Abstract
APP, amyloid β precursor protein, is linked to the onset of Alzheimer's disease (AD). We have here found that transforming growth factor β2 (TGFβ2), but not TGFβ1, binds to APP. The binding affinity of TGFβ2 to APP is lower than the binding affinity of TGFβ2 to the TGFβ receptor. On binding to APP, TGFβ2 activates an APP-mediated death pathway via heterotrimeric G protein Go, c-Jun N-terminal kinase, NADPH oxidase, and caspase 3 and/or related caspases. Overall degrees of TGFβ2-induced death are larger in cells expressing a familial AD-related mutant APP than in those expressing wild-type APP. Consequently, superphysiological concentrations of TGFβ2 induce neuronal death in primary cortical neurons, whose one allele of the APP gene is knocked in with the V642I mutation. Combined with the finding indicated by several earlier reports that both neural and glial expression of TGFβ2 was upregulated in AD brains, it is speculated that TGFβ2 may contribute to the development of AD-related neuronal cell death.
Transforming growth factor βs (TGFβs) have been implicated in a broad diversity of biological activities, including cell growth, cell death, cell differentiation, inflammation, and immunological reactions, by modifying the expression of specific sets of target genes (29, 30, 45). Three isoforms of TGFβs, TGFβ1, TGFβ2, and TGFβ3, bind to the constitutive active serine/threonine kinase TGFβ receptor II (TGFβRII). Upon ligand binding, the type I TGFβ receptor (TGFβRI) is recruited into a receptor signaling complex, and kinase activity of TGFβRI is activated by TGFβRII-mediated phosphorylation. The receptor complex then activates signaling cascades to target genes by phosphorylating Smad family transcription factors.
It has been generally accepted that functions of TGFβ family members may vary depending on cellular status and cell types. In neuronal tissues, it is clear that TGFβs play a neurotrophic role in some situations (5, 19, 27, 37), while they elicit cell-death-inducing effects in other situations (20, 42).
Accumulated evidence has revealed clear differences in biochemical and biological characteristics of TGFβ isoforms, although they share 71 to 76% identity in their amino acid sequence. It is especially noted that TGFβ2 has a lower affinity to the type II receptor than TGFβ1 and TGFβ3 (7, 28). In contrast, TGFβ2 has a higher affinity to the type III TGFβ receptor, which does not have the kinase domain and is considered to help TGFβ to bind to the type II receptor (29). It has also been reported that the TGFβ isoforms have their selective actions in certain systems. For example, TGFβ1 and TGFβ3, but not TGFβ2, strongly inhibit the growth of some glial cells (16).
Alzheimer's disease (AD), the most prevalent neurodegenerative disease, is characterized by three major pathological manifestations: neuronal loss, intracellular neurofibrillary tangles, and extracellular senile plaques. The major constituent of the plaques is amyloid β (Aβ), cleaved off from the transmembrane amyloid β precursor protein (APP) (33). Formation and accumulation of Aβ has been implicated in the development of AD (4, 8, 13, 44, 46). The removal of Aβ by anti-Aβ antibody from the brain improves the memory impairment of some AD model mice, indicating that upregulated Aβs contribute to the progressive memory impairment in vivo (11).
It has been suggested that TGFβ1 may be involved in the onset of AD. TGFβ1 enhanced the generation of Aβ in transgenic mice that constitutively overexpressed familial AD (FAD)-linked mutant APPs (21, 47). TGFβ1 also was shown to enhance the expression of APP in vitro (1, 12).
In addition, expression of TGFβ2 has been reported to increase in the FAD brain (10, 23, 36). Flanders et al. reported that the expression of TGFβ2 was markedly enhanced in glial cells as well as in neurons bearing neurofibrillary tangles in AD brains (10). Enzyme-linked immunosolvent assays also indicated that TGFβ2 levels were threefold higher in homogenates of AD brains than in those of controls. Despite these foregoing clinical findings, the biological significance of the upregulation of TGFβ2 in AD brains remains unknown.
APP structurally resembles a single transmembrane receptor (17). Multiple groups have found that overexpression of FAD-associated mutant APPs induces neuronal cell death by triggering intracellular death signals (14, 25, 31, 48, 49, 50). In addition, it has been shown that binding of an anti-APP antibody to APP or the artificial dimerization of the intracytoplasmic domain of APP triggers neuronal cell death mediated by a heterotrimeric G protein, Go, c-Jun N-terminal kinase (JNK), NADPH oxidase, and caspase 3 and/or related caspases (15, 39, 43). All these findings suggest that APP may be a putative receptor for a cell-death-inducing ligand(s), but direct evidence supporting this hypothesis has not been provided. In this regard, it is noted that TGFβ2 was shown to bind to the extracellular domain of APP (3), suggesting that TGFβ2 may be involved in APP-mediated cellular function.
We here demonstrate that TGFβ2 is a putative neuronal death-inducing ligand for APP. Our data suggest that TGFβ2 may be involved in the pathogenesis of AD-related neuronal loss.
MATERIALS AND METHODS
Cell lines, genes, recombinant proteins, and antibodies.
Neurohybrid F11 cells and F11/EcR cells were as described previously (14, 15). The wild-type APP (wtAPP), wtAPP lacking domain 20 (wtAPPΔ20), wtAPPΔ19, V642I-APP (the isoleucine mutation at valine 642 in APP695), and NL-APP (the asparagine/leucine mutation at Lys595/Met596 in APP695) cDNAs in the pcDNA3 vector were all as described previously (14, 15, 43). The APP isoform denoted as APP in this study is APP695. Carboxyl-terminally V5-tagged mouse amyloid precursor-like protein 2 (APLP2) cDNA in the pcDNA3.1/GS vector and horseradish peroxidase (HRP)-conjugated anti-V5 antibody were purchased from Invitrogen (Carlsbad, CA). The cDNA fragment encoding most parts of the extracellular domain of mouse APP695 (APP-ED) corresponding to amino acids 1 to 590, fused in frame to the cDNA encoding the Fc region of human immunoglobulin G (IgG) (26), was inserted into the pEF-BOS plasmid (32) and named pEF-APP-ED/Fc. The cDNA fragment encoding most parts of the extracellular domains of human ciliary neurotrophic factor receptor corresponding to amino acids 1 to 390 (CNTFR-ED), fused in frame to the cDNA encoding the Fc region of human IgG and C-terminally tagged with 6× histidine, was inserted into the pcDNA3 plasmid and named pcDNA3-CNTFR-ED/Fc. The vector encoding the extracellular domain and the transmembrane domain of the epidermal growth factor receptor (EGFR) fused with the cytoplasmic domain of mouse APP was as described previously (15). The human TGFβ receptor II expression vector was a kind gift of Xuedong Liu (Baylor College of Medicine). Lipofectamine, N2 supplement, and PLUS reagent were from Invitrogen (Carlsbad, CA). Recombinant human TGFβ1, TGFβ2, and TGFβ3 were from R&D Systems (Minneapolis, MN) and PeproTech EC Ltd. (London, United Kingdom). Recombinant human TGFα and an anti-TGFβ2 neutralizing polyclonal antibody were from R&D Systems. Antibodies to TGFβ1, TGFβ2, TGFβ3, and TGFβRII were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). 22C11 anti-APP antibody was from Chemicon (Temecula, CA). The His-tagged recombinant soluble APPα corresponding to amino acids 1 to 612 of APP695 was purchased from Sigma (St. Louis, MO). Other materials were all commercially available.
Transfection procedure, cell death assay, and cell viability assay.
The transient transfection procedures were as described previously (14, 15). At 24 h after transfection, F11 cells or F11/EcR cells were treated with recombinant TGFβ1, TGFβ2, TGFβ3, or TGFα in serum-free Ham's F-12 medium with N2 supplement. Transfection efficiency in these protocols has been determined to be invariably around 70%. At 72 h after transfection, the trypan blue exclusion assay was performed as a cell death assay, and the WST-8 assay and/or calcein fluorescence assay was performed as a cell viability assay (14, 15).
Immunofluorescence-based binding assays. (i) Method 1.
F11 cells (7 × 104 cells/well in 6-well plates) were replated onto 96-well plates (7 × 103 cells/well) at 24 h after transfection with the indicated amounts of wtAPP- or mutant APP-encoding plasmids or the TGFβRII-encoding plasmid. At 36 h after transfection, cells were combined with the indicated amounts of TGFβ1 or TGFβ2. After incubation for 6 h, they were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, followed by incubation at room temperature with antibody to TGFβ1 (1:50) or TGFβ2 (1:50) in PBS with 1% bovine serum albumin (BSA) for 2 h. After being washed with PBS three times, cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibody (Sigma) (1:150) in PBS with 1% BSA. After being washed three times with PBS, the immunofluorescence intensity was measured (excitation, 485 nm; emission, 535 nm) with a spectrofluorometer (Wallac 1420 ARVOsx Multi Label Counter). Cells not combined with TGFβs were immunostained with FITC-conjugated anti-rabbit IgG antibody without treatment with antibodies to TGFβs (indicated as “none”). This procedure was used in all binding assays except for the one shown in the experiments depicted in Fig. 2.
FIG. 2.
