Abstract
Burkholderia sacchari IPT101T induced the formation of 2-methylcitrate synthase and 2-methylisocitrate lyase when it was cultivated in the presence of propionic acid. The prp locus of B. sacchari IPT101T is required for utilization of propionic acid as a sole carbon source and is relevant for incorporation of 3-hydroxyvalerate (3HV) into copolyesters, and it was cloned and sequenced. Five genes (prpR, prpB, prpC, acnM, and ORF5) exhibited identity to genes located in the prp loci of other gram-negative bacteria. prpC encodes a 2-methylcitrate synthase with a calculated molecular mass of 42,691 Da. prpB encodes a 2-methylisocitrate lyase. The levels of PrpC and PrpB activity were much lower in propionate-negative mutant IPT189 obtained from IPT101T and were heterologously expressed in Escherichia coli. The acnM gene (ORF4) and ORF5, which are required for conversion of 2-methylcitric acid to 2-methylisocitric acid in Ralstonia eutropha HF39, are also located in the prp locus. The translational product of ORF1 (prpR) had a calculated molecular mass of 70,598 Da and is a putative regulator of the prp cluster. Three additional open reading frames (ORF6, ORF7, and ORF8) whose functions are not known were located adjacent to ORF5 in the prp locus of B. sacchari, and these open reading frames have not been found in any other prp operon yet. In summary, the organization of the prp genes of B. sacchari is similar but not identical to the organization of these genes in other bacteria investigated recently. In addition, this study provided a rationale for the previously shown increased molar contents of 3HV in copolyesters accumulated by a B. sacchari mutant since it was revealed in this study that the mutant is defective in prpC.
Polyhydroxyalkanoates (PHAs) are natural thermoplastics that occur in a wide variety of bacteria which synthesize these polyesters as carbon and energy storage compounds from renewable carbon sources. The thermoplastic properties of the copolyester poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [poly(3HB-co-3HV); Biopol] and its biodegradability are influenced mainly by the content of 3-hydroxyvalerate (3HV), which improves the properties of the molecule and also extends the range of its applications (21). Therefore, since PHAs can be obtained from renewable resources, commercial interest in these polyesters has increased a great deal during the last decade. Wild-type strains of different bacteria and mutants which are able to produce 3HV from unrelated carbon sources have been studied in the last few years (44, 45, 53).
The biotechnological processes now used for fermentative production of poly(3HB-co-3HV) by bacteria depend on a supply of propionic acid as a specific 3HV precursor substrate in addition to a carbohydrate or another main carbon source. This requirement allows workers to control the composition of the polyester by changing the ratio of propionic acid to glucose (carbohydrate) in the medium (12). The strain which is used for PHA accumulation is an important factor in the production of poly(3HB-co-3HV). Many bacteria exhibit only a low 3HV yield with propionic acid (Y3HV/Prop); in most cases this yield is approximately 0.1 g · g−1, whereas the theoretical value for the yield is 1.35 g · g−1 (17). The low yield may result from the predominance of oxidative pathways in propionic acid metabolism and the ability of the organisms to produce 3-hydroxybutyrate from this substrate. In fact, a number of pathways have been described for catabolism of propionic acid in different bacteria, and some of them seem to operate simultaneously (15, 56). The 2-methylcitric acid cycle (MCC) is one of these pathways, and at one time this pathway had been found only in yeasts and molds (31, 37). Recently, it was also found to occur in Escherichia coli and other non-PHA-producing microorganisms (16, 24, 50). The possibility that one or more oxidative propionic acid-degrading pathways operate in a PHA-producing strain could explain the low yield of 3HV in the copolyester.
Therefore, identification of the propionic acid catabolic pathways relevant for production of poly(3HB-co-3HV) in bacterial strains could improve such strains for industrial purposes. Burkholderia sacchari strain IPT101T was isolated from the soil of a sugarcane plantation in Brazil, and a polyphasic taxonomic study identified this strain as a member of a novel species of the genus Burkholderia (7). Recent studies with propionic acid-negative mutants of this PHA-producing strain indicated that propionic acid is catabolized by at least two pathways, an α-oxidation pathway and an alternative pathway that has not been identified yet. These two pathways could be responsible for more than 80% of the propionic acid utilization under PHA accumulation conditions and could account for the overall propionate catabolism (41). Mutant IPT189, which was unable to grow on propionic acid as a sole carbon source but still incorporated 3HV units from this substrate into the copolyester, was not affected in the α-oxidation pathway (41). This mutant exhibited an improved Y3HV/Prop, which ranged from 0.80 to 1.20 g · g−1 depending on the cultivation strategy used to produce poly(3HB-co-3HV) from sucrose and propionic acid (41). This study was performed to identify the genes in this mutant that are affected and to clarify propionic acid catabolism in B. sacchari IPT101T.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Wild-type strain B. sacchari IPT101T (Suc+ Prp+) (7, 17) and mutant IPT189 (Suc+ Prp−) obtained from IPT101T (41) were studied. E. coli S17-1 (42) and cosmids pVK100 (27) and pHC79 (20) were used in cloning experiments. E. coli XL-1 Blue (11) and the phagemids pBluescript SK− and pBCSK+ (Stratagene) were used for DNA sequencing. The propionate-negative Tn5 mutants SK 7286, VG17, and P2 of Ralstonia eutropha HF39 (8) were employed for phenotypic complementation.
Media and growth conditions.
Cells of E. coli S 17-1 and XL-1 Blue were cultivated in Luria-Bertani (LB) medium at 37°C; isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were added if appropriate (38). Strains of B. sacchari and R. eutropha were grown at 30°C in either nutrient broth or mineral salts medium (MM) (39) supplemented with carbon sources as indicated below. Solid media contained 15 to 20 g of agar per liter. Antibiotics were added to the media at the following final concentrations, if necessary: tetracycline, kanamycin, and ampicillin, 15, 50, and 100 μg · ml−1, respectively, for recombinant strains of B. sacchari; and kanamycin, streptomycin, and tetracycline, 160, 500, and 25 μg · ml−1, respectively, for mutants or recombinant strains of R. eutropha.
