Abstract
An RTK-Ras-mitogen-activated protein kinase (MAPK) signaling pathway plays a key role in vulval induction in Caenorhabditis elegans. We have previously carried out screens for suppressors of activated Ras to identify factors that play critical roles in the regulation of the pathway. ku258 was isolated as a semidominant allele that suppresses the Multivulva phenotype caused by activated let-60 ras. Our genetic and molecular analyses indicate that ku258 is a gain-of-function allele resulting from two point mutations in the C. elegans homolog of the transcriptional coactivator p300/CBP, cbp-1. Genetic data also suggest that cbp-1 may act downstream of the Ras signaling pathway, but not primarily downstream of the Wnt signaling pathway, to negatively regulate vulval cell fate specification. cbp-1 may function in concert with LIN-1, an Ets transcription factor family member that is one of the targets of MAPK. In vitro histone acetylation assays have revealed that together, the two point mutations cause a sevenfold increase in the histone acetyltransferase (HAT) activity of recombinant CBP-1. To our knowledge, this is the only such HAT activity mutation isolated in a CBP/p300 family protein, and this mutation may define a negative role of the HAT activity in antagonizing Ras function in a specific developmental event.
Through the study of genetically tractable developmental organisms such as Drosophila melanogaster and Caenorhabditis elegans, as well as work done in vertebrate cell culture, many of the major components and regulators of a canonical RTK-Ras-mitogen-activated protein kinase (MAPK) pathway have been identified (26, 39, 46). In C. elegans, activation of an RTK-Ras-MAPK pathway leads to the induction of three (P5.p to P7.p) of six equivalent vulval precursor cells (VPCs) to adopt a vulval cell fate (41). Gain-of-function mutations in components of the Ras pathway, such as activated let-60(n1046) Ras, cause a Multivulva (Muv) phenotype where more than three VPCs adopt the vulval cell fate (reviewed in reference 39). Conversely, loss-of-function mutations in Ras pathway components lead to a Vulvaless (Vul) phenotype where less than three VPCs are induced to form the vulva, resulting in an Egg-laying-defective (Egl) animal. LIN-1, an Ets domain transcription factor, and the vulva-specific HNF-3/Forkhead-like transcription factor, LIN-31, have both been proposed as negative regulators of vulval induction (5, 30). Biochemical evidence has suggested that these two transcription factors function as a heterodimer to negatively regulate induction (43). Upon activation, the worm MAPK homolog, MPK-1, phosphorylates LIN-1 and LIN-31, thereby disrupting the heterodimer.
CBP and p300 are two highly related proteins that are thought to act as transcriptional coactivators by coupling extracellular stimuli through their interactions with a large number of DNA binding nuclear factors to components of the basal transcriptional machinery (24). These two transcriptional coactivators are thought to play important roles in cell growth, proliferation, and differentiation (reviewed in reference 15). Although the precise function of CBP/p300 on these processes appears to be highly context dependent, an emerging body of evidence has demonstrated that they share many properties with tumor suppressors. Mice heterozygous for CBP have a significantly increased rate of malignancies than the rate for wild-type mice (28). Human patients with Rubinstein-Taybi syndrome, a result of CBP heterozygosity, also display an increased rate of malignancies (31). CBP and p300 have intrinsic histone acetyltransferase (HAT) activity (4, 33). A number of in vitro and cell culture studies have been carried out to address the importance of the intrinsic HAT activity of CBP/p300 for their function. It has been shown that the requirement of this HAT activity for transcription is highly context and promoter dependent with HAT activity being completely dispensable at some promoters. One recent in vivo study demonstrated that dCBP HAT activity is required to maintain an overexpression phenotype and global H4 acetylation in Drosophila (29). However, our understanding of the exact requirements for CBP/p300 HAT activity in specific signaling events and in the context of a developing organism has been slow in coming.
Much as in other systems, the C. elegans p300/CBP homolog, CBP-1, was shown to regulate the differentiation of a number of embryonic cell types and to generally inhibit proliferation (36). Subsequently, it was demonstrated that the HAT activity of CBP-1 was required for its proper embryonic function (44). The embryonic lethality of cbp-1 loss-of-function alleles has, up to this point, prevented the identification and analysis of CBP-1 function in the postembryonic worm. In this study we focus on genetic and biochemical analyses of a semidominant mutation in the C. elegans CBP/p300 homolog that reveal a role of this gene in antagonizing Ras function and vulval induction.