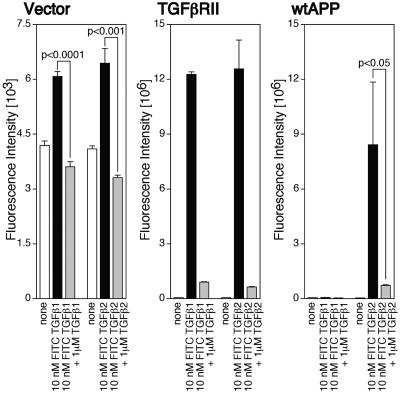
Specific association between TGFβ2 and F11 cells overexpressing wtAPP. F11 cells (7 × 104 cells/well in 6-well plates) were transfected with 0.25 μg of the TGFβRII-encoding plasmid or the wtAPP-encoding plasmid and were replated into 96-well plates coated with poly-l-lysine (7 × 103 cells/well) at 24 h after transfection. At 36 h after transfection, cells were combined with 10 nM FITC-labeled TGFβ1 or TGFβ2 in the presence or the absence of 1 μM unlabeled TGFβ1 or TGFβ2. To keep the total FITC amounts constant, proper amounts of free FITC were added. After incubation for 6 h, they were washed with PBS three times. The immunofluorescence intensity was measured (excitation, 485 nm; emission, 535 nm) with a spectrofluorometer (Wallac 1420 ARVOsx multilabel counter).
(ii) Method 2
In the experiments shown in Fig. 2, F11 cells (7 × 104 cells/well in 6-well plates) were replated onto 96-well plates coated with poly-l-lysine (7 × 103 cells/well) at 24 h after transfection with the indicated amounts of the wtAPP-encoding vector or the TGFβRII-encoding vector. At 36 h after transfection, cells were combined with the indicated amounts of FITC (purchased from Sigma)-labeled TGFβ1 or TGFβ2 in the presence or the absence of the indicated amounts of unlabeled TGFβ1 or TGFβ2. To keep the total FITC amounts in wells constant, proper amounts of free FITC were added. After 6 h of incubation, they were washed with PBS three times. The immunofluorescence intensity was then measured (excitation, 485 nm; emission, 535 nm) with a spectrofluorometer (Wallac 1420 ARVOsx Multi Label Counter).
FITC labeling of TGFβ1 and TGFβ2.
A mixture of 80 μl of 10 μM recombinant human TGFβ1 or TGFβ2 in 0.1 M Tris-HCl (pH 9.0) and 80 μl of 0.1 M Tris-HCl (pH 9.0) containing 100 μM FITC was incubated at 4°C in the dark. After labeling for 18 h, FITC-labeled TGFβs were purified by elution with ZipTip silica columns (Millipore, Tokyo, Japan) and were lyophilized according to the manufacture's instruction. Protein concentrations of each FITC-labeled TGFβ were measured using a bicinchoninic acid protein assay kit (Pierce).
Scatchard analysis.
Using binding data, we estimated amounts of free (unbound) TGFβ2 in each assay. When the increase of immunofluorescence intensity numbers corresponding to the amount of bound TGFβ2 is much smaller than the increase of the amount of added TGFβ2, we are able to estimate the amount of free TGFβ2 with very small errors. For example, based on the data that the immunofluorescence intensity numbers by treatment with 1 pM and 10 pM TGFβ2 are 90 and 180 arbitrary units, we are able to estimate that the amount of free TGFβ2 at 10 pM TGFβ2 is 80 to 100% of the amount of total TGFβ2. Kd was estimated with the software Cricket Graph III J.
Adenovirus vector-mediated expression.
The system of a replication-deficient adenovirus vector and adenoviruses encoding LacZ and V642I-APP were described previously (34). The cosmids for wtAPP were constructed by inserting the full-length wtAPP into the SwaI site of pAxCAwt.
Primary cortical neurons and cell viability assay.
Primary cortical neurons, obtained from embryonic day 14 (E14) ICR mice or V642I-APP knock-in mice, were seeded in poly-l-lysine-coated 24-well plates (Sumitomo Bakelite) at 1.25 × 105 cells/well in Neuron medium (Sumitomo Bakelite) (14, 34). Purity of neurons by this method was >98%. After incubation for 3 days, the culture medium was replaced with Dulbecco's modified Eagle medium containing N2 supplement. If indicated, on day 3 in vitro (DIV3), primary cortical neurons (PCNs) were infected by indicated multiplicities of infection (MOIs) of LacZ-, wtAPP-, or V642I-APP-encoded adenoviruses. On DIV4, indicated concentrations of TGFβ1, TGFβ2, or TGFβ3 were added, and cell viability was assessed by WST-8 assay and/or calcein fluorescence assay at 72 h after the onset of treatment.
Coimmunoprecipitation analysis.
F11 cells (7 × 104 cells/well in 6-well plates) were transfected with the pEF-APP-ED/Fc or pcDNA3-CNTFR-ED/Fc plasmid. At 24 h after transfection, cells were cultured in serum-free Ham's F-12 with N2 supplement. At 72 h after the onset of transfection, cultured-conditioned media were collected. Five-hundred microliters of the conditioned medium, 20 pmol of TGFβ2 or TGFβ1 (final concentration, 40 nM), and 40 μl of a 1:1 slurry of Protein G-Sepharose 4B were mixed and rotated at 4°C overnight before immunoblot analysis.
Immunoblot analysis.
Cell lysates (10 to 20 μg/lane) or pulled-down precipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and separated proteins were transferred onto polyvinylidene difluoride membranes as described previously (14). Visualization of the immunoreactive bands was performed by ECL (Amersham Pharmacia Biotech, Uppsala, Sweden). Densitometrical analysis of immunological signals were performed with NIH Image, version 1.62, software.
V642I-APP knock-in mice.
The targeted introduction of the V642I mutation in the mouse APP gene was performed as described previously (18). PCNs were prepared from 14-day-old embryos generated by crossing a heterozygous male mouse with a heterozygous female mouse. To identify homozygous, heterozygous, and wild-type PCNs, PCR analysis was performed as described previously (18).
Real-time PCR.
We performed real-time PCR to assess the amount of endogenous mRNA. Cells were harvested for the RNA extraction with an ISOGEN reagent (Nippon Gene, Toyama, Japan) followed by real-time PCR. The first-strand cDNAs were synthesized using Sensiscript reverse transcriptase (QIAGEN) with 0.5 μg total RNA. Real-time PCR analysis was performed using a QuantiTect SYBR Green PCR kit (QIAGEN), followed by analysis with an ABI PRISM 7700 (Applied Biosystems, Foster City, CA). We made sets of a sense primer and an antisense primer as follows: 5′-CACGCTACTTCCTCCTCAAG-3′ and 5′-CTCTGTCTTCATCAGCTGGC-3′ for mouse PAI-1 and 5′-TCCACCACCCTGTTGCTGTA-3′ and 5′-ACCACAGTCCATGCCATCAC-3′ for human and mouse G3PDH. Data analysis was performed using Sequence Detection System software, version 1.9.1 (Applied Biosystems). To adjust the expression level of each mRNA, G3PDH mRNA was used as an internal control.
Statistical analyses.
All cell death experiments, TGFβ binding assays, cell viability experiments, and real-time PCR experiments were done with n = 3 (n is number of determinations). All values in the experiments are means ± standard deviations. Statistical analyses were carried out with one-way analysis of variance followed by a post hoc test (Fisher's protected least significant difference test). P < 0.05 was assessed as significant.
RESULTS
Overexpression of APP in F11 cells results in the appearance of TGFβ2-specific receptors.
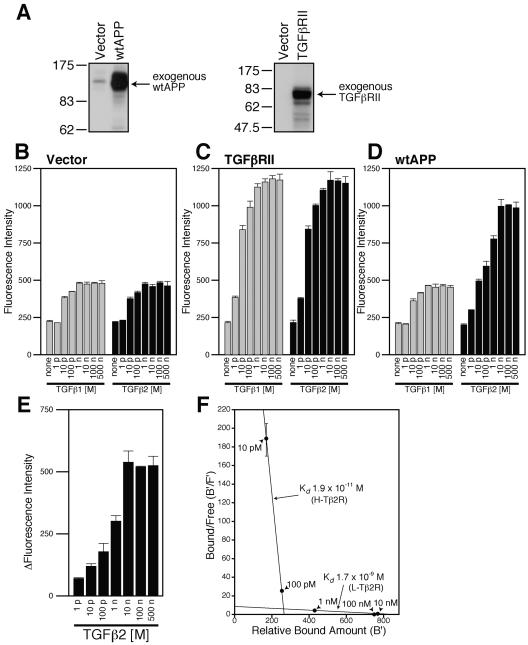
Based on findings that expression of TGFβ2 is upregulated in AD brains (10, 23, 36) and TGFβ2 binds to the extracellular domain of APP (3), we hypothesized that TGFβ2 displays some biological activities by binding to APP. To address this possibility, we first tried to look at the association between TGFβ2 and APP on the cell surface, using F11 neurohybrid cells. To this end, we developed immunofluorescence-based binding assays as described in Materials and Methods, using F11 cells or F11 cells that transiently overexpress wild-type APP (wtAPP) or TGFβ receptor type II (TGFβRII) (see the expression of wtAPP and TGFβRII in Fig. 1A). Overexpression levels of APP were roughly estimated to be 1 to 2, 3 to 5, 6 to 10, or 10 to 20 times more than the endogenous APP level when F11 cells were transfected with 0.1, 0.25, 0.5, or 1.0 μg of pcDNA3 expression vectors encoding APPs. Unexpectedly, these assays have turned out to be excellent in reproducibility. Errors generated in these assays were minimal, although it has been generally thought that the procedures to enhance signals with antibodies increase errors.
FIG. 1.