Isolation of DNA, transformation, and electrophoretic methods.
Genomic DNA and cosmids pVK100 and pHC79 were extracted by the methods described by Ausubel et al. (2), Marmur (29), and Birnboim and Doly (4), respectively. Transformation protocols, other molecular techniques, and agarose gel electrophoresis were performed as described by Sambrook et al. (38), unless indicated otherwise. DNA fragments were isolated from agarose gels with a Gene Clean kit (54). Enzymes, reagents, and kits were used according to the instructions provided by the manufacturers.
Construction of a genomic library of B. sacchari IPT101T.
Partially EcoRI-digested or completely HindIII-digested genomic DNA was ligated to EcoRI-restricted cosmid pVK100 or to HindIII-restricted cosmid pHC79 DNA. The ligation mixtures were packaged in λ phages and transduced to E. coli S17-1, and clones were selected on LB medium containing tetracycline. Propionic acid-negative mutant IPT189 of B. sacchari IPT101T (recipient) and E. coli S17-1 harboring hybrid plasmids (donor) were conjugated by spot agar mating, and transconjugants were selected on MM agar containing sucrose (0.1%, wt/vol) as the sole carbon source plus tetracycline.
DNA-DNA hybridization.
Southern hybridization was carried out by the method described by Oelmüller et al. (32) at 68°C. Colony hybridization was performed by the method of Grunstein and Hogness (18) at 68°C.
PCR amplification.
PCR amplification of DNA encoded on plasmids or genomic DNA was carried out as described in a laboratory manual (38) by using VENTR DNA polymerase (New England Biolabs, Schwalbach/Taunus, Germany) or Platinum Pfx DNA polymerase (Gibco BRL Life Technologies, Karlsruhe, Germany) and an Omnigene HBTR3CM DNA thermal cycler (Hybaid, Heidelberg, Germany).
Determination of DNA sequence.
DNA fragments of interest were ligated to the phagemid pBluescript SK− and transformed into competent cells of E. coli XL-1 Blue. Clones containing inserted DNA were selected on LB medium containing X-Gal and IPTG and cultivated in liquid LB medium with ampicillin and tetracycline, and hybrid plasmids were extracted in order to sequence the fragments. The primer-hopping strategy (47) was used to determine the DNA sequence of both DNA strands. Sequencing was performed by using a Sequi Therm EXCEL TM II long-read cycle sequencing kit (Epicentre Technologies, Madison, Wis.) and IRD 800-labeled oligonucleotides (MWG-Biotech, Ebersberg, Gemany). Sequencing was performed with a LI-COR 4000L automatic sequencing apparatus (MWG-Biotech).
Sequence data analysis.
Sequence data were compared with sequences in the GenBank and Prosite databases by using the programs BlastSearch 2.0.10 (1) and DBGET (3). A dendrogram was constructed with the program ClustalW (51).
Preparation of crude extracts.
Cells from 50-ml cultures were washed in 66.7 mM potassium phosphate buffer (pH 6.9) and resuspended in 5 ml of this buffer, and 10 μg of DNase I was added. The cells were disrupted by sonication with a Bandelin sonopuls GM200 (Bandelin Electronic, Berlin, Germany) by using an amplitude of 16 μm (1 min/ml). During ultrasonication the samples were chilled in an NaCl-ice bath. Soluble protein fractions of the crude extracts were obtained by centrifugation for 5 min at 13,000 rpm and 4°C (Minifuge; Heraeus, Osterode, Germany).
Cloning of prpB and prpC and heterologous expression in E. coli.
We constructed oligonucleotides to amplify prpB (prpBUP [5′-AAAGGATCCTTTGGAGACTCGCACACATGAACAG-3′] and prpBRP [5′-AAAGAATTCGTTACTTCTTCCGCGGCGAAC-3′]) and prpC (prpCUP [5′-AAATCTAGAGAGAGAAAGGACGACACATGAG-3′] and prpCRP [5′-AAACTGCAGACTGGCGTGCCTCGATCAG-3′]) from genomic DNA of B. sacchari IPT101T. The 920-bp DNA fragment obtained with prpBUP and prpBRP was cloned into pBluescript SK−, which resulted in pSK−/prpBBs. prpCUP contained an XbaI recognition sequence and prpCRP contained a PstI recognition sequence; this enabled forced cloning of prpC downstream of and colinear to the lacZ promoter with XbaI/PstI-restricted pBluescript SK− DNA, which resulted in pSK−/prpCBs.
Cells of E. coli harboring pSK−/prpBBs, pSK−/prpCBs, or pBluescript SK− were grown at 30°C for 16 h in 50 ml of LB medium containing 75 μg of ampicillin per ml plus 0.5 mM IPTG. The cells were harvested by centrifugation in a Minifuge RF (Heraeus, Osterode, Germany) (10 min, 4,500 rpm, 4°C), washed with 100 mM Tris-HCl (pH 8.0), resuspended in 5 ml of 100 mM Tris-HCl, and disrupted by two passages through a French press.
Determination of the activities of the MCC enzymes.
The activity of 2-methylcitrate synthase (EC 4.1.3.31) was measured by the method of Srere (43). The cuvette used (diameter, 1 cm) contained 1 ml (total volume) of a solution containing 2 mM oxaloacetate, 250 μM propionyl coenzyme A (propionyl-CoA), and 2 mM 5,5′-dithiobis-(2-nitrobenzoate) in 66.7 mM potassium phosphate buffer (pH 6.9). After addition of the crude extract, the increase in absorbance at 412 nm (ɛ = 13.6 cm2 · μmol−1) was determined with an Ultrospec 2000 photometer (Pharmacia, Uppsala, Sweden).