MATERIALS AND METHODS
Strains and genetic methods.
All strains other than those used for single-nucleotide polymorphism (SNP) mapping were derived from the wild-type Bristol strain, N2. Standard protocols for the handling, culture, and ethyl methanesulfonate mutagenesis of strains were used (7). Alleles and rearrangements used in this study have been described previously in reference 35 or as follows: cbp-1(ku258) (this study); let-60(n1046 gf), lin-15(n765 ts), lin-1(e1275), lin-31(n301), sur-6(ku123) (38); unc-29(e403), lin-1(n2515 gf) (22); unc-17(e113), unc-69(e587), sma-2(e502), ced-7(n1892), qC1, sqv-3(n2841), emb-9(cg56), unc-32(e189), dpy-19(e1259), glp-1(q231), tnDf2, and cbp-1(bm2) (44). An integrated ajm-1::gfp {pJS191 (ajm-1::gfp) plus pDP#MM016B [unc-119(+)]} II was used to visualize VPC fusion (32; unpublished results from our laboratory).
Mutant isolation.
The ku258 allele was isolated in a screen for temperature-sensitive (TS) suppressors of let-60(n1046), even though ku258 is a non-TS allele. The procedure is essentially the same as a TS mutant screen described in reference 17, except the let-60(n1046 gf) strain with a 76% Muv phenotype was mutagenized. Non-Muv F2 animals that gave less than 10% Muv progeny were outcrossed and the suppressor mutations mapped.
Phenotypic observations.
The scoring of phenotypes, such as Multivulva (Muv) or Egg laying defective (Egl), were performed as previously described (12). Vulval induction was scored by examining late L3 or early L4 stage larvae under Nomarski differential interference contrast microscopy (40). Sterility was determined by cloning L1 animals to individual plates and scoring for the presence or absence of progeny.
Genetic mapping.
We initially mapped ku258 to chromosome III between dpy-19 and unc-69. Additional data from four other mapping strains suggested ku258 was just left of sqv-3 and to the right of ced-7 at 0.80 map unit. SNP mapping was then performed as described previously (23) using the Hawaiian strain CB4856 (25). Two SNPs, kuP7 (R10E11 position 10779; CB4856 sequence, TTCAGCT [the nucleotide exhibiting SNP is underlined]) and kuP8 (R10E11 position 25695; CB4856 sequence, AGAATACA), were identified on cosmid R10E11. Two additional SNPs on cosmids K11H3 (13882) and C05B5 (4103) that were previously deposited in the Washington University C. elegans SNP database were also used. A ku258 unc-69(e587); let-60(n1046) strain was made and crossed with CB4856 to create ku258 unc-69(e587)/CB4856; let-60(n1046). Forty-two Unc-non-ku258 recombinants were isolated. Analysis of the SNPs, done by sequencing at least three combined PCR amplifications of genomic DNA from recombinant worms, determined that the last recombination event occurred between kuP7 and kuP8 on cosmid R10E11. The SNP data strongly suggested that ku258 was no more than 7 to 12 kb from kuP7.
PCR was performed to amplify all coding regions and splice junctions of cbp-1 and N2 (Bristol) genomic DNA. These products were then sequenced. ku258 lesions were confirmed by sequencing at least three independent PCR products. Reverse transcription-PCR of cbp-1 was also performed using ku258 mutant RNA as a template. Sequencing of this reverse transcription-PCR product confirmed the genomic DNA lesions. A gene nearby cbp-1, sqv-3, was also sequenced, and no mutations were identified.
Immunostaining of C. elegans larvae.
A mixed stage population of animals was fixed as described previously (6) and then stained overnight with the anti-CBP-1 antibody (1:500 dilution) (44). Animals were simultaneously stained with 4′,6′-diamidino-2-phenylindole (DAPI) to serve as a positive control for the identification of nuclei.
CBP-1 HAT domain purification and histone acetyltransferase assays.
A 2.4-kb XhoI fragment of cbp-1 (amino acids 803 to 1620 containing the bromodomain and HAT domain) was cloned into the mammalian glutathione S-transferase (GST) expression vector pEBG-3X-HV (3). Point mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Constructs were then transfected into HEK 293 cells using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were lysed in lysis/purification buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride), and fusion proteins were purified on glutathione Sepharose (Amersham Pharmacia Biotech). GST fusion proteins were analyzed by Western blotting using an anti-GST monoclonal antibody (Santa Cruz Biotechnology) and quantitated by silver staining.
