visualized using ethidium bromide. Pre-rRNA synthesis was measured by S1 nuclease protection assay (20) using a 32P-labeled oligonucleotide (5′-CCTCTCCAGCGACAGGTCGCCAGAGGACAGCGTGTCAGCAATAACCCGGCGGCCAAAATG-3′) (23). The protected nucleic acid was separated on a 10% polyacrylamide gel containing 8 M urea. The gel was exposed to X-ray film with an intensifying screen at −70°C for at least 1 h, and the film was analyzed using Kodak 1D Image software. The data were analyzed using one sample t test by comparing the means of each experiment to that of the control.
In vitro transcription assay.
In vitro transcription assay was performed as described previously (23). Purified proteins, including Pol I, human SL1, FLAG-UBF, FLAG-UBF1-670, or human recombinant FGF-2, were mixed with 100 μg/ml α-amanitin and 100 ng of prHu3 (a plasmid containing the rRNA gene promoter and part of the human rRNA gene) (23). The transcripts controlled by the rRNA gene promoter were examined using S1 nuclease protection assay (20).
Nuclear and nucleolar chromatin immunoprecipitation.
The nuclear chromatin immunoprecipitation method previously described (9) was modified. Ten 150-mm dishes containing HeLa cells (80% confluent) were treated with 0.25% formaldehyde and incubated in buffer C (10 mM Tris-HCl [pH 8.0], 85 mM KCl, 0.5% NP-40) containing protease inhibitor cocktail. Nuclei were collected by centrifugation (microcentrifuge, 20 s) and then resuspended in buffer D (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 1% SDS) containing protease inhibitor cocktail. Nuclear chromatin was sonicated at 4°C (Sonic Dismembrator, Model 300, Fisher) (15 s at 60% full power, 16 bursts). Disrupted nuclear chromatin was collected using centrifugation (2 min at 9,000 × g). Nucleolar chromatin was prepared, and the immunoprecipitation was performed as previously described (26). Two micrograms of affinity-purified goat immunoglobulin G (IgG) (Jackson ImmunoResearch Laboratories, Inc.), mouse monoclonal hemagglutinin antibody (Roche), mouse monoclonal nucleolin antibody (Upstate), mouse monoclonal UBF antibody (Santa Cruz Biotechnology, Inc.), or a goat polyclonal FGF-2 antibody (R & D Systems) was used to immunoprecipitate complexes.
PCR analyses.
Chromatin immunoprecipitates (8 μl) were mixed with 0.5 U of Vent polymerase and 2 mM deoxynucleotide triphosphate mix. After denaturation at 95°C for 30 s, 30 cycles of PCR were performed, each of which consisted of 30 s at 95°C, 30 s at 45°C (for rRNA gene regions) (26) or 55°C (for the γ-actin promoter region) (9), and 2 min at 72°C. PCR products (10 μl) were analyzed on an 8% polyacrylamide gel and visualized using ethidium bromide. Primer sequences for rRNA (26) and γ-actin (9) genes were previously described.
RESULTS
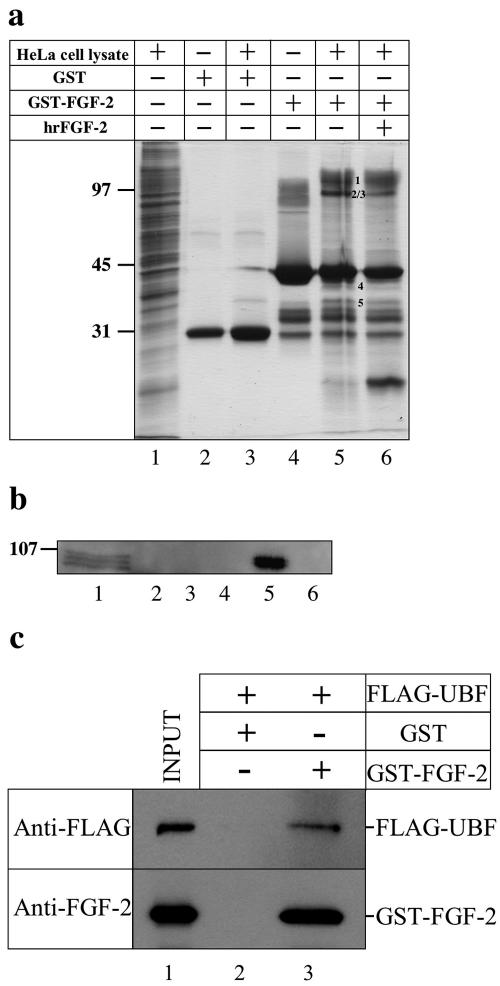