The activity of 2-methylisocitrate lyase was assayed in a 0.5-ml (total volume) mixture containing 66.7 mM potassium phosphate buffer (pH 6.9), 3.3 mM phenylhydrazine, 2.5 mM cysteine, 5 mM MgCl2, and cell extract. The reaction was started by adding 1.25 mM K3-methylisocitrate, which was obtained by alkaline cleavage of the methylisocitric acid lactone (10). The increase in absorbance at 324 nm was measured. The molar extinction coefficient of pyruvate phenylhydrazone was 12 mM−1 · cm−1 (28).
One unit of enzyme activity was defined as the amount of activity that converted 1 μmol of substrate per min. The amount of soluble protein was determined by the method of Bradford (6); crystalline bovine serum albumin was used as the standard.
Nucleotide sequence accession number.
The nucleotide and amino acid sequence data determined in this study have been deposited in the GenBank nucleotide sequence database under accession no. AY033092.
RESULTS
Complementation of a propionate-negative mutant.
UV-induced mutant IPT189 of B. sacchari IPT101T is not able to grow on propionic acid as a sole carbon source but still incorporates 3HV into PHA when it is starved for nitrogen (41). We decided to use this mutant for identification of the genes required for propionate catabolism in B. sacchari by phenotypic complementation. Four clones of the gene library prepared from EcoRI-restricted genomic DNA of IPT101T and pVK100 were identified by minicomplementation of mutant IPT189. In these clones the ability to grow on propionic acid was restored.
The hybrid plasmid of one clone (p17227), which consisted of four different EcoRI restriction fragments, was digested with EcoRI, and the fragments were ligated to EcoRI-digested pVK100 DNA. The ligation products were transformed into E. coli S 17-1, and clones were selected on LB agar plates containing tetracycline. Phenotypic complementation of IPT189 resulted in identification of six transconjugants in which the Prp+ phenotype was restored. All hybrid plasmids harbored a 2-kbp EcoRI fragment; four plasmids (pB6, pB10, pB27, and pC14) harbored only this fragment, whereas one (pB7) also harbored a 3-kbp EcoRI fragment and one (pA26) also harbored the 3-kbp fragment plus a 5-kbp EcoRI fragment. When the 2-kbp EcoRI fragment was digested with SalI, a 1.1-kbp SalI restriction fragment was obtained, and this 1.1-kbp fragment was ligated to SalI-digested pVK100 DNA, which still complemented mutant IPT189. Analysis of the 2-kbp EcoRI fragment revealed partial sequences of two open reading frames, whose putative translational products exhibited high levels of similarity to the 2-methylcitrate synthases (prpC gene product) of E. coli (50) and Salmonella enterica (24) or to aconitate hydratases of several organisms (see below).
Characterization of the Burkholderia prp locus.
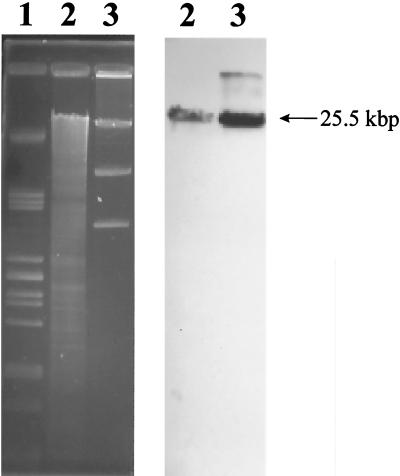
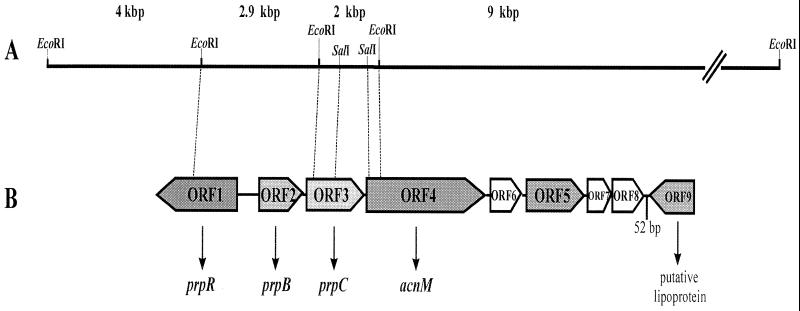
In order to clone a larger DNA fragment comprising the entire prp region, genomic DNA of B. sacchari IPT101T was digested with different restriction enzymes and used for Southern hybridization with the 2-kbp EcoRI fragment as a probe. Fragments that were approximately 25 to 30 kbp long (HindIII-digested DNA), 20 kbp long (BamHI-digested DNA), and 7 kbp long (ClaI-digested DNA) gave hybridization signals. Subsequently, HindIII-digested DNA was ligated to HindIII-linearized pHC79 and transduced into E. coli S17-1. Tetracycline-resistant clones were screened by colony hybridization by using the 2-kbp EcoRI fragment as a homologous probe. Five positive clones (clones A to E), each harboring a 25.5-kbp HindIII fragment, were identified (Fig. 1). The hybrid cosmid of clone D, which harbored only the 25.5-kbp HindIII fragment, was restricted with EcoRI, which resulted in eight subfragments (1.4, 2, 2.4, 2.9, 3.8, 4, 6.5 [pHC79], and 9 kbp). All of these fragments were subcloned into the vector pBCSK+ and were at least partially sequenced. The 2-kbp EcoRI fragment was identical to the fragment that was initially cloned, whereas the 2.9-kbp fragment was adjacent to and upstream of the 2-kbp EcoRI fragment and the 9-kbp fragment was adjacent to and downstream of the 2-kbp EcoRI fragment (Fig. 2A). The 4-kbp EcoRI fragment was localized upstream of the 2.9-kbp fragment (Fig. 2A). On the last four fragments five open reading frames were identified, and these open reading frames exhibited similarities to MCC structural genes (Fig. 2B).
FIG. 1.