Histone acetyltransferase assays were performed as described previously (33) with some modifications. Reactions were carried out at 25°C for 60 min. Fifty nanograms of purified GST-CBP-1 HAT domain was used in each HAT reaction.
RESULTS
ku258 is a semidominant suppressor of activated Ras.
Previously, a genetic screen for suppressors of the Muv phenotype of an activated ras allele, let-60 ras (n1046), were carried out in our laboratory to identify a number of genes acting in a Ras-mediated signaling pathway (e.g., 37, 38, 42, 47). One allele, ku258, was isolated in a modified version of such a screen, in which the suppressor screen was combined with a TS screen (see Materials and Methods) (17). The observed suppression of the Muv phenotype (from 76% to 5%) was due to a reduction in ectopic induction of the VPCs (Table 1). Vulval induction of ku258 alone is near that of the wild type (Table 1). ku258 is also a semidominant suppressor of let-60(n1046); ku258 heterozygotes significantly suppressed the Muv phenotype of let-60 ras (n1046) (Table 1)
TABLE 1.
cbp-1(ku258) suppression of let-60 (n1046 gf)a
| Genotypeb | Muvc (%) (n) | Inductiond (n) | Vule (%) (n) | Eglf (%) (n) | Sterilityg (%) (n) | Lethality (%) (n)
|
|
|---|---|---|---|---|---|---|---|
| Embryonich | Larvali | ||||||
| Wild type (N2) | 0 (many) | 3.0 (29) | 0 (29) | 0 (many) | 0 (many) | 1.3 (525) | 0 (518) |
| cbp-1(ku258) | 0 (233) | 2.94 (40) | 8 (40) | 8 (53) | 19 (65) | 4 (428) | 10 (411) |
| let-60(n1046 gf) | 76 (212) | 4.29 (46) | 0 (46) | 0 (42) | 0 (42) | 1.6 (613) | 1.3 (603) |
| cbp-1(ku258)/+; let-60(n1046 gf) | 10 (504) | 3.33 (33) | 0 (33) | 5 (38) | 12 (43) | ND | ND |
| cbp-1(ku258); let-60(n1046 gf) | 5 (456) | 3.18 (49) | 0 (49) | 12 (42) | 18 (51) | 13 (381) | 22 (330) |
| cbp-1(bm2)/cbp-1(bm2) | ND | 100 (many) | |||||
| cbp-1(bm2)/+ | 0 (295) | 3.0 (15) | 0 (15) | ||||
| cbp-1(bm2)/+; let-60(n1046 gf) | 83 (503) | 4.5 (26) | 0 (26) | ||||
| cbp-1(bm2)/cbp-1(ku258); let-60(n1046 gf) | 3 (380) | 3.09 (21) | 0 (21) | ||||
| tnDf2/+; let-60(n1046 gf) | 96 (307) | ||||||
| nDf20; let-50(n1046 gf); nDp2[unc-86(e1416)] | 36 (205) | ||||||
All strains were grown at 20°C. cbp-1 (bm2 lf) is linked to dpy-18 in all of the relevant strains. tnDf2 was linked to unc-32(e189). The number of animals scored (n) is indicated in parentheses. ND, not determined.
gf indicates gain of function. lf indicates loss of function.
Percentage of animals displaying one or more ectopic pseudovulvae (ventral protrusions).
Average number of VPCs adopting a vulval cell fate relative to the number in the wild-type animal, as scored with Nomarski optics. In the wild-type animal, 3 of 6 VPCs are induced.
Percentage of animals in which <3 of the VPCs normally induced in wild-type animals (P5.p to P7.p) is not induced, as scored with Nomarski optics.
Egl animals were scored as the percentage of animals that were unable to lay eggs and had a Bag phenotype (larvae that hatched within the worm). Does not include sterile animals.
Percentage of animals that reached the adult stage but were unable to produce progeny.
Percentage of eggs that failed to hatch.
Percentage of animals that died during any of the larval stages.