We recently deciphered the targeting signals that direct 18-kDa FGF-2 to the nucleus and nucleolus in HeLa and COS-7 cells (30). To identify potential nuclear targets, we isolated FGF-2-interacting proteins from HeLa cell lysates using GST-FGF-2 (31) (Fig. 1a). Five protein bands were pulled down with GST-FGF-2 but not with GST (Fig. 1a, compare lanes 3 and 5). The binding was reduced in the presence of hrFGF-2 (Fig. 1a, compare lanes 5 and 6). Using mass spectrometry, we identified the FGF-2-interacting proteins as human nucleolin, upstream binding factor, ribosomal protein P0, and histone H1 (Table 1). Of the two reported UBF isoforms (UBF1 and UBF2), UBF1 stimulates rRNA transcription, whereas the function of UBF2 remains unknown (11). The identity of UBF was confirmed using a UBF antibody (Fig. 1b). UBF was found in HeLa cell lysate (Fig. 1b, lane 1) and was pulled down with GST-FGF-2 (lane 5) but not with GST (lane 3) or GST-FGF-2 in the presence of hrFGF-2 (lane 6), demonstrating that UBF interacts specifically with FGF-2 in vitro. To determine whether UBF and FGF-2 directly interact, we used purified FLAG-UBF and GST-FGF-2 (Fig. 1c, lane 1). FLAG-UBF bound to GST-FGF-2 (Fig. 1c, lane 3) but not to GST (lane 2), demonstrating that FGF-2 directly interacts with UBF in vitro. The FLAG peptide did not bind to GST-FGF-2 (data not shown).
FIG. 1.
Identification of FGF-2 interacting proteins. (a) Proteins in nuclear extracts of HeLa cells that bound to GST (lane 3), GST-FGF-2 (lane 5), or GST-FGF-2 in the presence of a 10-fold excess of hrFGF-2 (lane 6) were separated on a SDS-PAGE gel and stained with Coomassie blue. HeLa cell lysate (lane 1), purified GST (lane 2), and GST-FGF-2 (lane 4) were also separated on the gel. Numbers in lane 5 correspond to proteins bound specifically to FGF-2. (b) Proteins in lanes 1 to 6 of panel a were transferred to a polyvinylidene difluoride membrane and probed with a UBF antibody. Three bands (lane 1) in the HeLa cell lysate likely correspond to UBF1, UBF2, and phosphorylated UBF (35). (c) Purified FLAG-UBF was mixed with either GST (lane 2) or GST-FGF-2 (lane 3). Bound proteins were eluted using glutathione and then detected on immunoblots by a FLAG antibody (anti-FLAG panel) or an FGF-2 antibody (anti-FGF-2 panel). Inputs (10% of total) were loaded in lane 1.
TABLE 1.
FGF-2 interacting proteins identified from HeLa cell lysate
| Band | Deduced sequence | Identification |
|---|---|---|
| 1 | GFGFVDFNSEEDAK | Nucleolin |
| 2 | GFGFVDFNSEEDAK | Nucleolin |
| SISLYYTGEK | ||
| GIAYIEFK | ||
| GYAFIEFASFEDAK | ||
| 3 | HPLNISEEGITK | Upstream binding factor |
| TLTELILDAQEHVK | ||
| VHLDLSWK | ||
| FSQELLISNGELNHLPLK | ||
| 4 | IILQLLDDYPK | Ribosomal protein P0 |
| TSFFQALGITTK | ||
| VLALSVETDYTFPLAEK | ||
| 5 | ASGPPVSELLTK | Histone H1 |
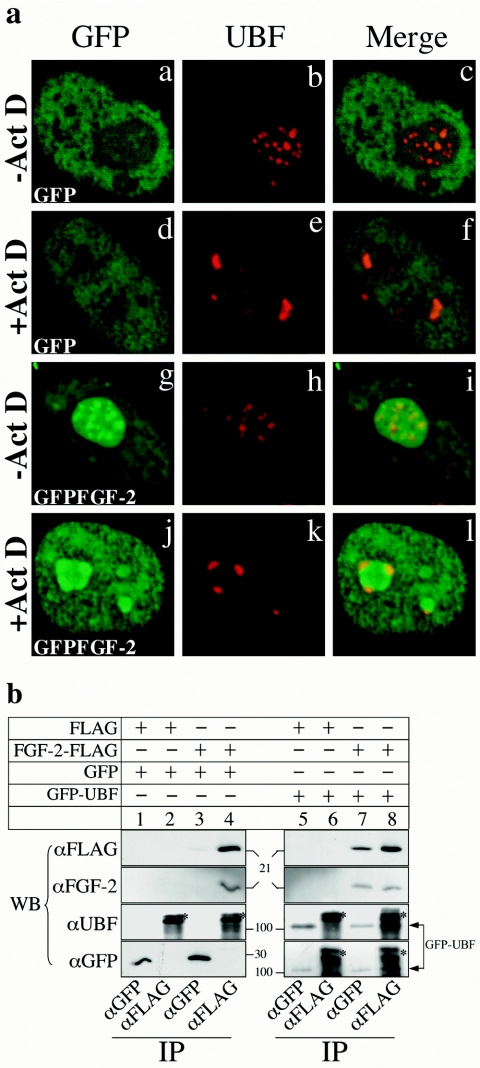
To investigate the interaction between UBF and 18-kDa FGF-2 in vivo, we transfected HeLa cells with a plasmid encoding GFP-FGF-2 (30). GFP-FGF-2, but not GFP alone, accumulated in the nucleolus (Fig. 2a, panels a and g). UBF was concentrated in patches in the nucleolus (Fig. 2a, panels b and h). Merged images showed that GFP-FGF-2 (Fig. 2a, panel i), but not GFP (Fig. 2a, panel c), colocalized with UBF. Actinomycin D inhibits rRNA transcription and redistributes components involved in rRNA transcription, including UBF, to the periphery of nucleolus (Fig. 2a, panel e) (21). Interestingly, GFP-FGF-2 also redistributed to the periphery of the nucleolus and localized with UBF in the presence of actinomycin D (Fig. 2a, panels j through l).