Overexpression of wtAPP induces the expression of TGFβ2-specific receptors. (A) F11 cells (7 × 104 cells/well in 6-well plates) were transfected with 0.5 μg of the pcDNA3 vector, pcDNA3-TGFβRII, or pcDNA3-wtAPP. Cell lysates (10 μg in each lane) were immunoblotted with antibody to APP (22C11) or TGFβRII. (B to D) F11 cells (7 × 104 cells/well in 6-well plates) were transfected with 0.25 μg of the pcDNA3 vector (B), pcDNA3-TGFβRII (C), or pcDNA3-wtAPP (D). At 36 h after transfection, the cells were combined with the indicated concentrations of TGFβ1 or TGFβ2. Immunofluorescence-based binding assays were performed as described in Materials and Methods. (E) The mean of immunofluorescence intensity numbers representing the association between TGFβ2 and F11 cells for each concentration of TGFβ2 shown in panel B were subtracted from immunofluorescence intensity numbers representing the association between TGFβ2 and F11 cells for the same concentration of TGFβ2 shown in panel D. Resulting intensity numbers were considered to correspond to the association between TGFβ2 and wtAPP-induced TGFβ2-specific receptors. (F) A simulated Scatchard plot of the association between TGFβ2 and wtAPP-induced TGFβ2-specific receptors based on the analysis shown in Table 1. A vertical bar for each point indicates an estimated range of bound/free numbers. p, pico; n, nano; B′, relative amount of bound TGFβ2; F′, estimated amount of free TGFβ2.
First of all, to test the feasibility of these assays, we looked at the association between TGFβs and F11 cells or F11 cells ectopically overexpressing TGFβRII. As shown in Fig. 1B, treatment with either TGFβ1 or TGFβ2 increased the fluorescence intensity in a dose-dependent fashion. The pattern and the extent of the TGFβ-dose-dependent increase in the fluorescence intensity by treatment with TGFβ1 were very similar to those by treatment with TGFβ2. It also is noted that the increase in the fluorescence intensity reached a plateau at 1 nM TGFβ, suggesting that the increase in the fluorescence intensity represented the saturatable association between TGFβ1 or TGFβ2 and endogenous TGFβ-binding proteins, including the TGFβ receptor. In addition, 50% effective concentrations of the association between TGFβ1 or TGFβ2 and F11 cells appeared to be around 10 pM (Fig. 1B). Considering that the Kd for the association between TGFβ1 or TGFβ2 and the TGFβ receptor was estimated to be 5 to 50 pM, this number is quite reasonable. Combined with an additional finding that the increase in the fluorescence intensity by TGFβ treatment was enhanced by overexpression of TGFβRII (Fig. 1C), we have concluded that the association between TGFβ1 or TGFβ2 and the receptors is faithfully examined by the binding assay.
We then looked at the association between TGFβ1 or TGFβ2 and F11 cells overexpressing wtAPP. As shown in Fig. 1D, overexpression of wtAPP upregulated the association between TGFβ2 and F11 cells but not the association between TGFβ1 and F11 cells. It is especially noted that the association between TGFβ2 and F11 cells overexpressing wtAPP reached a plateau at a TGFβ2 concentration of 10 nM (Fig. 1D, right panel), indicating that this binding was also saturated. Similar to the association between TGFβ1 and F11 cells, the association between TGFβ3 and F11 cells remained unchanged by overexpression of wtAPP (data not shown).
To evaluate the extent of the association between TGFβ2 and exogenously overexpressed APP (or APP-induced proteins) (TGFβ2 concentrations, 1 pM to 500 nM), we subtracted the mean immunofluorescence intensity numbers shown in the right panel of Fig. 1B, representing the associations between TGFβ2 and the endogenous TGFβ-binding proteins, from the mean immunofluorescence intensity numbers shown in the right panel of Fig. 1D that indicated the association between TGFβ2 and F11 cells overexpressing APP (Fig. 1E). Using these binding data, we further tried to roughly estimate amounts of free TGFβ2 in each binding assay (Table 1). When the increase of the immunofluorescence intensity numbers corresponding to the amounts of bound TGFβ2 is much smaller than the increase of the amount of added TGFβ2, we are able to estimate the amounts of free TGFβ2 with a very small error in each binding assay. For example, if immunofluorescence intensity numbers obtained at TGFβ2 concentrations of 1 pM and 10 pM are 90 and 180 arbitrary units, we are able to estimate that 80 to 100% of added TGFβ2 remains unbound at 10 pM of TGFβ2, because bound amounts of TGFβ2 are calculated to be 20% if we hypothesize that all TGFβ2 polypeptides bind to the receptors at the TGFβ2 concentration of 1 pM. We estimated approximate amounts of free TGFβ2 at 10 pM, 100 pM, 1 nM, 10 nM, and 100 nM of TGFβ2 in this way (Table 1). Using these data, we simulated Scatchard analysis (Fig. 1F) and found that APP overexpression seemed to induce two kinds of receptors specifically bound by TGFβ2. The roughly estimated Kd for the high-affinity receptor (H-Tβ2R) and the low-affinity receptor (L-Tβ2R) are 1.9 × 10−11 M and 1.7 × 10−9 M, respectively.
TABLE 1.
Estimation of amounts of free TGFβ2 in binding assaysa
| TGFβ2 concn | Total TGFβ2 amt (fmol) | Mean change in fluorescence intensity (arbitrary units) | Relative bound amt (B′) | Estimated free amt (fmol) (F′) | B′/F′ |
|---|---|---|---|---|---|
| 1 pM | 0.1 | 70 | 100 | 0.0-0.1 | |
| 10 pM | 1 | 119 | 170 | 0.83-1 | 170-205 |
| 100 pM | 10 | 178 | 254 | 9.75-10 | 25.4-26.0 |
| 1 nM | 100 | 301 | 430 | 99.6-100 | ≈4.32 |
| 10 nM | 1,000 | 538 | 769 | 999-1,000 | ≈0.77 |
| 100 nM | 10,000 | 519 | 750 | 9,999-10,000 | ≈0.075 |
TGFβ2 concentrations and mean change in fluorescence intensity intensity numbers were derived from Fig. 1E. Relative amounts of bound TGFβ2 (B′) were calculated under the condition that the amount of bound TGFβ2 was 100 when the TGFβ2 concentration was 1 pM.
To confirm that the increase in the fluorescence intensity caused by treatment with TGFβ1 or TGFβ2 was really derived from the association of TGFβ1 or TGFβ2 with F11 cells, we further examined whether the addition of an excess amount of unlabeled TGFβ1 or TGFβ2 inhibited the binding between FITC-labeled TGFβ1 or TGFβ2 and F11 cells (Fig. 2). To this end, we developed another immunofluorescence-based binding assay using FITC-labeled TGFβ1 or TGFβ2 instead of unlabeled TGFβ1 or TGFβ2 as the ligand. To increase sensitivity in detection of the binding, the fixation procedure with paraformaldehyde was omitted and attachment of F11 cells to culture dishes was enhanced by precoating with poly-l-lysine as shown in Materials and Methods. Using this binding assay, we again demonstrated that treatment with 10 nM FITC-TGFβ1 as well as FITC-TGFβ2 significantly increased the immunofluorescence intensity in F11 cells (Fig. 2, vector). As expected, the addition of an excess amount of unlabeled TGFβ1 or TGFβ2 completely suppressed the increase in the immunofluorescence intensity. Furthermore, overexpression of TGFβRII markedly increased the immunofluorescence intensity (Fig. 2, TGFβRII). Addition of an excess amount of unlabeled TGFβ1 or TGFβ2 again almost completely suppressed the increase in the immunofluorescence intensity. In accordance with data shown in Fig. 1D, treatment with 10 nM FITC-TGFβ2, but not 10 nM FITC-TGFβ1, increased the immunofluorescence intensity in F11 cells overexpressing wtAPP (Fig. 2, wtAPP). The increase was also inhibited by treatment with an excess amount of unlabeled TGFβ2. These findings clearly indicated that the TGFβ-mediated increase in the immunofluorescence intensity in F11 cells really reflected the specific binding between TGFβs and F11 cells and confirmed that overexpression of APP in F11 cells resulted in the appearance of TGFβ2-specific receptors.
The low-affinity APP-induced TGFβ2-specific receptor is APP.
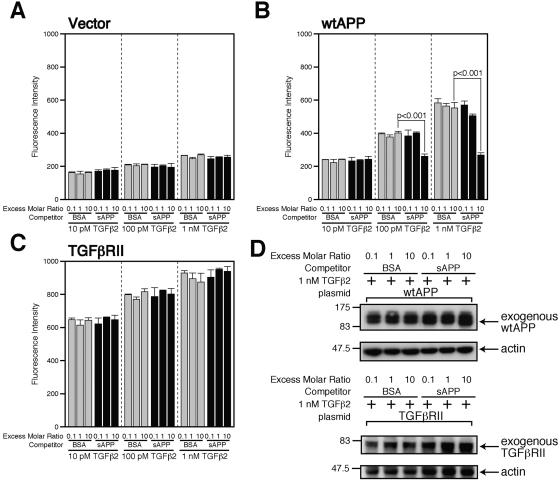
To address whether the TGFβ2-specific receptor is APP itself or other unidentified proteins whose expression are induced by APP, we performed competition experiments by adding as a competitor larger amounts of soluble APPα of human APP695 (sAPP), corresponding to amino acids 1 to 612 of APP695 (the numbering follows that of Kang et al. and Santiago-Garcia et al. [17, 41]), to binding assays (Fig. 3). The addition of sAPP did not reduce the association between TGFβ2 and F11 cells (Fig. 3A) or F11 cells overexpressing TGFβRII (Fig. 3C), even when the amount of added sAPP was 10 times more than the amount of TGFβ2 (Fig. 3A and C). Expression levels of cotransfected TGFβRII were not affected by addition of sAPP (Fig. 3D, bottom panel). It is speculated that competition did not occur in these cases because TGFβ2 has a far higher affinity to TGFβRII or the TGFβ receptor than to these APP-induced TGFβ2-specific receptors.
FIG. 3.