Animals carrying the ku258 mutation were generally sick and displayed a number of pleiotropic defects in addition to suppressing let-60(n1046). Sterility was observed in 19% of ku258 homozygotes. A low percentage of ku258 animals also displayed embryonic lethality (4%) and larval lethality (10%) (Table 1). Inviable embryos appeared to be arrested shortly after gastrulation, and the observed larval lethality occurred at various larval stages. Lastly, both a protruding vulva (Pvul) and a slow growth phenotype were also observed for some ku258 animals.
The ku258 allele results from two point mutations in cbp-1, the C. elegans homolog of p300/CBP.
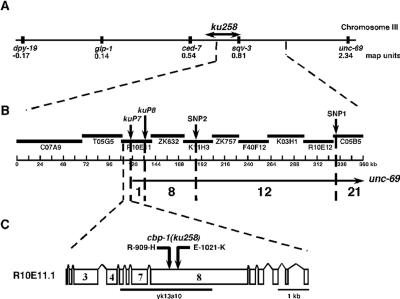
We mapped ku258 to the middle of chromosome III between ced-7 and unc-69 (Fig. 1A). SNP mapping was employed to obtain a high-resolution map position for ku258 (23) (see Materials and Methods). Our data indicated that ku258 lay within 7 to 12 kb on either side of SNP kuP7on cosmid R10E11 (Fig. 1B). The only predicted open reading frame (ORF) within 12 kb to the left of SNP kuP7 is R10E11.1. The predicted ORF was amplified and sequenced from ku258 genomic DNA, and two lesions were found (Fig. 1C).
FIG. 1.
Mapping and cloning of cbp-1(ku258). (A) ku258 was mapped to 0.80 map unit between sqv-3 and ced-7 on chromosome III. (B) ku258 was then linked to unc-69, and a ku258 unc-69(e587)/CB4856; let-60(n1046) SNP mapping strain was constructed. Forty-two Unc-non-ku258 recombinant chromosomes were isolated. Using the identified SNPs, kuP7, kuP8, SNP2, and SNP1, the region in which these recombination events occurred was determined. Using these data, ku258 is predicted to lie within 7 to 12 kb on either side of kuP7. R10E11.1/cbp-1 is the only open reading frame predicted within a 7- to 12-kb interval to the left of kuP7. (C) ku258 genomic DNA was sequenced, and two mutations, R-909-H and E-1021-K, were identified within the R10E11.1/cbp-1 ORF.
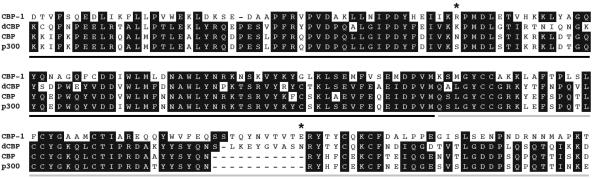
R10E11.1 encodes the worm homolog of the transcriptional coactivators p300 and CBP, cbp-1 (36). cbp-1 has been shown to be required for the proper differentiation and control of proliferation of a variety of tissues in the developing embryo (36, 44); however, a role for cbp-1 in Ras-mediated vulval fate specification has not been previously reported. CBP-1 shares a high degree of sequence conservation and a similar sequence arrangement with its human homologs, CBP/p300 (36). Both the bromodomain, which has been implicated in the binding of acetylated lysine residues, as well as the HAT domain, which is responsible for the intrinsic acetyltransferase activity of CBP/p300, are present in the C. elegans gene (Fig. 2) (15).
FIG. 2.
Multiple-sequence alignment of the CBP-1 bromodomain and partial HAT domain from amino acid 861 to amino acid 1054. CBP-1 (C. elegans), dCBP (Drosophila), CBP (human), and p300 (human) bromodomains are underlined with a black line. The partial HAT domains (remainder of sequence) are underlined with a gray line. cbp-1(ku258) mutant residues are marked by asterisks. Gaps introduced to maximize alignment are indicated by dashes.