FIG. 2.
FGF-2 interacts with UBF in vivo. (a) Confocal images of HeLa cells expressing GFP (panels a through f) or GFP-FGF-2 (panels g through l) and incubated with actinomycin D (Act D) (panels d through f and j through l) or without actinomycin D (panels a through c and g through i). GFP and GFP-FGF-2 were localized by autofluorescence (panels a, d, g, and j). UBF was detected by using a UBF monoclonal antibody and a Texas Red conjugated secondary antibody (panels b, e, h, and k). Merged images are shown in panels c, f, i, and l. (b) COS-7 cells were cotransfected with p3XFLAG-CMV-14 and pEGFP-N1 (lanes 1 and 2), pFGF-2-3XFLAG-CMV-14 and pEGFP-N1 (lanes 3 and 4), p3XFLAG-CMV-14 and pEGFP-C1-UBF (lanes 5 and 6), or pFGF-2-3XFLAG-CMV-14 and pEGFP-C1-UBF (lanes 7 and 8), and then proteins were immunoprecipitated (IP) with GFP (lanes 1, 3, 5, and 7) or FLAG (lanes 2, 4, 6, and 8) antibodies. Immunoprecipitated proteins were identified using Western blots (WB) probed with FLAG, FGF-2, UBF, or GFP antibodies. The position of GFP-UBF is indicated by arrows. Asterisks in lane 2, 4, 6, and 8 indicate background bands.
We also examined the interactions between FGF-2 and UBF in vivo by coimmunoprecipitating them from cells expressing both FGF-2-FLAG and GFP-UBF (Fig. 2b). FGF-2-FLAG was immunoprecipitated from cells expressing both FGF-2-FLAG and GFP-UBF (Fig. 2b, lane 7) using a GFP antibody but not from cells expressing both FGF-2-FLAG and GFP (lane 3). Similarly, GFP-UBF was immunoprecipitated from cells expressing both FGF-2-FLAG and GFP-UBF using a FLAG antibody (Fig. 2b, lane 8) but not from cells expressing only GFP-UBF (lane 6). Control experiments indicated that the GFP antibody has no affinity for endogenous FGF-2 or UBF (Fig. 2b, lanes 1 and 3). Similarly, the FLAG antibody has no affinity for endogenous FGF-2, endogenous UBF, or GFP (Fig. 2b, lane 2). Together, these results suggest that FGF-2 and UBF interact in vivo.