Southern hybridization of E. coli S17-1 clones, harboring HindIII-digested genomic DNA of B. sacchari IPT101T in cosmid pHC79, identified by colony hybridization by using the 2-kbp EcoRI fragment as a probe. Lane 1, PstI-digested λ DNA; lanes 2, HindIII-digested genomic DNA of B. sacchari IPT101T; lanes 3, HindIII-restricted hybrid cosmids of the E. coli S17-1 clone.
FIG. 2.
(A) Arrangement of four EcoRI subfragments on the 25.5-kbp HindIII fragment of B. sacchari IPT101T; (B) organization of the prp locus in B. sacchari IPT101T.
ORF2 encodes a 2-methylisocitrate lyase (PrpB).
The 892-bp open reading frame ORF2 starts with an ATG codon at nucleotide 2140 that is the most probable translational initiation codon of the structural gene which was designated prpB. This was concluded on the basis of the amino acid alignment and on the basis of the tentative ribosome-binding site which preceded this ATG. The prpB gene product consisted of 297 amino acids, and it had a calculated molecular mass of 31,732 Da and a pI of 4.99. The primary structure of PrpB revealed a level of identity of 64 mol% to the prpB gene product of R. eutropha HF39 (Table 1), which catalyzes cleavage of 2-methylisocitric acid to succinate and pyruvate in this strain (8). Furthermore, an isocitrate lyase signature (K-[KR]-C-G-H-[LMQR]) was present, as it is in all known methylisocitrate lyase sequences; this signature includes highly conserved cysteine (C), glutamate (G), and histidine (H) residues which are important for the catalytic activity of isocitrate lyases (http://www.expasy.ch/cgi-bin/nicesite.pl?PS00161).
TABLE 1.
Comparison of the translational products of genes located in the prp cluster of several organisms with the translational products of prpB, prpC, acnM, and ORF5 of B. sacchari IPT101T
| Strain | mol% identity with products of B. sacchari IPT101T genesa
|
Referenceb | |||
|---|---|---|---|---|---|
| prpB | prpC | acnM | ORF5 | ||
| R. eutropha HF39 | 64 | 82 | 91 | 84 | 8 |
| B. cepacia | NDc | 88 | 83 | 80 | |
| N. meningitidis serogroup A | 63 | 74 | 79 | 67 | 34 |
| N. meningitidis serogroup B | 63 | 74 | 78 | 67 | 49 |
| P. aeruginosa PAO1 | 61 | 54 | 83 | 71 | 46 |
| P. putida KT2440 | 64 | 53 | 86 | 83 | |
| P. fluorescens | 60 | 54 | 84 | 77 | |
| P. syringae | 64 | 52 | —d | 79 | |
| E. coli K-12 | 58 | 76 | ND | ND | 50 |
| E. coli O157:H17 | 57 | 76 | ND | ND | 35 |
| S. enterica serovar Typhimurium | 58 | 75 | ND | ND | 22 |
| L. pneumophila | ND | 51 | ND | ND | |
| V. cholerae | 57 | 51 | 75 | 69 | 19 |
| S. putrefaciens | 66 | 54 | 84 | 80 | |
| R. metallidurans CH34 | 60 | 84 | 89 | 80 | |
Levels of amino acid identity.
The data for some of the organisms were obtained from the following sources: B. cepacia, P. fluorescens, and R. metallidurans, http://www.expasy.ch/cgi-bin/nicesite.p1?PS00161; P. putida, P. syringae, and S. putrefaciens, http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism; and L. pneumophila, accession no. AAD42885.
ND, not detected in the database.
—, only incomplete sequences were available.
To confirm the presence of a 2-methylisocitrate lyase in B. sacchari, cells of B. sacchari IPT101T and IPT189 were grown in 50 ml of MM containing 1.0% (wt/vol) sodium gluconate for 30 h at 30°C. The cells were then harvested, washed with sterile saline, and transferred into fresh MM containing 0.2% (wt/vol) sodium propionate (IPT189 and IPT101T) or 0.2% (wt/vol) sodium gluconate (IPT101T) as the sole carbon source. After 24 h of incubation at 30°C, we measured the 2-methylisocitrate lyase activities in crude extracts of the propionate-induced and uninduced cells of IPT101T and the propionate-induced cells of IPT189, as shown in Table 2. The results revealed that formation of 2-methylisocitrate lyase is induced by propionic acid in B. sacchari IPT101T, as shown previously for the MCC in R. eutropha HF39 (8). The activity of this enzyme was strongly reduced in the propionic acid-negative mutant IPT189. Furthermore, a 2-methylisocitrate lyase activity of 16.6 mU/mg of protein was observed in the recombinant E. coli(pSK−/prpBBs), compared to an activity of only 2.6 mU/mg of protein in an E. coli strain harboring pBluescript SK− as a control.
TABLE 2.
Activities of 2-methylcitrate lyase and 2-methylcitrate synthase in crude extracts of B. sacchari wild-type strain IPT101T and mutant Prp189
| Crude extract | Carbon sourcea | Sp act (U/mg of protein)
|
|
|---|---|---|---|
| 2-Methylcitrate synthase | 2-Methylisocitrate lyase | ||
| IPT101T | Sodium gluconate | 0.076 | 0.017 |
| Sodium propionate | 0.410 | 0.112 | |
| Prp189 | Sodium propionate | 0.076 | 0.006 |
The cells were cultivated on MM containing different carbon sources after growth in MM containing sodium gluconate.
Thirty-four nucleotides upstream of prpB a consensus σ54 promoter like that in S. enterica serovar Typhimurium LT2 (33) and a putative upstream activator sequence (GATTTCAAATTTGAAATCNCAATTTCAGACAGGAAATTG [the different typefaces and underlining indicate the sequences which correspond]) like that in R. eutropha (8) were identified. The putative translational product of ORF1 exhibited 37 mol% amino acid identity to the prpR gene product of Pseudomonas aeruginosa and to other transcriptional activators belonging to the σ54 (rpoN) family (33). The molecular mass of PrpR, 70,598 Da, corresponded to the molecular mass of the putative prpR gene product of R. eutropha (8). It is therefore probably a transcriptional activator of the prp operon in B. sacchari and is referred to in this paper as prpR.