The two mutations identified in cbp-1(ku258) were only 336 bp apart and therefore tightly linked genetically. Since neither mutation was found in the starting strain used for the screen, both guanine-to-adenosine transitions were likely introduced during ethyl methanesulfonate mutagenesis. The first mutation in the cbp-1(ku258) allele results in a conservative substitution of a histidine for an arginine in the bromodomain at amino acid position 909 (Fig. 2). The crystal structures for the bromodomains of the p300/CBP-related proteins Gcn5 and PCAF have been previously determined (8, 34). On the basis of these structures, the residue corresponding to position 909 in CBP-1 sits within the ZA loop that creates a hydrophobic pocket and appears to contact acetylated lysines. The second mutation identified in cbp-1(ku258) results in a nonconservative substitution of a glutamic acid for a lysine at amino acid position 1021 (Fig. 2). E-1021-K occurs in a 10- to 11-residue region present in cbp-1 and dCBP but absent in human p300/CBP. These extra residues reside within the originally reported minimal domain of p300/CBP that still possesses histone acetyltransferase activity (4, 33).
ku258 is likely a gain-of-function allele of cbp-1.
cbp-1(ku258) displays semidominant suppression of activated let-60 Ras in the vulva. We carried out dosage analysis on cbp-1(ku258) to better understand the genetic nature of this dominance and the function of wild-type cbp-1 in the vulva. Animals homozygous for a genetically null allele of cbp-1, bm2 (a deletion), are fully arrested at an early embryonic stage (44). This null allele was used to create a hemizygous strain for cbp-1 in a let-60 (n1046) ras background. This strain displayed an increased penetrance of the Muv phenotype (83%) compared to let-60(n1046) alone (76%) (Table 1), suggesting that cbp-1 normally functions to negatively regulate Ras signaling in the vulva. This in turn suggests that cbp-1(ku258) is a gain-of-function allele, as it is able to suppress activated ras. Additionally, cbp-1(ku258), when in trans to the deletion allele, appears to suppress let-60 ras (n1046) more effectively than does cbp-1(ku258)/+ and to the same extent as the cbp-1(ku258) homozygote (Table 1). These data suggest that wild-type protein may compete with the mutant protein for interactions with other proteins, thereby slightly reducing the potency of ku258-activated ras suppression. Additionally, a duplication, nDp2, that covers cbp-1 as well as a number of other genes in the region is able to partially suppress the Muv phenotype caused by let-60(n1046) (Table 1). This suggests that additional copies of cbp-1 can suppress let-60(n1046) and again indicates that CBP-1 normally functions to antagonize Ras signaling in the VPCs.
Lastly, the cbp-1(bm2) null mutants have a very distinct early embryonic arrest phenotype characterized by a complete lack of differentiation (44). As mentioned earlier, cbp-1(ku258) mutants arrest at a much later embryonic stage with obvious differentiation of a number of tissues. If cbp-1(ku258) were a straightforward dominant-negative mutation, cbp-1(ku258) homozygotes could also be expected to display an embryonic arrest similar to that of the null mutation. Failure to see this embryonic arrest is also consistent with cbp-1(ku258) being a gain-of-function mutation. Due to technical challenges, we have not been able to conclusively examine, utilizing mosaic analysis and stage- and tissue-specific RNA interference, the effect of a strong loss of cbp-1 function on vulval development (data not shown).
cbp-1 may act with the transcriptional machinery of the Ras pathway in the vulva.
The Ets transcription factor family member LIN-1 is thought to act at the transcriptional terminus of the Ras pathway as a negative regulator of vulval induction together with the HNF-3/Forkhead-like protein LIN-31 (5, 30, 43). cbp-1(ku258) partially suppressed the Muv phenotype produced by the lin-1(e1275 ts) hypomorphic allele from 88% Muv to 39% Muv at 20°C (Table 2). Although the number of ectopic VPC inductions was clearly reduced and the percentage of Vul animals was increased when they were examined under differential interference contrast microscopy, these double-mutant animals were very sick, and the position and morphology of the VPCs made it impossible to accurately score the exact level of VPC induction. The increase in Vul animals and the abnormal morphology and position of the VPCs in cbp-1(ku258); lin-1(e1275) animals is reflected in the increase of Egg-laying-defective (Egl) animals from 53% for lin-1(e1275) alone to 100% for the double mutant (Table 2). As one might predict from the increased sickness observed in the cbp-1(ku258); lin-1(e1275 ts) double mutant, cbp-1(ku258) was lethal in the background of the strong loss-of-function or null allele, lin-1(n1047). This suggests that cbp-1 might be functioning with lin-1 in events other than vulval induction. Although lin-1(e1275 ts) is a hypomorphic allele resulting from a nonsense mutation (5), loss-of-function mutations of genes acting upstream of lin-1 in the Ras-MAPK pathway are unable to suppress this allele (16, 21, 47). Therefore, the suppression of the lin-1(e1275) allele by the ku258 mutation is more consistent with cbp-1 functioning downstream of or parallel to lin-1.