Soluble APP antagonized TGFβ2-induced death. (A to C) F11 cells (7 × 104 cells/well in 6-well plates) were transfected with 0.25 μg of the pcDNA3 vector (A), pcDNA3-wtAPP (B), or pcDNA3-TGFβRII (C). At 36 h after transfection, cells were combined with the indicated concentrations of TGFβ1 or TGFβ2 in the presence of the indicated amounts of sAPP or BSA as a competitor. Immunofluorescence-based binding assays were performed as described in Materials and Methods. (D) F11 cells (7 × 104 cells/well in 6-well plates) were transfected with 0.25 μg of pcDNA3-wtAPP (upper panel) or pcDNA3-TGFβRII (lower panel). At 36 h after transfection, cells were combined with 1 nM TGFβ2 in the presence of 10 nM sAPP or BSA. Cell lysates (10 μg in each lane) were immunoblotted with antibody to APP (22C11) (upper) or TGFβRII (lower) as well as actin (upper and lower).
In contrast, sAPP reduced the association between TGFβ2 and F11 cells expressing wtAPP only when the concentration of added TGFβ2 was 100 pM or 1 nM (but not 10 pM TGFβ2) and the amount of added sAPP was 10 times more than the amount of TGFβ2 (Fig. 3B). What was concluded by this observation was not affected by another observation that the addition of sAPP slightly increased expression of cotransfected wtAPP (Fig. 3D, top panel), because the increase in expression of wtAPP is considered to enhance the association between TGFβ2 and F11 cells. Based on the fact that TGFβ2 bound to L-Tβ2R as well as H-Tβ2R at 1 nM and 10 pM while almost all TGFβ2 bound to H-Tβ2R at 10 pM as shown in Fig. 1E and F, we have concluded that sAPP competes with L-Tβ2R, but not with H-Tβ2R, for the association with TGFβ2. This finding strongly suggests that L-Tβ2R is APP itself or an APP derivative, while H-Tβ2R may be a protein or proteins with a specific affinity to TGFβ2 whose expression is induced by APP.
TGFβ2 binds to wtAPP and mutant APPs.
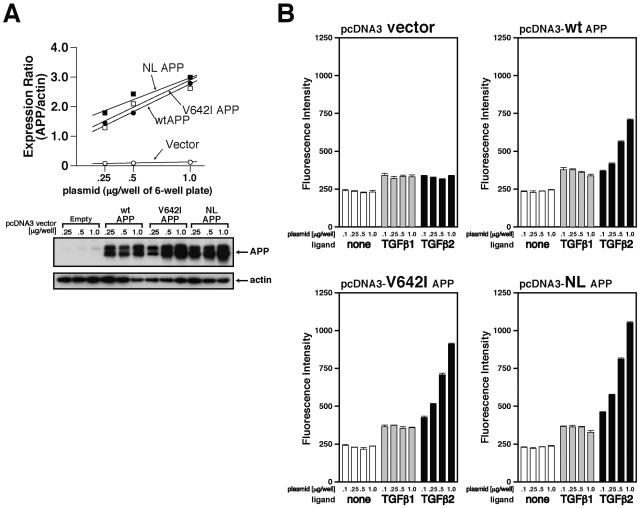
We further examined whether TGFβ2 binds to various FAD-linked mutant APPs in a similar fashion as well as whether the association between TGFβ2 and a mutant APP occurs in a manner dependent on the expression levels of APP. As shown in the top panel of Fig. 4A, there is a good positive correlation between the amount of the transfected plasmid and protein expression of APP. Treatment with 100 pM TGFβ1 or TGFβ2 increased the immunofluorescence intensity of F11 cells which had been transfected with the empty vector by about 100 U compared to that of nontreated cells in this particular experiment (Fig. 4B, top left panel, vector). In a similar fashion, TGFβ1 treatment increased by about 100 U the fluorescence intensity of F11 cells which had been transfected with the wtAPP-coding vector independently of the APP expression level (range of plasmid amounts, 0.25 to 1.0 μg) (Fig. 4B, top right panel, TGFβ1). In contrast, TGFβ2 treatment (100 pM) increased by more than 100 U the fluorescence intensity of F11 cells which had been transfected with the wtAPP-coding vector in a fashion dependent on the APP expression level (Fig. 4B, top right panel, TGFβ2). It maximally increased the fluorescence intensity by 500 U when F11 cells had been transfected with 1.0 μg of the wtAPP-coding plasmid. Binding experiments using F11 cells have further indicated that the association between TGFβ2 and V642I-APP (the isoleucine mutation at valine 642 in APP695) or NL-APP (the asparagine/leucine mutation at Lys595/Met596 in APP695) occurs in a similar dose-responsive fashion (Fig. 4B, bottom panels).
FIG. 4.
TGFβ2 binds to wtAPP and FAD-related APP mutants. (A) F11 cells were transfected with stepwise-increasing amounts (0.25, 0.5, and 1.0 μg) of the pcDNA3 vector, pcDNA3-wtAPP, pcDNA3-V642I-APP, or pcDNA3-NL-APP. Cell lysates (10 μg in each lane) were immunoblotted with antibody to APP (22C11) or actin. These experiments were simultaneously performed. (B) Immunofluorescence-based binding assays were performed as described in Materials and Methods. The concentration of added TGFβs was 100 pM.
TGFβ2 binds to the extracellular domain of APP.
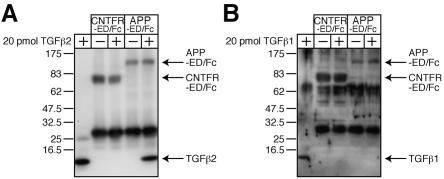
We next examined by a coimmunoprecipitation method whether there is an association between the extracellular domain of APP (APP-ED) and TGFβ2 (Fig. 5). To this end, we constructed a plasmid coding for APP-ED fused in frame to the Fc region of human IgG1 (APP-ED/Fc) that is secreted from transfected cells into media. As a negative control, we constructed another plasmid encoding the extracellular domain of human ciliary neurotrophic factor receptor fused to the IgG1 Fc portion (CNTFR-ED/Fc). Conditioned media containing APP-ED/Fc or CNTFR-ED/Fc were then mixed with 20 pmol of TGFβ2 (concentration, 40 nM) together with Protein G-Sepharose for coimmunoprecipitation analysis, which indicated that TGFβ2 bound to APP-ED/Fc while it did not bind to CNTFR-ED/Fc (Fig. 5A). A similar binding experiment further indicated that, in contrast to TGFβ2, either TGFβ1 (Fig. 5B) or TGFβ3 (data not shown) did not coprecipitate with APP-ED/Fc or CNTFR-ED/Fc, confirming the specificity of the association between TGFβ2 and APP-ED.
FIG. 5.
TGFβ2 binds to the extracellular domain of APP. (A and B) Twenty picomoles of recombinant TGFβ2 (A) or TGFβ1 (B) (final concentration, 40 nM) was mixed with APP-ED/Fc protein or CNTFR-ED/Fc protein for coimmunoprecipitation analysis. Immunoprecipitates were immunoblotted with antibodies to TGFβ1, TGFβ2, 6× histidine (to detect CNTFR/Fc), and APP.
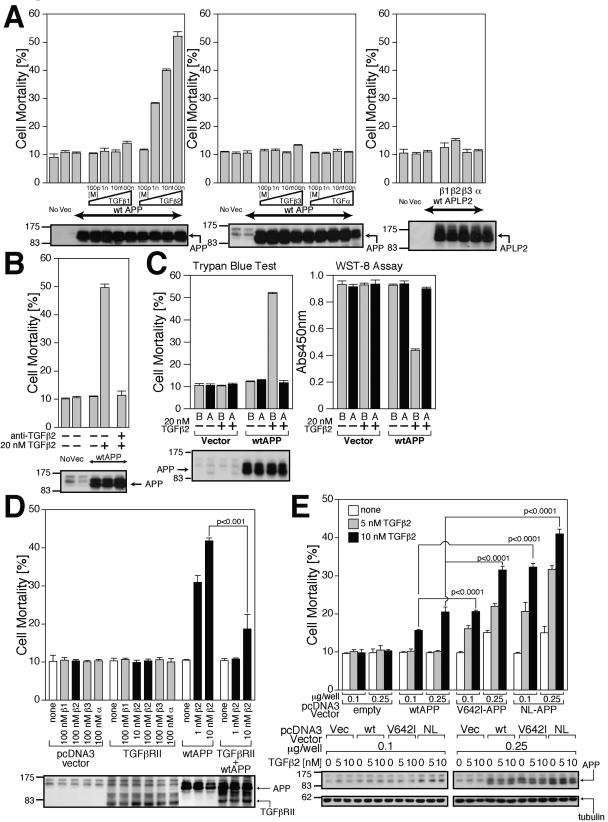
TGFβ2 induces death in F11 cells overexpressing wtAPP.
We then asked what the biological consequence of the association between TGFβ2 and APP was. To answer this question, we examined whether TGFβ2 treatment induces death in F11 cells transfected with 0.5 μg of the wtAPP-encoding plasmid, which was not enough to induce death in F11 cells. We then found that cell death was induced by treatment with TGFβ2 in a dose-responsive manner (Fig. 6A). In contrast, treatment with TGFβ1, TGFβ3, or TGFα did not induce death. Neither TGFβ1, TGFβ2, TGFβ3, nor TGFα induced death in F11 cells overexpressing amyloid precursor-like protein 2 (APLP2), suggesting that cell death occurred only when TGFβ2 was added in the presence of overexpressed wtAPP in F11 cells. We simultaneously recognized that 1 nM or higher concentrations of TGFβ2 were necessary for substantial induction of death (Fig. 6A). Combined with the finding that TGFβ2 concentrations of 100 pM or more are necessary for the substantial association between TGFβ2 and L-Tβ2R (APP) in F11 cells while the association between TGFβ2 and H-Tβ2R is saturated at 100 pM TGFβ2 (Fig. 1F), it is speculated that TGFβ2 induced death in F11 cells by binding to APP.