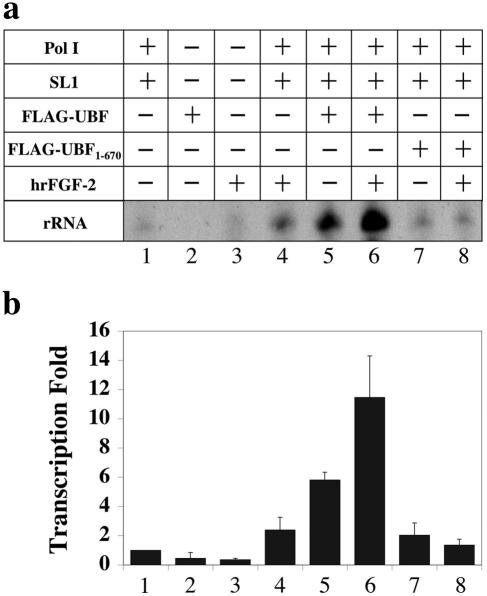
Bovine endothelial cells can internalize FGF-2 and translocate it into the nucleus and nucleolus. Nucleolar localization of FGF-2 coincides with an increase in rRNA transcription (5). The interaction between FGF-2 and UBF described above suggests that, together, they directly and cooperatively regulate rRNA transcription in the nucleolus. To examine this possibility, we measured the effect of FGF-2 on rRNA transcription in vitro using purified Pol I, SL1, and UBF (Fig. 3a and b). SL1, which is essential for rRNA transcription, is a complex of proteins containing TBP and three TBP-associated factors (42). In the presence of Pol I and SL1, UBF stimulated transcription sixfold (Fig. 3a and b, compare lanes 1 and 5). Together FGF-2 and UBF stimulated rRNA transcription 12-fold (Fig. 3a and b, compare lanes 1 and 6). Alone, FGF-2 (Fig. 3a and b, lanes 2) and UBF (lanes 3) had no activity. In the presence of Pol I and SL1, FGF-2 stimulated transcription 2.5-fold (Fig. 3a and b, compare lanes 1 and 4), suggesting either that FGF-2 has some ability to stimulate transcription directly or that the Pol I or SL1 preparation contains trace amounts of UBF. A UBF mutant (FLAG-UBF1-670) lacking the C-terminal acidic tail required for transcriptional activation failed to stimulate transcription (Fig. 3a and b, lanes 7) as previously described (35). FGF-2 stimulation of UBF-dependent rRNA transcription was defective in the presence of FLAG-UBF1-670 (Fig. 3a and b, lanes 8). Together, these results suggest that maximal stimulation of rRNA transcription by FGF-2 is dependent on UBF.
FIG. 3.
FGF-2 stimulates rRNA transcription in vitro. (a) The amount of rRNA transcription in the presence of the indicated combinations of purified proteins was measured using S1 nuclease protection. (b) Band intensities in panel a were quantified and normalized to the amount of rRNA transcription in the presence of RNA Pol I and SL1 (lane 1). Bars represent the standard errors of four determinations. Relative to lane 1, P values of lanes 2 to 8 were 0.07, 0.0009, 0.05, 0.0004, 0.005, 0.09, and 0.2, respectively.
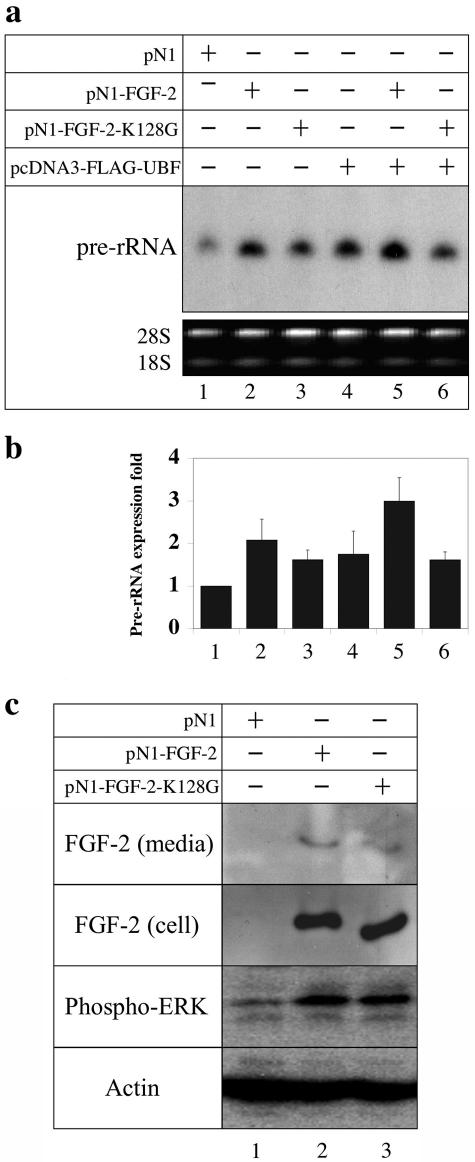
To determine the effect of FGF-2 on the synthesis of pre-rRNA in vivo, we expressed wild-type 18-kDa FGF-2 and FLAG-UBF in HEK293 cells and measured the synthesis of pre-rRNA (45S) using an S1 nuclease protection assay. Because the processing of 45S pre-rRNA is much faster than its synthesis, the expression level of pre-rRNA at steady state will primarily represent the rate of pre-rRNA synthesis (12). HEK293 cells were chosen because they express a very low level of FGF-2 [Fig. 4c, FGF-2 (cell) panel, lane 1]. Furthermore, pre-rRNA synthesis and ERK activity in HEK293 cells can be regulated by exogenously added FGF-2 (32; data not shown). Pre-rRNA synthesis was stimulated 2.1-fold in cells expressing FGF-2 (Fig. 4a and b, lanes 1 and 2). Expressing FLAG-UBF alone in HEK293 cells increased the pre-rRNA synthesis 1.8-fold (Fig. 4a and b, lanes 4), whereas coexpressing FLAG-UBF and FGF-2 stimulated pre-rRNA synthesis about 3-fold (Fig. 4a and b, lanes 5). To define the contribution of nucleolar FGF-2, we took advantage of an FGF-2 mutant (FGF-2-K128G) that is excluded from the nucleolus (30). Expressing FGF-2-K128G in HEK293 cells increased pre-rRNA synthesis 1.8-fold (Fig. 4a and b, lanes 1 and 3). However, coexpressing FLAG-UBF and FGF-2-K128G did not fully stimulate pre-rRNA synthesis (Fig. 4a and b, lanes 5 and 6). Therefore, the failure of this mutant to fully stimulate pre-rRNA synthesis is consistent with its exclusion from the nucleolus and loss of access to UBF. Together, these results demonstrate that FGF-2 directly regulates rRNA transcription in the nucleolus.