ORF3 encodes a 2-methylcitrate synthase (PrpC).
The deduced amino acid sequence of ORF3 (length, 1,167 bp), which started with the ATG at position 3082, exhibited levels of identity ranging from 51 mol% (Vibrio cholerae, Legionella pneumophila) to 88 mol% (Burkholderia cepacia) to prpC gene products of several organisms (Table 1) and had a molecular mass of 42,691 Da and a theoretical pI of 7.27. The prpC gene encodes a 2-methylcitrate synthase in R. eutropha HF39 (8), E. coli (50), and S. enterica serovar Typhimurium (22). A comparison of the amino acid sequences of all of the available prpC translational products of several organisms resulted in identification of two motifs with 100% identity to the citrate synthase signature (http://www.expasy.ch/cgi-bin/prosite-search-ac?PS00480) and to the putative active sites of citrate synthases, both of which are highly conserved in all putative methylcitrate synthases (Fig. 3).
FIG. 3.
Comparison of 2-methylcitrate synthase motifs with the corresponding citrate synthase signature. The shaded amino acids are the putative active sites (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html), which were identified on the basis of similarity. References, accession numbers (if available) and contig numbers are given in parentheses. Data for some of the organisms were obtained from the following websites: B. cepacia, R. metallidurans CH34, and Pseudomonas fluorescens, http://www.expasy.ch/cgi-bin/nicesite.pl?PS00161; and P. putida KT2440, P. syringae, and S. putrefaciens, http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism.
To confirm the presence of a 2-methylcitrate synthase in B. sacchari, cells were grown in the way described above for the analysis of the 2-methylisocitrate lyase. Measurement of 2-methylcitrate synthase activity revealed that formation of this enzyme was also induced by propionic acid, and cells of mutant IPT189 exhibited much less activity of this enzyme than the wild type when they were cultivated in the presence of propionic acid (Table 2). Furthermore, a level of 2-methylcitrate synthase activity of 0.91 U/mg of protein was determined in recombinant E. coli(pSK−/prpCBs), compared to a level of activity of 0.006 U/mg of protein in an E. coli strain harboring only pBluescript SK−.
In addition, the prpC gene of R. eutropha (8) was ligated into the vector pBRR1-JO2 (J. Overhage, personal communication) and transformed into E. coli XL-1 Blue. Hybrid plasmids pBRR1-JO2/prpCRe and pBRR1-JO2 were then transferred via E. coli S17-1 by conjugation into B. sacchari propionate-negative mutant IPT189. Hybrid plasmid pBRR1-JO2/prpCRe restored the ability of mutant IPT189 to grow on MM agar plates containing 0.1% (wt/vol) sodium propionate as the sole carbon source, whereas IPT189 harboring only the vector did not grow.
Putative gene product of ORF4.
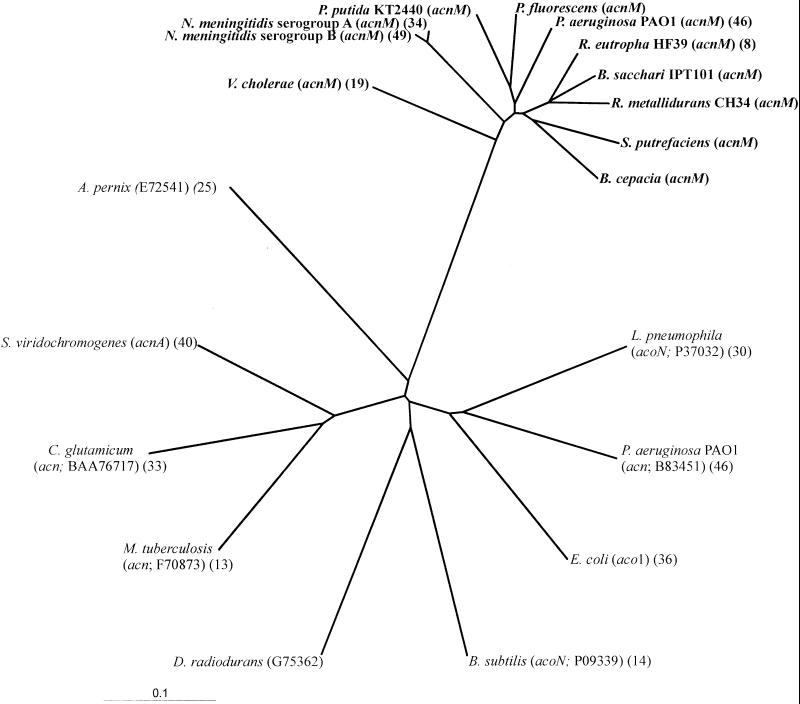
Downstream and 136 bp from ORF3, ORF4 started with a GTG codon at position 4385, which was preceded by a putative ribosome-binding site. The putative translational product of ORF4 had a calculated molecular mass of 94,376 Da and a theoretical pI of 5.27. A comparison of the deduced amino acid sequence of ORF4 with the primary structures of other proteins revealed that the highest levels of identity, 91 and 89 mol%, were the levels of identity to the aconitate hydratases of R. eutropha HF39 (8) and Ralstonia metallidurans CH34 (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html), respectively, as shown in Table 1; the genes for these enzymes are located downstream of the prpC gene in a cluster (8). In contrast, the levels of identity of the gene product of ORF4 to aconitases in the tricarboxylic acid cycle were rather low (40 to 45 mol%). A dendrogram based on the amino acid sequences of several aconitate hydratases revealed that the putative acnM translational products form a cluster, which separates them from citric acid cycle aconitases (Fig. 4). The amino acid sequences of aconitate hydratases include three highly conserved cysteine residues, which represent the ligands for the 4Fe-4S cluster. These three cysteine residues also occur in all known acnM gene products, as shown in Fig. 5. A difference in the vicinity of these cysteine residues was found when the acnM translational products and the other aconitate hydratases were compared. Whereas all acnM translational products have a polar asparagine residue next to the cysteine residue (Fig. 5), in most cases the other aconitate hydratases have the nonpolar amino acid isoleucine at this position; in some cases an enzyme has valine or leucine at this position (http://www.expasy.ch/cgi-bin/prosite-search-ac?PS00450; http://www.expasy.ch/cgi-bin/prosite-search-ac?PS01244).