FIG. 6.
TGFβ2 triggers death in F11 cells overexpressing APPs. (A) F11 cells, transfected with 0.5 μg of pcDNA3-wtAPP or pcDNA3.1/GS-mouse APLP2, were treated with 100 pM, 1 nM, 10 nM, or 100 nM of TGFβ1, TGFβ2, TGFβ3, or TGFα. Cell mortality was determined by trypan blue exclusion assays at 48 h after the onset of TGF treatment. Cell lysates (20 μg in each lane) were immunoblotted with antibody to APP or HRP-conjugated anti-V5 monoclonal antibody for APLP2. (B) The effect of anti-TGFβ2 neutralizing antibody on TGFβ2/wtAPP-induced death in F11 cells. F11 cells, transfected with 0.5 μg of pcDNA3-wtAPP, were treated with 20 nM TGFβ2 in the presence of 10 μg/ml anti-TGFβ2 neutralizing antibody or control rabbit IgG. Cell lysates (20 μg in each lane) were immunoblotted with antibody to APP. (C) The effect of sAPP on TGFβ2/wtAPP-induced death in F11 cells. F11 cells, transfected with 0.5 μg of pcDNA3-wtAPP, were treated with 10 nM TGFβ2 in the presence or the absence of 100 nM of sAPP (labeled “A”) or BSA (labeled “B”). Cell mortality and cell viability were determined by trypan blue exclusion assay and WST-8 assay at 48 h after the onset of TGFβ2 treatment. Cell lysates (20 μg in each lane) were immunoblotted with antibody to APP. (D) F11 cells, transfected with 0.25 μg of the pcDNA3 vector, pcDNA3-TGFβRII, or pcDNA3-wtAPP or 0.25 μg of pcDNA3-TGFβRII and 0.25 μg of pcDNA3-wtAPP, were treated with indicated concentrations of TGFβ1 (β1), TGFβ2 (β2), TGFβ3 (β3), or TGFα (α). To keep the amounts of transfected vectors constant, proper amounts of the backbone vector were added. Cell mortality was determined by trypan blue exclusion assay at 48 h after the onset of TGF treatment. Cell lysates (20 μg in each lane) were immunoblotted with antibody to APP or TGFβRII. (E) TGFβ2 induces higher grade death in F11 cells ectopically expressing FAD-related APPs. F11 cells, transfected with 0.1 or 0.25 μg of pcDNA3-wtAPP, pcDNA3-V642I-APP, and pcDNA3-NL-APP, were treated with or without 5 or 10 nM TGFβ2. Cell mortality was determined by trypan blue exclusion assay at 48 h after the onset of TGFβ2 treatment. Cell lysates (20 μg in each lane) were immunoblotted with antibody to APP and tubulin. Vec, vector; Abs450nm, absorbance at 450 nm.
To confirm the involvement of TGFβ2 in neuronal cell death, we performed a neutralizing experiment using anti-TGFβ2 antibody. The addition of the neutralizing anti-TGFβ2 antibody completely abolished TGFβ2-induced neuronal cell death (Fig. 6B). In addition, we confirmed that addition of an excess amount of sAPP nullified TGFβ2-induced cell death without alteration of wtAPP expression (Fig. 6C), strongly supporting the notion that TGFβ2 induces neuronal cell death by binding to APP.
We then asked if the association between TGFβ2 and the endogenous TGFβ receptor (TGFβR) affects TGFβ2-induced death mediated by wtAPP in F11 cells overexpressing wtAPP. To address this question, we actually examined whether overexpression of TGFβRII altered TGFβ2-induced neuronal cell death. As shown in Fig. 6D, TGFβ2 did not induce death in F11 cells overexpressing TGFβRII alone. Conversely, coexpression of TGFβRII markedly reduced TGFβ2-induced death in F11 cells overexpressing wtAPP (Fig. 6D), probably because overexpressed TGFβRII inhibited the association between TGFβ2 and wtAPP by preferentially trapping TGFβ2. Taken altogether, we have concluded that TGFβ2-induced death in F11 cells, mediated by APP, is not affected by the TGFβ receptor-mediated signal.
TGFβ2 treatment results in higher-grade death in F11 cells expressing FAD-related mutant APPs.
We further characterized TGFβ2-induced neuronal cell death from the standpoint of FAD-linked APP mutation and APP expression levels (Fig. 6E). Very low-level or low-level ectopic expression of wtAPP, V642I-APP, and NL-APP was obtained by transiently transfecting 0.1 or 0.25 μg of each expression vector in F11 cells (bottom panel). In accordance with our earlier findings (14), low-grade death was induced by low-level expression of V642I-APP or NL-APP alone but not by low-level expression of wtAPP alone (see transfection with 0.25 μg of vectors) in F11 cells (Fig. 6E). Treatment with TGFβ2 induced or accelerated death in F11 cells ectopically expressing wtAPP, V642I-APP, or NL-APP in a manner dependent on the APP expression level and the TGFβ2 concentration. Apparently, treatment with TGFβ2 resulted in higher grade cell death in F11 cells expressing V642I-APP or NL-APP than in those expressing wtAPP.
Characterization of TGFβ2-induced neuronal cell death.
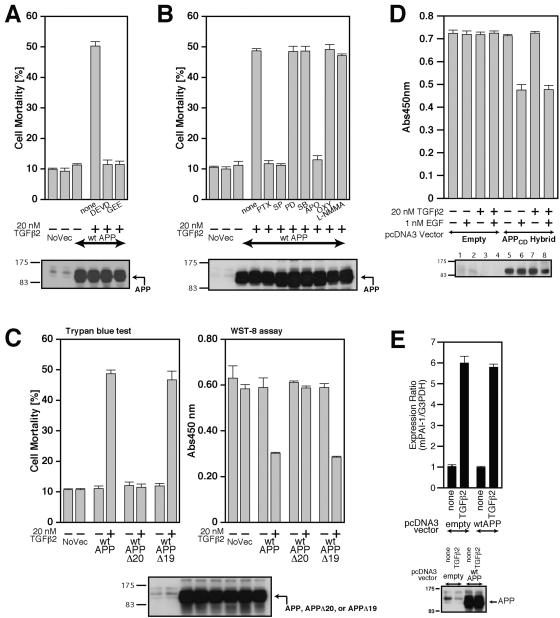
To obtain information about intracellular mediators for TGFβ2-induced cell death, we performed pharmacological analysis using a cell-permeable caspase 3 and/or related caspase inhibitor, Ac-DEVD-CHO (DEVD), and an established cell-permeable antioxidant, glutathione-ethyl-ester (GEE). Both of them completely inhibited TGFβ2-induced cell death (Fig. 7A), indicating that caspase 3 and/or related caspases and reactive oxygen species (ROS) source enzyme are involved in this neuronal cell death.
FIG. 7.
Characterization of TGFβ2-triggered death. (A and B) F11 cells, seeded on 6-well plates at 7 × 104 cells/well, were transfected with 0.5 μg of pcDNA3-wtAPP and then treated with 20 nM TGFβ2 in the presence or the absence of either 100 μM Ac-DEVD-CHO (DEVD), 1 mM GEE (A), 1 μg/ml PTX, 1 μM SP600125 (SP), 50 μM PD98059 (PD), 20 μM SB203580 (SB), 300 μM apocynin (APO), 100 μM oxypurinol (OXY), or 1 mM L-NMMA (B). Cell lysates (20 μg in each lane) were submitted to immunoblot analysis with 22C11 for APP. (C) F11 cells, seeded on 6-well plates at 7 × 104 cells/well, were transfected with 0.5 μg of pcDNA3-wtAPP, pcDNA3-wtAPPΔ20, or pcDNA3-wtAPPΔ19 and then treated with or without 20 nM TGFβ2. Cell mortality (trypan blue exclusion assay) and cell viability (WST-8 assay) were determined at 48 h. (D) TGFβ2 treatment does not enhance EGFR-ED+TM/APP-CD (labeled APPCD-Hybrid)-mediated death of F11 cells induced by treatment with 1 nM EGF. F11 cells, transfected with 1.0 μg of pcDNA3-EGFR-ED+TM/APP-CD or the backbone pcDNA3 vector, were treated with 1 nM EGF together with or without 20 nM TGFβ2. Cell viability was determined by WST-8 assays at 48 h after the onset of TGFβ2 treatment. Cell lysates (20 μg in each lane) were immunoblotted with antibody to EGFR to detect the APPCD hybrid. (E) Enforced expression of wtAPP did not result in the enhancement of the TGFβ2-induced activation of plasminogen activator inhibitor-1 (PAI-1) mRNA expression. F11 cells, transfected with 0.5 μg of the pcDNA3 vector or pcDNA3-wtAPP, were treated with 100 pM of TGFβ2 at 24 h after transfection. After incubation for 48 h, they were harvested for real-time PCR-based determination of mRNA amounts. NoVec, no vector; Abs450nm, absorbance at 450 nm; mPAI-1, mouse PAI-1.