FIG. 4.
FGF-2 enhances pre-rRNA synthesis in HEK293 cells. (a) The amount of pre-rRNA in HEK293 cells transfected with pN1 (lane 1), pN1-FGF-2 (lane 2), pN1-FGF-2-K128G (lane 3), pcDNA3-FLAG-UBF (lane 4), pN1-FGF-2 and pcDNA3-FLAG-UBF (lane 5), and pN1-FGF-2-K128G and pcDNA3-FLAG-UBF (lane 6) was determined using S1 nuclease protection and normalized to the protected oligonucleotide in lane 1. The loading controls (18S and 28S RNA) were visualized using ethidium bromide. (b) The protected oligonucleotides in panel c were quantified, and band intensities were normalized to the amount of pre-rRNA in cells transfected with control plasmid (lane 1). Bars represent the standard error of four determinations. Relative to lane 1, P values of lanes 2 to 6 were 0.02, 0.01, 0.07, 0.006, and 0.007, respectively. (c) HEK293 cells were transfected with control plasmid pN1 (lane 1), pN1-FGF-2 (lane 2), or pN1-FGF-2-K128G (lane 3), and then extracts were immunoblotted using antibodies against either FGF-2, phospho-ERK1/ERK2 (T202/Y204), or actin.
FGF-2 may stimulate pre-rRNA synthesis intracellularly in the nucleolus or extracellularly through the ERK pathway after secretion and binding to an FGF receptor (41). To define the contributions of each pathway to pre-rRNA synthesis, we first determined whether HEK293 cells secrete FGF-2. HEK293 cells were transfected with pN1-FGF-2, and the amount of FGF-2 in the medium and cell lysates was determined. About 6% of the total FGF-2 was found in the medium, indicating that HEK293 cells can secrete FGF-2 [Fig. 4c, FGF-2 (cell) and FGF-2 (media) panels lane 2]. To determine whether secreted FGF-2 activated the ERK pathway, we measured the level of phosphorylated ERK. The level of phosphorylated ERK was elevated in HEK293 cells that were secreting FGF-2, suggesting that this pathway was activated (Fig. 4c, phospho-ERK panel, lanes 1 and 2). The FGF-2-K128G mutant was secreted, and it activated the ERK pathway (Fig. 4c, lane 3). These results suggest that both extracellular and intracellular FGF-2 contribute to the full stimulation of rRNA transcription.