FIG. 4.
Dendrogram based on aconitases and acnM translational products of several bacteria. The numbers in parentheses are accession numbers and references. Data for some of the organisms were obtained from the following websites: P. fluorescens and B. cepacia, http://www.expasy.ch/cgi-bin/nicesite.pl?PS00161; and S. putrefaciens and P. putida, http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism.
FIG. 5.
Comparison of motifs of the acnM translational products of several organisms with the aconitase signature. The shaded amino acids are the putative cysteine residues which bind the Fe-S-cluster (identified on the basis of similarity) (http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism). The boldface N is a highly conserved asparagine residue in acnM translational products; in aconitases in the tricarboxylic acid cycle this amino acid is isoleucine (indicated by boldface I). The references and accession numbers (if available), as well as contig numbers, are given in parentheses. Data for some of the organisms were obtained from the following websites: B. cepacia, R. metallidurans CH34, and P. fluorescens, http://www.expasy.ch/cgi-bin/nicesite.pl?PS00161; and P. putida KT2440, P. syringae, and S. putrefaciens, http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism.
Complementation of Tn5-induced prpC and acnM mutants of R. eutropha HF39.
Phenotypic complementation of Tn5-induced mutants VG17 and P2 of R. eutropha HF39 with a defect in a putative aconitate hydratase gene and phenotypic complementation of prpC mutant SK7286 of R. eutropha HF39 with the 2-kbp EcoRI or 1.1-kbp SalI fragment in cosmid pVK100 failed. Since neither fragment harbored the complete prpC gene or the complete aconitate hydratase gene, complementation of B. sacchari mutant IPT189 most probably resulted from homologous recombination of the incomplete genes present on the plasmids with the corresponding defective parts in the mutant genome, a process that can occur only between highly homologous or identical DNA sequences. Therefore, no complementation of the R. eutropha mutants could occur.
Putative gene product of ORF5.
The 396-amino-acid translational product of 1,191-bp ORF5 had a calculated molecular mass of 41,443 Da and a theoretical pI of 5.95 and exhibited the highest levels of identity (84 and 83 mol%) to the ORF5 translational products of R. eutropha HF39 (8) and Pseudomonas putida KT2440 (Table 1). Analysis of mutants has shown previously that in R. eutropha HF39 ORF5 is required for conversion of 2-methylcitrate into 2-methylisocitrate (8).
Putative gene products of ORF6, ORF7, ORF8, and ORF9.
Three additional small open reading frames that are 366 bp long (ORF6), 198 bp long (ORF7), and 372 bp long (ORF8) are located in the prp cluster of B. sacchari IPT101T. The translational product of ORF6 exhibited 50 mol% identity to a hypothetical 14.0-kDa protein encoded in the fabI-sapF region of E. coli (5), whereas the translational products of ORF7 and ORF8 exhibited 53 and 44 mol% identity to hypothetical proteins Rv1113 and Rv1114 of Mycobacterium tuberculosis, respectively (13). ORF9 was located 52 bp downstream of and antilinear with respect to the prp locus, and the translational product of ORF9 exhibited identity to a hypothetical gene product of Rubrivivax gelatinosus, probably a lipoprotein (accession no. T50918).
DISCUSSION
Although several pathways for propionic acid catabolism in bacteria have been described, it has been suggested that in E. coli, especially at a higher substrate concentration, the main route is α-oxidation of propionate via acrylyl-CoA and lactyl-CoA to pyruvate (56). However, this suggestion was based on some misleading enzyme activity measurements (50) and radiolabeling experiments (56). Nevertheless, results obtained in radiolabeling experiments did not allow the researchers to distinguish between α-oxidation and the 2-methylcitrate pathway, since the patterns of the labeled products formed were identical (i.e., decarboxylation of propionate at C-1). Therefore, the results obtained previously should be carefully reviewed. However, in the last few years there has been evidence that propionic acid is oxidized to pyruvate by the 2-methylcitrate pathway (MCC) in E. coli. In addition, activity of 2-methylcitric acid synthase, which is one of the key enzymes of the 2-methylcitrate pathway, has also been detected in E. coli (50), S. enterica (23), R. eutropha (8), P. aeruginosa (55), and Pyrococcus furiosus (16, 55). The other enzymes of this pathway, encoded by the prpE and prpB genes, were identified as a propionyl-CoA synthetase (PrpE) in S. enterica serovar Typhimurium LT2 (23) and a 2-methylisocitrate lyase (PrpB) in E. coli and Aspergillus nidulans (10), respectively. Although it is evident that the MCC is probably used most often for catabolism of propionic acid in bacteria and that this pathway also occurs in some eukaryotes, the MCC has been studied in detail only in a few bacteria and in the eukaryote A. nidulans (9, 10) at a biochemical and molecular level. For most bacteria evidence for this pathway has been obtained only from genome sequence data.
This study revealed that the 2-methylcitric acid pathway is the predominant pathway for catabolism of propionic acid in B. sacchari IPT101T. An understanding of this pathway and the availability of the genes of this pathway should allow workers to obtain improved strains of B. sacchari that are more suitable for fermentative production of poly(3HB-co-3HV) from sucrose and propionic acid.