We further treated cells with the pertussis toxin (PTX) to define the involvement of PTX-sensitive heterotrimeric G proteins with mitogen-activated protein kinase (MAPK) family inhibitors, including SP600125 (SP) for a JNK-specific inhibitor, SB203580 (SB) for a p38 MAPK-specific inhibitor, or PD98059 (PD) for a MEK/MAPK inhibitor to define the involvement of MEK/MAPK family proteins and with specific inhibitors of ROS source enzymes, including apocynin (APO) for an NADPH oxidase inhibitor, oxypurinol (OXY) for a xanthine oxidase inhibitor, or NG-methyl-l-arginine methylesterhydrochloride (L-NMMA) for a nitric oxide synthase (NOS) inhibitor to specify the source enzyme of ROS (Fig. 7B). Treatment with DEVD, GEE, PTX, SP, and APO inhibited TGFβ2-induced cell death, indicating that TGFβ2-induced cell death is mediated by a PTX-sensitive heterotrimeric G protein, JNK, NADPH oxidase, and caspase 3 and/or related caspases.
Involvement of the intracellular Go-interacting domain, but not the Aβ-corresponding domain, of APP in TGFβ2/APP induces cell death.
To further confirm the involvement of the PTX-sensitive heterotrimeric G protein, we tried to identify the domain in the intracytoplasmic region of APP essential for TGFβ2-induced cell death (Fig. 7C). F11 cells, transfected with the full-length wtAPP, wtAPP lacking domain 20 (wtAPPΔ20), or wtAPP lacking domain 19 (wtAPPΔ19) (14), were treated with 20 nM TGFβ2. We found that domain 20, corresponding to the His657-Lys676 region, is essential for TGFβ2-induced cell death. This finding indicates that Go is the signal transducer, because it was shown that Go is functionally coupled with domain 20 of APP (14, 35, 48, 49), and Aβ is not involved in TGFβ2-induced cell death, because wtAPPΔ20 does not mediate TGFβ2-induced cell death. wtAPPΔ20, which contains the Aβ-corresponding region, should mediate TGFβ2-induced cell death if Aβ plays a major role in TGFβ2-induced cell death.
Thus, we concluded that TGFβ2/APP-induced cell death is mediated by Go, JNK, NADPH oxidase, and caspase 3/related caspases. We confirmed that this pathway also is triggered in F11 cells expressing FAD-related APP mutants upon TGFβ2 binding (data not shown).
TGFβ2 does not induce neuronal cell death by triggering other neurotoxic signals unrelated to APP.
We next tried to completely rule out the possibility that TGFβ2 indirectly enhanced APP-mediated cell death by binding to other molecules unrelated to APP, which is linked to a certain cell-death-enhancing signal. To this end, we examined the effect of TGFβ2 treatment on APP-mediated cell death by using a system expressing a fusion construct consisting of the extracellular and transmembrane domains of the epidermal growth factor receptor and the full cytoplasmic domain of APP (APP-CD hybrid) (15). Treatment with 1 nM EGF induces death in F11 cells transfected with the vector encoding APP-CD hybrid (Fig. 7D). This cell death progresses via Go, c-Jun N-terminal kinase, NADPH oxidase, and caspase 3 and/or related caspases (15). In this cell death system, the addition of TGFβ2 did not enhance EGF-induced cell death (Fig. 7D), confirming that TGFβ2 does not increase APP-mediated cell death by activating another cell-death-enhancing signal through receptors other than APP.
Expression of APP does not enhance the TGFβ2-induced TGFβ receptor-mediated signal.
We also tried to exclude the possibility that enforced expression of APP proteins increases the TGFβ2-induced TGFβ receptor-mediated signal by modifying the function of various signal transducers. To address this issue, we examined with real-time PCR how TGFβ2-induced upregulation of expression of plasminogen activator inhibitor 1 (PAI-1) mRNA (24) is modified by the expression of wtAPP. PAI-1 is a representative target of TGFβ-induced TGFβ receptor-mediated signals. Consequently, we found that enforced expression of wtAPP did not alter TGFβ2-induced upregulation of PAI-1 mRNA, indicating that overexpression of APP does not affect the TGFβ2-induced TGFβ receptor-mediated signal (Fig. 7E). This experiment additionally suggests that H-Tβ2R is not TGFβ receptor type III (TGFβRIII) or an alternatively spliced TGFβRII, TGFβRII-B, both of which have special affinities to TGFβ2 (40), because upregulation of these proteins should enhance the TGFβ2-induced TGFβ receptor-mediated signals.
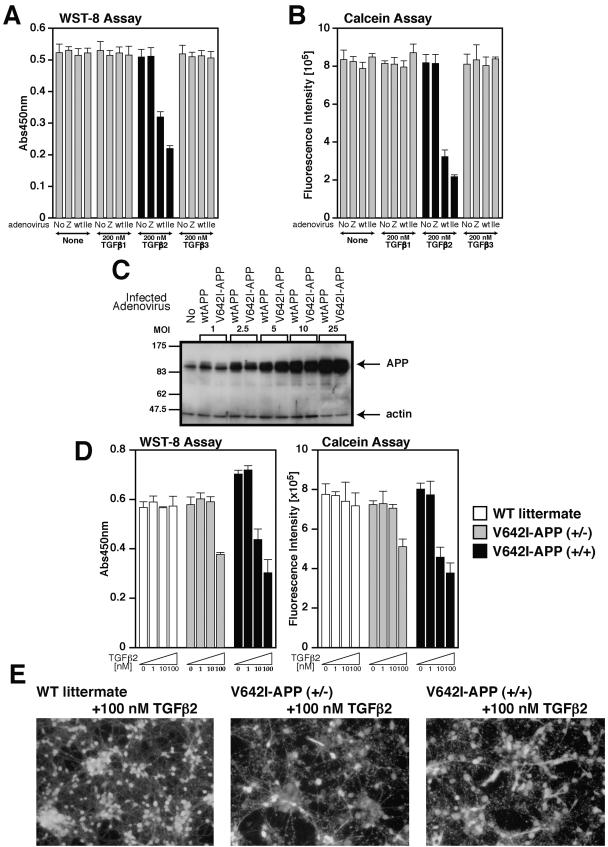
TGFβ2 induces death in primary cortical neurons.
We then asked whether TGFβ2 induces death in primary cortical neurons (PCNs), cells that are more physiologically postmitotic neuronal than F11 cells. To express APP proteins efficiently, we used an adenovirus-mediated expression system. In our earlier study, we showed that adenovirus-mediated overexpression of V642I-APP induces death in PCNs (34). In order to minimize death in PCNs induced by expression of V642I-APP itself, we selected a multiplicity of infection (MOI) of 5, a very low MOI, for adenoviral infection. The overexpression levels of APPs at the MOI of 5 were estimated to be five times higher than the endogenous APP expression level. In contrast to the fact that lipofection-based overexpression of V642I-APP alone induces low-grade death in F11 cells (Fig. 6E) (14), adenovirus-mediated overexpression of V642I-APP and wtAPP at this MOI did not induce death in PCNs. At 24 h after infection with the control LacZ-encoding virus, the wtAPP-encoding virus, or the V642I-APP-encoding virus, we treated cells with 200 nM TGFβ1, TGFβ2, or TGFβ3. We found that TGFβ2, but neither TGFβ1 nor TGFβ3, reduced cell viability of PCNs that adenovirally overexpressed wtAPP or V642I-APP (Fig. 8A and B). Expression levels of wtAPP and V642I-APP at each MOI were similar (Fig. 8C). Note that higher grade cell death was induced in PCNs ectopically expressing V642I-APP than in those expressing wtAPP.
FIG.8.
TGFβ2 triggers death in primary cortical neurons. (A and B) On day 3 in vitro (DIV3), PCNs were infected by LacZ (Z)-, wtAPP (wt)-, or V642I-APP (Ile)-encoding adenoviruses at an MOI of 5. On DIV4, PCNs were treated with 200 nM TGFβ1, TGFβ2, or TGFβ3. At 72 h after the onset of TGFβ treatment, cell viability was measured by WST-8 assay (A) and calcein assay (B). (C) wtAPP and V642I-APP were adenovirally overexpressed in PCNs at various MOIs. PCN lysates were subjected to immunoblot analysis with antibody to APP or actin. (D) PCNs derived from homozygous or heterozygous V642I-APP knock-in mice at E14 or wild-type littermate mice at E14 were seeded on poly-l-lysine-coated 96-well plates at 5 × 104 cells/well. On DIV4, they were treated with 0, 1, 10, and 100 nM TGFβ2. At 72 h after the onset of TGFβ treatment, cell viability was measured by WST-8 assay (left panel) and calcein assay (right panel). (E) Representative fluorescent microscopic views of calcein acetoxymethylester-stained PCN shown in panel D. Abs450nm, absorbance at 450 nm; WT, wild type.
TGFβ2 induces death in PCNs prepared from mice knocked in with the V642I-APP mutation.
We next tested whether TGFβ2 induces death in PCNs even in the absence of ectopic overexpression of APP. To this end, we used “V642I-APP knock-in mice,” mice that contain a V642I-APP mutant allele (or alleles) under the physiological promoter (18). The protein expression level of V642I-APP is equal to that of wtAPP in vivo, as described in our earlier study (18). We prepared PCNs from V642I-APP+/+ (homozygous), V642I-APP+/− (heterozygous), and wild-type littermate mice. Treatment with TGFβ2 at concentrations of 1 nM or less did not decrease viability in any type of PCN. Treatment with 10 nM TGFβ2 decreased viability in PCNs from the homozygous mouse but not in those from the heterozygous mouse or in those from the wild-type littermate mouse. Treatment with 100 nM TGFβ2 decreased viability in PCNs from both homozygous and heterozygous mice but not in those from the wild-type littermate mouse (Fig. 8D and E).