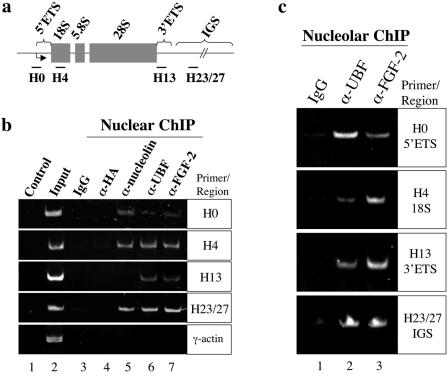
The interaction between FGF-2 and UBF suggests that FGF-2 can associate with rRNA genes. We explored this possibility using nuclear chromatin immunoprecipitation. A single rRNA gene repeat, about 43 kb in humans, is composed of the intergenic spacer (IGS) and the rRNA precursor, including 5′ and 3′ external-transcribed spacers (ETS), three rRNA genes (18S, 5.8S, and 28S), and two internal-transcribed spacers (26) (Fig. 5a). UBF was previously shown to associate with these regions of DNA (26). The different rRNA gene regions and the promoter region of γ-actin (control) were amplified from a nuclear chromatin fraction (Fig. 5b, lane 2) but not control buffer (Fig. 5b, lane 1). The 5′ ETS, including the rRNA gene promoter (Fig. 5b, H0 panel), 18S gene (Fig. 5b, H4 panel), 3′ ETS (Fig. 5b, H13 panel), and IGS (Fig. 5b, H23/27 panel) were immunoprecipitated with either a UBF (lane 6) or an FGF-2 antibody (lane 7). However, the γ-actin promoter (Fig. 5b, γ-actin panel) was not immunoprecipitated with these two antibodies (lanes 6 and 7), indicating that association of UBF and FGF-2 with rRNA genes is specific. Nucleolin binds to rRNA genes in vitro, but no in vivo evidence has been reported (17). A nucleolin antibody immunoprecipitated the regions of 5′ ETS, 18S gene, and IGS but not those of 3′ ETS and the γ-actin promoter (lane 5). No DNA was precipitated with either rabbit IgG or a mouse hemagglutinin antibody (lanes 2 and 3). Other studies have demonstrated that UBF is associated with rRNA genes using nucleolar chromatin immunoprecipitation (26). We then further investigated the binding activities of FGF-2 to rRNA genes using nucleolar chromatin fractions. Regions of 5′ ETS, including the rRNA gene promoter, 18S gene, 3′ ETS, and IGS from nucleolar chromatin fractions were immunoprecipitated using a UBF (Fig. 5c, lane 2) or an FGF-2 antibody (lane 3) but not IgG (lane 1). These results indicate that FGF-2 is localized to the rRNA transcription complex within the nucleolus.
FIG. 5.
FGF-2 localizes to rRNA genes. (a) A human rRNA gene repeat contains 18S, 5.8S, and 28S units and the ETS and IGS. The regions targeted in PCR analyses are also indicated. (b) Control PCRs contained the total chromatin (lane 2) or water (lane 1). PCRs of immunoprecipitates using the indicated antibodies are shown in lanes 3 to 7. (c) PCRs of immunoprecipitates from nucleolar chromatin using the indicated antibodies are shown in lanes 1 to 3.
DISCUSSION
Using purified GST-FGF-2 (31), we identified nucleolin, histone H1, UBF, and ribosomal protein P0 as FGF-2 interacting proteins. We demonstrated that FGF-2 interacts with UBF, associates with rRNA genes, and regulates rRNA transcription in vitro and in vivo. Several other growth factors have been found in the nucleolus (27). To our knowledge, this is the first demonstration of a growth factor regulating transcription in the nucleolus by interacting with a transcription factor. Perhaps other nucleolar growth factors (27) regulate transcription by an analogous mechanism.
UBF is a transcription factor that plays a key role in the initiation of rRNA transcription (11). Using high-mobility group (HMG) boxes and a dimerization domain located in its N terminus, UBF dimerizes and bends the DNA helix, drawing the elements essential for rRNA transcription together (11). UBF also recruits SL1 and Pol I to the promoter (11). The activity of UBF can be modulated by phosphorylation (15, 18, 32, 33, 35-37, 40), expression (19, 28), and direct interactions with effectors (7, 38, 39).
Cell surface receptor-mediated signaling pathways have been shown to regulate rRNA transcription through UBF (15, 32). Activation of the ERK pathway resulted in the phosphorylation of threonine residues (117 and 201) in HMG boxes 1 and 2 of UBF (32). The phosphorylation of these residues diminished binding of UBF to DNA and increased rRNA transcription perhaps by facilitating promoter escape or Pol I clearance (32). Insulin receptor substrate 1 has been shown to translocate to the nucleolus and activate rRNA transcription by interacting with phosphatidylinositol 3-kinase (PI3-K) and UBF (15, 33). Insulin receptor substrate 1 stimulates the phosphorylation of UBF's C terminus by PI3-K. Increased phosphorylation of UBF's C terminus by p70 ribosomal S6 kinase 1 (S6K1) (18) or by CK2 (35-37) enhances SL1 recruitment thereby stimulating rRNA transcription. Thus, phosphorylation of UBF at HMG boxes 1 and 2 regulates its interaction with DNA, while phosphorylation at the C terminus regulates SL1 binding.
The cellular level of UBF also influences Pol I transcription. For example, overexpressing UBF enhances rRNA synthesis (19, 28) (Fig. 4).
Direct interactions between UBF and its modulators often attenuate rRNA transcription (7, 38, 39). Tumor suppressors, such as the retinoblastoma susceptibility gene product (pRb) (7, 38) and p53 (39), inhibit rRNA transcription by interacting with UBF and SL1, respectively. In contrast, the interaction between FGF-2 and UBF reported here stimulates rRNA transcription. Given that both proteins are associated with the rRNA gene promoter, it is tempting to speculate that direct interactions between FGF-2 and UBF stimulate transcription by enhancing a post-preinitiation complex assembly step, such as promoter escape or Pol I clearance. Alternatively, FGF-2 may stimulate Pol I transcription by facilitating phosphorylation of UBF through its interaction with CK2.