In a previous study two classes of propionic acid-negative mutants of B. sacchari IPT101T were obtained. Representatives of the first class might be affected in an unknown pathway, whereas representatives of the second class were defective in the MCC (41). Mutant IPT189, which belongs to the second class, exhibited the highest increase in the yield of 3HV units in the copolyester poly(3HB-co-3HV) from propionate. The results of growth experiments performed in a bioreactor, compared to the results of shaken flask experiments, revealed that the pathway affected in mutant IPT189 is important if low concentrations (0.02 to 0.04%, wt/vol) of propionic acid are present in the medium, whereas the other pathway of propionic acid oxidation is more important if a higher concentration (0.1%, wt/vol) of propionic acid is present in the medium. More than 90% of the mutants obtained after enrichment, which was performed with high concentrations of this organic acid, were deficient in utilization of intermediates of the α-oxidation pathway (41).
Phenotypic complementation of such a mutant led to cloning of a 25.5-kbp HindIII fragment which carried a cluster of genes whose putative gene products exhibited strong similarities to the gene products of genes of the prp operon of R. eutropha and several other gram-negative bacteria (8). The gene products of this prp operon constitute the 2-methylcitric acid pathway in this mutant strain. In B. sacchari IPT101T a putative aconitate hydratase gene (acnM) is also located downstream of prpC, as shown for the propionate cluster in several organisms. The prpD gene, which is probably involved in the isomerization reaction in the MCC of S. enterica serovar Typhimurium, as shown by the accumulation of 2-methylcitric acid (24) and in vitro conversion of 2-methylcitrate into 2-methylisocitrate (25), is missing at the prp locus of B. sacchari, as also observed for R. metallidurans CH34, B. cepacia, Neisseria meningitidis serogroups A and B, Shewanella putrefaciens, and Pseudomonas syringae. If the acnM and ORF5 gene products are also involved in conversion of 2-methylcitrate into 2-methylisocitrate, as in R. eutropha HF39, the prpD gene product is not required for a functional MCC in B. sacchari IPT101T. If ORF5, ORF7, and ORF8 are also not required for a functionally active MCC, the prp locus of this strain consists of the minimal set of structural genes (prpB, prpC, acnM, and ORF5) which are required for the function of this cyclic pathway.
As shown by the reduced specific activities of 2-methylcitrate synthase and 2-methylisocitrate lyase in propionate-induced mutant IPT189 and by the complementation of mutant IPT189 by the R. eutropha HF39 prpC gene, the formation of 2-methylcitrate might influence the transcription of the prp locus in B. sacchari IPT101T, as supposed for S. enterica serovar Typhimurium (33).
A number of authors have analyzed (by nuclear magnetic resonance) amino acids or other products formed in radiolabeling experiments to evaluate the relevance of propionate catabolism pathways. However, labeling tests allow simple distinctions between randomizing pathways (which lead to succinate) and nonrandomizing pathways (which lead to acetyl-CoA). The poly(3HB-co-3HV) production system could be used in a similar way. The poly(3HB-co-3HV) production system is a useful system for revealing the relative contributions of the various pathways to the catabolism of propionate since it generates large amounts of products, in contrast to the amounts of amino acids obtained in labeling studies, which make it difficult to analyze the labeled products. Independent of radiolabeling assays, the poly(3HB-co-3HV) production system can indicate the relevance of the different pathways if mutants are analyzed and if their Y3HV/Prop values are compared. Use of this system in previous studies (41) showed that in B. sacchari the 2-methylcitrate pathway operates better at low propionate concentrations (0.2 to 0.4 g · liter−1; Y3HV/Prop = 1.2 g · g−1) than at higher propionate concentrations (1 g · liter−1; Y3HV/Prop = 0.8 g · g−1); i.e., a mutation in the MCC resulted in more propionate available for 3HV synthesis and therefore increased Y3HV/Prop in the presence of a low propionate concentration.
Moreover, the system provided evidence that a second minor pathway is present since the maximum theoretical yield (1.35 g · g−1) was not achieved (41).
Acknowledgments
We are in indebted to W. Buckel and D. Darley (Philipps-Universität Marburg, Marburg, Germany) for the gift of methylisocitric acid lactone.
This work was supported by FAPESP and PADCT/Finep-MCT.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Rent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Bairoch, A., P. Bucher, and K. Hoffmann. 1997. The PROSITE database, its status in 1997. Nucleic Acids Res. 25:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. I. I. L. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, F. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 7.Brämer, C. O., P. Vandamme, L. F. da Silva, J. G. C. Gomez, and A. Steinbüchel. 2001. Burkholderia sacchari sp. nov., a polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Bacteriol. Evol. Microbiol. 51:1709–1713. [DOI] [PubMed] [Google Scholar]
- 8.Brämer, C. O., and A. Steinbüchel. 2001. The methylcitric acid pathway in Ralstonia eutropha: new genes identified involved in propionate metabolism. Microbiology 147:2203–2214. [DOI] [PubMed] [Google Scholar]
- 9.Brock, M., R. Fischer, D. Linder, and W. Buckel. 2000. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol. Microbiol. 35:961–973. [DOI] [PubMed] [Google Scholar]
- 10.Brock, M., D. Darley, S. Textor, and W. Buckel. 2001. 2-Methylisocitrate lyases from the bacterium Escherichia coli and the filamentous fungus Aspergillus nidulans. Eur. J. Biochem. 268:3577–3586. [DOI] [PubMed] [Google Scholar]
- 11.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid-transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5:376–379. [Google Scholar]
- 12.Byrom, D. 1990. Industrial production of copolymers from Alcaligenes eutrophus, p.113–117. In E. A. Dawes (ed.), Novel biodegradable microbial polymers. Kluwer, London, United Kingdom.