DISCUSSION
This study has shown that TGFβ2, but not TGFβ1 or TGFβ3, acts as a ligand for APP. It has revealed a novel function unique to TGFβ2. TGFβ2 triggers the APP-mediated cell death cascade and induces neuronal cell death. It is especially noted that TGFβ2 induces death in PCNs without ectopic overexpression of APP, prepared from heterozygous V642I-APP knock-in mice whose single allele contains the V642I-APP gene under the physiological APP promoter (Fig. 8D and E), leading to the speculation that various FAD-related point mutations contribute to the development of neuronal cell death in the presence of superphysiological concentrations of TGFβ2 by increasing the sensitivity of APP to TGFβ2-induced cell death in vivo.
At present, however, we do not have direct evidence that TGFβ2 induces neuronal cell death through APP in vivo. We need to add 10 nM TGFβ2 or more, which is supposed to be 100-fold higher than the levels observed in normal sera and cerebrospinal fluids, in order to induce neuronal cell death in vitro. However, it is natural that physiological concentrations of cell-death-inducing ligands are far lower than the effective concentrations, because death should not occur easily. We have found that TGFβ2 is substantially secreted from neurons as well as from glial cells (Y. Hashimoto, M. Nawa, and M. Matsuoka, unpublished observations). This finding implies that TGFβ2, secreted by neurons and glial cells, binds to and affects neurons by the autocrine mechanism in cooperation with the paracrine mechanism. In this situation, it is possible that the local TGFβ2 concentration in the fluid around neurons increases to a level sufficient for induction of neuronal cell death in vivo when expression of TGFβ2 is enhanced. In AD brains, it has been reported that upregulated expression of TGFβ2 is seen not only in glial cells but also in neurons themselves (6, 9, 10, 23, 36, 38). In agreement with this finding, we have recently found that toxic Aβ42 upregulates expression of TGFβ2 (Y. Hashimoto, M. Nawa, and M. Matsuoka, unpublished), supporting the idea that expression of TGFβ2 is upregulated in AD brains.
It has been reported that some cytokines, such as tumor necrosis factor α, interleukin-1β, and gamma interferon, stimulate γ-secretase-mediated cleavage of APP (22). Similarly, TGFβ2 may stimulate the γ-secretase activity. If this is the case, concentrations of toxic Aβ and the amount of the APP intracellular domain (AICD) may get sufficiently upregulated by TGFβ2-mediated putative stimulation of the γ-secretase activity to induce neuronal death. We did not accurately test whether TGFβ2 treatment increases γ-secretase activity in our cell death system. However, we estimated that concentrations of toxic Aβ42 secreted in the culture media were at the level of <1 μM by immunoblot analysis (Y. Hashimoto and M. Matsuoka, unpublished data), which is far less than the concentration necessary for induction of death in PCNs or F11 cells. It also is indicated in Fig. 7C that wtAPPΔ19, but not wtAPPΔ20, can mediate TGFβ2-induced cell death. Considering that wtAPPΔ19 lacks the 19-amino-acid-long cytoplasmic domain essential for the transcriptional activity of AICD while wtAPPΔ20 contains the full extracellular domain as well as the full transmembrane domain of APP from which toxic Aβ is generated (14), we can speculate that neither Aβ nor AICD is involved in TGFβ2-induced cell death.
Most sporadic AD patients do not have APP mutations. However, this study indicates that TGFβ2 induces death in neuronal cells without APP mutations only when they overexpress wtAPP. Therefore, an important issue to be addressed is whether TGFβ2 is involved in the development of AD without APP mutations. Currently, we do not have evidence directly confirming or disproving that expression of APP is upregulated in brains of AD patients in vivo. In contrast to such sporadic AD patients, APP has been shown to be overexpressed in patients with Down's syndrome, who develop AD at a higher incidence rate (2), supporting the possibility that TGFβ2-induced death signals mediated by overexpressed wtAPP contribute to the onset of AD associated with Down's patients.
In summary, we provide evidence that superphysiological concentrations of TGFβ2, whose expression has been demonstrated to be upregulated in AD brains, induces neuronal cell death by triggering a cell death signal through APP. The findings shown in this study may contribute to the further clarification of the precise mechanism of AD-related neuronal cell death.
Acknowledgments
We are indebted to Masaki Kitajima for essential support to this study; Mark C. Fishman for F11 neuronal hybrid cells; John T. Potts, Jr., Etsuro Ogata, Yoshiomi Tamai, and Yumi Tamai for indispensable support; and Dovie Wylie for expert technical assistance. We especially thank Takako Hiraki and Tomo Yoshida-Nishimoto for essential assistance.
This work was supported in part by grants from the Takeda Science Foundation (Y.H. and M.M.), a Keio University Grant-in-Aid for Encouragement of Young Medical Scientists (T.C., M.Y., and H.S.), and The Japan Society for the Promotion of Science.
REFERENCES
- 1.Amara, F. M., A. Junaid, R. R. Clough, and B. Liang. 1999. TGF-β1, regulation of Alzheimer amyloid precursor protein mRNA expression in a normal human astrocyte cell line: mRNA stabilization. Brain Res. Mol. Brain Res. 71:42-49. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, R. A. 1994. Differences in beta-amyloid (β/A4) deposition in human patients with Down's syndrome and sporadic Alzheimer's disease. Neurosci. Lett. 169:133-136. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer, S., M. B. Podlisny, D. J. Selkoe, I. Heid, and A. Fontana. 1990. Transforming growth factor-β bound to soluble derivatives of the β amyloid precursor protein of Alzheimer's disease. Biochem. Biophys. Res. Commun. 171:890-897. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, N. J., A. Chadwick, P. L. Lantos, R. Levy, and M. N. Rossor. 1993. βA4 protein deposition in familial Alzheimer's disease with the mutation in codon 717 of the βA4 amyloid precursor protein gene and sporadic Alzheimer's disease. Neurosci. Lett. 149:137-140. [DOI] [PubMed] [Google Scholar]
- 5.Chalazonitis, A., J. Kalberg, D. R. Twardzik, R. S. Morrison, and J. A. Kessler. 1992. Transforming growth factor β has neurotrophic actions on sensory neurons in vitro and is synergistic with nerve growth factor. Dev. Biol. 152:121-132. [DOI] [PubMed] [Google Scholar]
- 6.Chao, C. C., S. Hu, W. H. Frey, T. A. Ala, W. W. Tourtellotte, and P. K. Peterson. 1994. Transforming growth factor β in Alzheimer's disease. Clin. Diagn. Lab. Immunol. 1:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheifetz, S., J. A. Weatherbee, M. L. Tsang, J. K. Anderson, J. E. Mole, R. Lucas, and J. Massagué. 1987. The transforming growth factor-β system, a complex pattern of cross-reactive ligands and receptors. Cell 48:409-415. [DOI] [PubMed] [Google Scholar]
- 8.Citron, M., T. Oltersdorf, C. Haass, L. McConlogue, A. Y. Hung, P. Seubert, C. Vigo-Pelfrey, I. Lieberburg, and D. J. Selkoe. 1992. Mutation of the β-amyloid precursor protein in familial Alzheimer's disease increases β-protein production. Nature 360:672-674. [DOI] [PubMed] [Google Scholar]
- 9.Flanders, K. C., G. Lüdecke, S. Engels, D. S. Cissel, A. B. Roberts, P. Kondaiah, R. Lafayatis, M. B. Sporn, and K. Unsicker. 1991. Localizations and actions of transforming growth factor-βs in the embryonic nervous system. Development 113:183-191. [DOI] [PubMed] [Google Scholar]
- 10.Flanders, K. C., C. F. Lippa, T. W. Smith, D. A. Pollen, and M. B. Sporn. 1995. Altered expression of transforming growth factor-β in Alzheimer's disease. Neurology 45:1561-1569. [DOI] [PubMed] [Google Scholar]
- 11.Gelinas, D. S., K. DaSilva, D. Fenili, P. St. George-Hyslop, and J. McLaurin. 2004. Immunotherapy for Alzheimer's disease. Proc. Natl. Acad. Sci. USA 101:14657-14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, C. W., and A. J. Patel. 1993. Regulation of β-amyloid precursor protein isoform mRNAs by transforming growth factor-β1 and interleukin-1β in astrocytes. Brain Res. Mol. Brain Res. 19:251-256. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, J., and D. J. Selkoe. 2002. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297:353-356. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, Y., T. Niikura, Y. Ito, and I. Nishimoto. 2000. Multiple mechanisms underlie neurotoxicity by different types of Alzheimer's disease mutations of amyloid precursor protein. J. Biol. Chem. 275:34541-34551. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, Y., T. Niikura, T. Chiba, E. Tsukamoto, H. Kadowaki, H. Nishitoh, Y. Yamagishi, M. Ishizaka, M. Yamada, M. Nawa, K. Terashita, S. Aiso, H. Ichijo, and I. Nishimoto. 2003. The cytoplasmic domain of Alzheimer's amyloid precursor protein causes sustained ASK1/JNK-mediated neurotoxic signal via dimerization. J. Pharmacol. Exp. Ther. 306:889-902. [DOI] [PubMed] [Google Scholar]