Our findings raise the possibility that the high-molecular-weight forms of FGF-2 may also directly regulate transcription in the nucleus. Although the high-molecular-weight isoforms of FGF-2 contain N-terminal extensions (14), they retain the core elements of the 18-kDa form containing motifs required for UBF interaction and rRNA transcriptional control. They are distributed throughout the nucleus and nucleolus (10, 27). Further investigation will clarify the functional relationship between members of the FGF-2 family and their roles, if any, in rRNA transcription.
FGF-2 can act extracellularly through cell surface receptors (41) and intracellularly in the nucleolus through UBF, as shown here, to stimulate rRNA transcription. The FGF-2 K128G mutant, which is restricted from the nucleolus, underscores the importance of the subcellular localization of FGF-2. Such a dual mode of action may allow cells to regulate the effects of FGF-2 by controlling expression, secretion, internalization, and the amount or the activity of nuclear/nucleolar FGF-2. In cells expressing little or no FGF-2, regulation of rRNA transcription by FGF-2 may depend mostly on extracellular FGF-2 signaling initiated at the cell surface. FGF-2 plays an important role in angiogenesis, cardiac hypertrophy, and certain types of cancer (3, 22). The interactions of FGF-2 in the nucleolus described here may serve as novel targets for attenuating FGF-2's intracrine pathway without affecting its extracellular paracrine and autocrine pathways.
Acknowledgments
We thank Wenquan Wang (University of Alabama) for assistance in statistical analysis of data, Sonia Navarro (University of Southern California) for technical assistance, Jimmy Calaycay and Julie Rushbrook for the mass spectrometry analysis at the Protein Structure Laboratory at SUNY Downstate, Sui Huang for providing pEGFP-C1-UBF, and Vincent Garofalo (SUNY Downstate) for assistance with digital images.
This work was supported by an award (0151261T) from the American Heart Association to William J. Chirico.
REFERENCES
- 1.Arnaud, E., C. Touriol, C. Boutonnet, M. C. Gensac, S. Vagner, H. Prats, and A. C. Prats. 1999. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol. Cell. Biol. 19:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailly, K., F. Soulet, D. Leroy, F. Amalric, and G. Bouche. 2000. Uncoupling of cell proliferation and differentiation activities of basic fibroblast growth factor. FASEB J. 14:333-344. [PubMed] [Google Scholar]
- 3.Bikfalvi, A., S. Klein, G. Pintucci, and D. B. Rifkin. 1997. Biological roles of fibroblast growth factor-2. Endocr. Rev. 18:26-45. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet, H., O. Filhol, I. Truchet, P. Brethenou, C. Cochet, F. Amalric, and G. Bouche. 1996. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 271:24781-24787. [DOI] [PubMed] [Google Scholar]
- 5.Bouche, G., N. Gas, H. Prats, V. Baldin, J. P. Tauber, J. Teissie, and F. Amalric. 1987. Basic fibroblast growth factor enters the nucleolus and stimulates the transcription of ribosomal genes in ABAE cells undergoing G0→G1 transition. Proc. Natl. Acad. Sci. USA 84:6770-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouche, G., V. Baldin, P. Belenguer, H. Prats, and F. Amalric. 1994. Activation of rDNA transcription by FGF-2: key role of protein kinase CKII. Cell. Mol. Biol. Res. 40:547-554. [PubMed] [Google Scholar]
- 7.Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosomal biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and γ-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claus, P., F. Doring, S. Gringel, F. Muller-Ostermeyer, J. Fuhlrott, T. Kraft, and C. Grothe. 2003. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J. Biol. Chem. 278:479-485. [DOI] [PubMed] [Google Scholar]
- 11.Comai, L. 2004. Mechanism of RNA polymerase I transcription. Adv. Protein Chem. 67:123-155. [DOI] [PubMed] [Google Scholar]
- 12.Craig, N., S. Kass, and B. Sollner-Webb. 1987. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc. Natl. Acad. Sci. USA 84:629-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dailey, L., D. Ambrosetti, A. Mansukhani, and C. Basilico. 2005. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 16:233-247. [DOI] [PubMed] [Google Scholar]
- 14.Delrieu, I. 2000. The high molecular weight isoforms of basic fibroblast growth factor (FGF-2): an insight into an intracrine mechanism. FEBS Lett. 468:6-10. [DOI] [PubMed] [Google Scholar]
- 15.Drakas, R., X. Tu, and R. Baserga. 2004. Control of cell size through phosphorylation of upstream binding factor 1 by nuclear phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 101:9272-9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florkiewicz, R. Z., J. Anchin, and A. Baird. 1998. The inhibition of fibroblast growth factor-2 export by cardenolides implies a novel function for the catalytic subunit of Na+,K+-ATPase. J. Biol. Chem. 273:544-551. [DOI] [PubMed] [Google Scholar]