- 13.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, S. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 39:537–544. [DOI] [PubMed] [Google Scholar]
- 14.Dingman, D. W., and A. L. Sonenshein. 1987. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citB gene. J. Bacteriol. 169:3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Briera, A., and A. Garrido-Pertierra. 1988. A degradation pathway of propionate in Salmonella typhimurium LT-2. Biochimie 70:757–768. [DOI] [PubMed] [Google Scholar]
- 16.Gerike, U., W. H. David, N. J. Russel, M. L. Dyall-Smith, and J. D. Michael. 1998. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology 144:929–935. [DOI] [PubMed] [Google Scholar]
- 17.Gomez, J. G. C., M. F. A. Rodrigues, R. C. P. Alli, B. B. Torres, C. L. B. Netto, M. S. Oliveira, and L. F. daSilva. 1996. Evaluation of soil Gram-negative bacteria yielding polyhydroxyalkanoic acids from carbohydrates and propionic acid. Appl. Microbiol. Biotechnol. 45:785–791. [Google Scholar]
- 18.Grunstein, M., and D. S. Hogness. 1975. Colony hybridization: a method for the isolation of cloned DNA that contains a specific gene. Proc. Natl. Acad. Sci. USA 72:3961–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. A. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291–298. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, P. A. 1985. Applications of PHB—a microbially produced biodegradable thermoplastic. Phys. Technol. 16:32–36. [Google Scholar]
- 22.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381–1388. [DOI] [PubMed] [Google Scholar]
- 24.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181:5615–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horswill, A. R., and J. C. Escalante-Semerena. 2001. In vitro conversion of propionate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40:4703–4713. [DOI] [PubMed] [Google Scholar]
- 26.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyperthermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83–101. [DOI] [PubMed] [Google Scholar]
- 27.Knauf, V. C., and E. W. Nestor. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45–54. [DOI] [PubMed] [Google Scholar]
- 28.Luttik, M. A. H., P. Kötter, F. A. Salomons, I. van der Klei, J. P. van Dijken, and J. T. Pronk. 2000. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. J. Bacteriol. 182:7007–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acids from microorganisms. J. Mol. Biol. 3:208–218. [Google Scholar]
- 30.Mengaud, J. M., and M. A. Horwitz. 1993. The major iron-containing protein of Legionella pneumophila is an aconitase homologous with the human iron-responsive element-binding protein. J. Bacteriol. 175:5666–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyakoshi, S., H. Uchiyama, T. Someya, T. Satoh, and T. Tabuchi. 1987. Distribution of the methylcitric acid cycle and β-oxidation for propionate in fungi. Agric. Biol. Chem. 51:2381–2387. [Google Scholar]
- 32.Oelmüller, U., N. Krüger, A. Steinbüchel, and C. G. Friedrich. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73–84. [Google Scholar]
- 33.Palacios, S., and J. C. Escalante-Semerena. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502–506. [DOI] [PubMed] [Google Scholar]
- 35.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529–533. [DOI] [PubMed] [Google Scholar]
- 36.Prodromou, C., P. J. Artymiuk, and J. R. Guest. 1992. The aconitase of Escherichia coli. Nucleotide sequence of the aconitase gene and amino acid sequence similarity with mitochondrial aconitases, the iron-responsive-element-binding protein and isopropylmalate isomerases. Eur. J. Biochem. 204:599–609. [DOI] [PubMed] [Google Scholar]
- 37.Pronk, J. T., A. van der Linden-Beuman, C. Verduyn, W. A. Scheffers, and van J. P. Dijken. 1994. Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology 140:717–722. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultivierung wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209–222. [PubMed] [Google Scholar]
- 40.Schwartz, D., S. Kaspar, G. Kienzlen, K. Muschko, and W. Wohlleben. 1999. Inactivation of the tricarboxylic acid cycle aconitase gene from Streptomyces viridochromogenes Tu494 impairs morphological and physiological differentiation. J. Bacteriol. 181:7131–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva, L. F., J. G. C. Gomez, M. S. Oliveira, and B. B. Torres. 2000. Propionic acid metabolism and poly-3-hydroxybutyrate-co-3-hydroxyvalerate (P3HB-co-3HV) production by Burkholderia sp. J. Biotechnol. 76:165–174. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784–791. [Google Scholar]
- 43.Srere, P. A. 1966. Citrate-condensing enzyme-oxaloacetate binary complex. J. Biol. Chem. 241:2157–2165. [PubMed] [Google Scholar]
- 44.Steinbüchel, A., and U. Pieper. 1992. Production of a copolyester of 3-hydroxybutyric acid and 3-hydroxyvaleric acid from unrelated carbon sources by a mutant of Alcaligenes eutrophus. Appl. Microbiol. Biotechnol. 37:1–6. [Google Scholar]
- 45.Steinbüchel, A., and H. F. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219–228. [Google Scholar]
- 46.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrook-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. M. Lim, K. A. Smith, D. H. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959–964. [DOI] [PubMed] [Google Scholar]
- 47.Strauss, E. C., J. A. Kobori, G. Siu, and L. E. Hood. 1986. Specific-primer-directed DNA sequencing. Anal. Biochem. 154:353–360. [DOI] [PubMed] [Google Scholar]
- 48.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809–1815. [DOI] [PubMed] [Google Scholar]
- 50.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Müller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428–436. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thony-Meyer, L., and P. Kunzler. 1996. The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for an effective root nodule symbiosis. J. Bacteriol. 178:6166–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valentin, H. F., and D. Dennis. 1996. Metabolic pathway for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) formation in Nocardia corallina: inactivation of mutB by chromosomal integration of a kanamycin resistance gene. Appl. Environ. Microbiol. 62:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogelstein, B., and D. Gillespie. 1979. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. USA 76:615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson, D., D. L. Lindel, and R. Fall. 1983. Pseudomonas aeruginosa contains an inducible methylcitrate synthase. Curr. Microbiol. 8:17–21. [Google Scholar]
- 56.Wegener, W. S., H. C. Reeves, R. Rabin, and S. J. Ajl. 1968. Alternate pathways of metabolism of short-chain fatty acids. Bacteriol. Rev. 32:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]