- 16.Jennings, M. T., and J. A. Pietenpol. 1998. The role of transforming growth factor β in glioma progression. J. Neurooncol. 36:123-140. [DOI] [PubMed] [Google Scholar]
- 17.Kang, J., H. G. Lemaire, A. Unterbeck, J. M. Salbaum, C. L. Masters, K. H. Grzeschik, G. Multhaup, K. Beyreuther, and B. Muller-Hill. 1987. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 325:733-736. [DOI] [PubMed] [Google Scholar]
- 18.Kawasumi, M., T. Chiba, M. Yamada, M. Miyamae-Kaneko, M. Matsuoka, J. Nakahara, T. Tomita, T. Iwatsubo, S. Kato, S. Aiso, and I. Nishimoto, and K. Kouyama. 2004. Targeted introduction of V642I mutation in amyloid precursor protein gene causes functional abnormality resembling early stage of Alzheimer's disease in aged mice. Eur. J. Neurosci. 19:2826-2838. [DOI] [PubMed] [Google Scholar]
- 19.Krieglstein, K., C. Suter-Crazzolara, W. H. Fischer, and K. Unsicker. 1995. TGF-β superfamily members promote survival of midbrain dopaminergic neurons and protect them against MPP+ toxicity. EMBO J. 14:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieglstein, K., S. Richter, L. Farkas, N. Schuster, N. Dunker, R. W. Oppenheim, and K. Unsicker. 2000. Reduction of endogenous transforming growth factors β prevents ontogenetic neuron death. Nat. Neurosci. 3:1085-1090. [DOI] [PubMed] [Google Scholar]
- 21.Lesné, S., F. Docagne, C. Gabrie, G. Liot, D. K. Lahiri, L. Buée, L. Plawinski, A. Delacourte, E. T. MacKenzie, A. Buisson, and D. Vivien. 2003. Transforming growth factor-β1 potentiates amyloid-β generation in astrocytes and in transgenic mice. J. Biol. Chem. 278:18408-18418. [DOI] [PubMed] [Google Scholar]
- 22.Liao, Y.-F., B. J. Wang, H. T. Cheng, L. H. Kuo, and M. S. Wolf. 2004. Tumor necrosis factor-α, interleukin-1β, and interferon-γ stimulate γ-secretase-mediated cleavage of amyloid precursor protein through a JNK-dependent MAPK Pathway. J. Biol. Chem. 279:49523-49532. [DOI] [PubMed] [Google Scholar]
- 23.Lippa, C. F., K. C. Flanders, E. S. Kim, and S. Croul. 1998. TGF-β receptors-I and -II immunoexpression in Alzheimer's disease: a comparison with aging and progressive supranuclear palsy. Neurobiol. Aging 19:527-533. [DOI] [PubMed] [Google Scholar]
- 24.Lund, L. R., A. Riccio, P. A. Andreasen, L. S. Nielsen, P. Kristensen, M. Laiho, O. Saksela, F. Blasi, and K. Dano. 1987. Transforming growth factor-β is a strong and fast acting positive regulator of the level of type-1 plasminogen activator inhibitor mRNA in WI-38 human lung fibroblasts. EMBO J. 6:1281-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, J. J., W. Wallace, T. Riccioni, D. K. Ingram, G. S. Roth, and J. W. Kusiak. 1999. Death of PC12 cells and hippocampal neurons induced by adenoviral-mediated FAD human amyloid precursor protein gene expression. J. Neurosci. Res. 55:629-642. [DOI] [PubMed] [Google Scholar]
- 26.Lyman, S. D., L. James, T. Vanden Bos, P. de Vries, K. Brasel, B. Gliniak, L. T. Hollingsworth, K. S. Picha, H. J. McKenna, R. R. Splett, F. A. Fletcher, E. Maraskovsky, D. E. Williams, and M. P. Beckman. 1993. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: a proliferative factor for primitive hematopoietic cells. Cell 75:1157-1167. [DOI] [PubMed] [Google Scholar]
- 27.Martinou, J. C., A. Le Van Thai, AValette, and M. J. Weber. 1990. Transforming growth factor β1 is a potent survival factor for rat embryo motoneurons in culture. Brain Res. Dev. Brain Res. 52:175-181. [DOI] [PubMed] [Google Scholar]
- 28.Massagué, J., J. Andres, L. Attisano, S. Cheifetz, F. Lopez-Casillas, M. Ohtsuki, and J. L. Wrana. 1992. TGF-β receptors. Mol. Reprod. Dev. 32:99-104. [DOI] [PubMed] [Google Scholar]
- 29.Massagué, J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 30.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 31.McPhie, D. L., R. Coopersmith, A. Hines-Peralta, Y. Chen, K. J. Ivins, S. P. Manly, M. R. Kozlowski, K. A. Nevé, and R. L. Nevé. 2003. DNA synthesis and neuronal apoptosis caused by familial Alzheimer's disease mutants of the amyloid precursor protein are mediated by the p21 activated kinase PAK3. J. Neurosci. 23:6914-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevé, R. L., D. L. McPhie, and Y. Chen. 2000. Alzheimer's disease: a dysfunction of the amyloid precursor protein. Brain Res. 886:54-66. [DOI] [PubMed] [Google Scholar]
- 34.Niikura, T., M. Yamada, T. Chiba, S. Aiso, M. Matsuoka, and I. Nishimoto. 2004. Characterization of V642I-AβPP-induced cytotoxicity in primary neurons. J. Neurosci. Res. 77:54-62. [DOI] [PubMed] [Google Scholar]
- 35.Nishimoto, I., T. Okamoto, Y. Matsuura, S. Takahashi, T. Okamoto, Y. Murayama, and E. Ogata. 1993. Alzheimer amyloid protein precursor complexes with brain GTP binding protein Go. Nature 362:75-79. [DOI] [PubMed] [Google Scholar]
- 36.Peress, N. S., and E. Perillo. 1995. Differential expression of TGF-β 1, 2 and 3 isotypes in Alzheimer's disease: a comparative immunohistochemical study with cerebral infarction, aged human and mouse control brains. J. Neuropathol. Exp. Neurol. 54:802-811. [DOI] [PubMed] [Google Scholar]
- 37.Poulson, K. T., A. P. Armanini, R. D. Klein, M. A. Hynes, H. S. Phillips, and A. Rosenthal. 1994. TGF-β2 and TGF-β3 are potent survival factors for midbrain dopaminergic neurons. Neuron 13:1245-1252. [DOI] [PubMed] [Google Scholar]
- 38.Pratt, B. M., and J. M. McPherson. 1997. TGF-β in the central nervous system: potential roles in ischemic injury and neurodegenerative diseases. Cytokine Growth Factor Rev. 8:267-292. [DOI] [PubMed] [Google Scholar]
- 39.Rohn, T. T., K. J. Ivins, B. A. Bahr, C. W. Cotman, and D. H. Cribbs. 2000. A monoclonal antibody to amyloid precursor protein induces neuronal apoptosis. J. Neurochem. 74:2331-2342. [DOI] [PubMed] [Google Scholar]
- 40.Rotzer, D., M. Roth, M. Lutz, D. Lindemann, W. Sebald, and P. Knaus. 2000. Type III TGF-β receptor-independent signalling of TGF-β2 via TβRII-B, an alternatively spliced TGF-β type II receptor. EMBO J. 20:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago-Garcia, J., J. Mas-Oliva, T. L. Innerarity, and R. E. Pitas. 2001. Secreted forms of the amyloid-β precursor protein are ligands for the class A scavenger receptor. J. Biol. Chem. 276:30655-30661. [DOI] [PubMed] [Google Scholar]
- 42.Schuster, N., and K. Krieglstein. 2002. Mechanisms of TGF-β-mediated apoptosis. Cell Tissue Res. 307:1-14. [DOI] [PubMed] [Google Scholar]
- 43.Sudo, H., Y. Hashimoto, T. Niikura, Z. Shao, T. Yasukawa, Y. Ito, M. Yamada, M. Hata, T. Hiraki, M. Kawasumi, K. Kouyama, and I. Nishimoto. 2001. Secreted Aβ does not mediate neurotoxicity by antibody-stimulated amyloid precursor protein. Biochem. Biophys. Res. Commun. 282:548-556. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, N., T. T. Cheung, X. D. Cai, A. Odaka, L. Otvos, Jr., C. Eckman, T. E. Golde, and S. G. Younkin. 1994. An increased percentage of long amyloid β protein secreted by familial amyloid β protein precursor (βAPP717) mutants. Science 264:1336-1340. [DOI] [PubMed] [Google Scholar]
- 45.ten Dijke, P., and C. S. Hill. 2004. New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 29:265-273. [DOI] [PubMed] [Google Scholar]
- 46.Tomita, T., K. Maruyama, T. C. Saido, H. Kume, K. Shinozaki, S. Tokuhiro, A. Capell, J. Walter, J. Grunberg, C. Haass, T. Iwatsubo, and K. Obata. 1997. The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid β protein ending at the 42nd (or 43rd) residue. Proc. Natl. Acad. Sci. USA 94:2025-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyss-Coray, T., E. Masliah, M. Mallory, L. McConlogue, K. Johnson-Wood, C. Lin, and L. Mucke. 1997. Amyloidogenic role of cytokine TGF-β1 in transgenic mice and in Alzheimer's disease. Nature 389:603-606. [DOI] [PubMed] [Google Scholar]
- 48.Yamatsuji, T., T. Okamoto, S. Takeda, Y. Murayama, N. Tanaka, and I. Nishimoto. 1996. Expression of V642 APP mutant causes cellular apoptosis as Alzheimer trait-linked phenotype. EMBO J. 15:498-509. [PMC free article] [PubMed] [Google Scholar]
- 49.Yamatsuji, T., T. Okamoto, S. Takeda, H. Fukumoto, T. Iwatsubo, N. Suzuki, A. Asami-Odaka, S. Ireland, T. Kinane, U. Giambarella, and I. Nishimoto. 1996. G protein-mediated neuronal DNA fragmentation by familial Alzheimer's disease associated V642 mutants of APP. Science 272:1349-1352. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, B., F. J. Chrest, W. E. Horton, Jr., S. S. Sisodia, and J. W. Kusiak. 1997. Expression of mutant amyloid precursor proteins induces apoptosis in PC12 cells. J. Neurosci. Res. 47:253-263. [PubMed] [Google Scholar]