- 17.Hanakahi, L. A., H. Sun, and N. Maizels. 1999. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 274:15908-15912. [DOI] [PubMed] [Google Scholar]
- 18.Hannan, K. M., Y. Brandenburger, A. Jenkins, K. Sharkey, A. Cavanaugh, L. Rothblum, T. Moss, G. Poortinga, G. A. McArthur, R. B. Pearson, and R. D. Hannan. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23:8862-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannan, R. D., V. Stefanovsky, L. Taylor, T. Moss, and L. I. Rothblum. 1996. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 93:8750-8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, M. J., and J. C. B. Zomerdijk. 2004. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 279:8911-8918. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, P., M. Mannervik, L. Tora, and M. Carmo-Fonseca. 1996. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133:225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kardami, E. Z. S. Jiang, S. K. Jimenez, C. J. Hirst, F. Sheikh, P. Zahradka, and P. A. Cattini. 2004. Fibroblast growth factor 2 isoforms and cardiac hypertrophy. Cardiovasc. Res. 63:458-466. [DOI] [PubMed] [Google Scholar]
- 23.Lin, C. Y., J. Tuan, P. Scalia, T. Bui, and L. Comai. 2002. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr. Biol. 12:2142-2146. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi, Y., K. Kihara, K. Mizuno, Y. Masamune, Y. Yoshitake, and K. Nishikawa. 1992. Direct effect of basic fibroblast growth factor on gene transcription in a cell-free system. Proc. Natl. Acad. Sci. USA 89:5216-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ornitz, D. M., and N. Itoh. 2001. Fibroblast growth factors. Genome Biol. 2:reviews3005.1-3005.12. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Sullivan, A. C., G. J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulating sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pederson, T. 1998. Growth factors in the nucleolus? J. Cell Biol. 143:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poortinga, G., K. M. Hannan, H. Snelling, C. R. Walkley, A. Jenkins, K. Sharkey, M. Wall, Y. Brandenburger, M. Palatsides, R. B. Pearson, G. A. McArthur, and R. D. Hannan. 2004. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 23:3325-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quarto, N., F. P. Finger, and D. B. Rifkin. 1991. The NH2-terminal extension of high molecular weight bFGF is a nuclear targeting signal. J. Cell. Physiol. 147:311-318. [DOI] [PubMed] [Google Scholar]
- 30.Sheng, Z., J. A. Lewis, and W. J. Chirico. 2004. Nuclear and nucleolar localization of 18-kDa fibroblast growth factor-2 is controlled by C-terminal signals. J. Biol. Chem. 279:40153-40160. [DOI] [PubMed] [Google Scholar]
- 31.Sheng, Z., S. B. Chang, and W. J. Chirico. 2003. Expression and purification of a biologically active basic fibroblast growth factor fusion protein. Protein Expr. Purif. 27:267-271. [DOI] [PubMed] [Google Scholar]
- 32.Stefanovsky, V. Y., G. Pelletier, D. P. Bazett-Jones, C. Crane-Robinson, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Nucleic Acids Res. 29:3241-3247.11470882 [Google Scholar]
- 33.Sun, H., X. Tu, M. Prisco, A. Wu, I. Casiburi, and R. Baserga. 2003. Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol. Endocrinol. 17:472-486. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, S. A., A. A. Protter, L. Bitting, J. C. Fiddes, and J. A. Abraham. 1991. Cloning, recombinant expression, and characterization of basic fibroblast growth factor. Methods Enzymol. 198:96-116. [DOI] [PubMed] [Google Scholar]
- 35.Tuan, J. C., W. Zhai, and L. Comai. 1999. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxyl-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19:2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voit, R., A. Kuhn, E. E. Sander, and I. Grummt. 1995. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23:2593-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voit, R., A. Schnapp, A. Kuhn, H. Rosenbauer, P. Hirschmann, H. G. Stunnenberg, and I. Grummt. 1992. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 11:2211-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voit, R., K. Schafer, and I. Grummt. 1997. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 17:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai, W., and L. Comai. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai, W., and L. Comai. 1999. A kinase activity associated with simian virus 40 large T antigen phosphorylates upstream binding factor (UBF) and promotes formation of a stable initiation complex between UBF and SL1. Mol. Cell. Biol. 19:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, J., X. Yuan, M. Frodin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405-413. [DOI] [PubMed] [Google Scholar]
- 42.Zomerdijk, J. C., H. Beckmann, and R. Tjian. 1994. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 266:2015-2018. [DOI] [PubMed] [Google Scholar]