TABLE 2.
Genetic interactions with cbp-1(ku258)a
| Genotypeb | Muv (%) (n) | Induction (n) | Vul (%) (n) | Egl (%) (n) | Sterility (%) (n) |
|---|---|---|---|---|---|
| Wild type (N2) | 0 (many) | 3.0 (29) | 0 (29) | 0 (many) | 0 (many) |
| cbp-1(ku258) | 0 (233) | 2.94 (40) | 8 (40) | 8 (53) | 19 (65) |
| lin-1(e1275 ts) | 88 (357) | 4.65 (31) | 0 (31) | 53 (36) | 12 (41) |
| cbp-1(ku258); lin-1(e1275 ts) | 39 (492)e | NDc | NDc | 100 (34) | 24 (45) |
| lin-31(n301 lf) | 86 (314) | NDd | 33 (21) | 21 (33) | 0 (33) |
| lin-31(n301 lf); cbp-1(ku258) | 83 (197) | NDd | 40 (22) | 39 (36) | 36 (56) |
| lin-1(n2515 gf) | 0 (131) | 2.97 (27) | 4 (27) | 11 (36) | 0 (36) |
| lin-1(n2515 gf); cbp-1(ku258) | 1 (212) | 2.43 (25) | 56 (25) | 79 (28) | 24 (37) |
| sur-6(ku123 lf) | 0 (117) | 2.97 (26) | 2 (26) | 6 (34) | 0 (34) |
| sur-6(ku123 lf); cbp-1(ku258) | 0 (129) | 2.91 (21) | 10 (21) | 10 (31) | 11 (35) |
| pry-1(mu38) 20°C | 21 (106) | 3.21 (14) | 0 (14) | ND | ND |
| pry-1(mu38); cbp-1(ku258) 20°C | 8 (127)e | 3.09 (22) | 5 (22) | ND | ND |
| pry-1(mu38) 15°C | 64 (108) | 3.96 (19) | 0 (19) | ND | ND |
| pry-1(mu38); cbp-1(ku258) 15°C | 19 (108)e | 3.18 (18) | 0 (18) | ND | ND |
| huls1 + HS | 66 (76) | 4.53 (15) | 0 (15) | ND | ND |
| cbp-1(ku258); huls1 + HS | 65 (78) | 4.32 (18) | 0 (18) | ND | ND |
| lin-15(n765 ts) | 100 (343) | 5.82 (24) | 0 (24) | 13 (32) | 3 (33) |
| cbp-1(ku258); lin-15(n765 ts) | 98 (229) | 5.67 (12) | 0 (12) | 19 (26) | 30 (37) |
Unless otherwise indicated, all strains were grown at 20°C. Phenotypic definitions are as defined in Table 1, footnotes c to g. See Gleason et al. (14) for a detailed description of huls1 and heat shock conditions used. The number of animals scored (n) is indicated in parentheses. ND, not determined.
gf indicates gain of function. lf indicates loss of function. ts indicates temperature sensitive. lin-1(n2515) was linked to unc-17(e113) in these strains, and sur-6(ku123 lf) was linked to unc-29(e403) in these strains. huIs1 contains a gain-of-function mutation in bar-1 under the control of a heat shock promoter (hs::delNTbar-1).
Abnormal morphology and location of the VPCs and their descendants made accurate scoring of induction difficult.
The presence of extra VPCs as well as an abnormal morphology and location of the VPCs and their descendents made accurate scoring of induction difficult.
Significantly different from the value for single-mutant controls (P < 0.01 by Fisher's exact test).
We wanted to know whether cbp-1(ku258) was capable of enhancing the vulval defects caused by a gain-of-function mutation in lin-1 or loss-of-function mutations in genes that positively regulate Ras-MAPK signaling. lin-1(n2515) is a gain-of-function mutation that is thought to result from an inability of LIN-1 to be negatively regulated by MPK-1 phosphorylation and causes a weak VPC underinduction phenotype (22). cbp-1(ku258); lin-1(n2515) double mutants display a significant enhancement of the vulval underinduction defect (2.43 VPCs induced, 56% Vul) compared to the phenotypes of either cbp-1(ku258) (2.94 VPCs induced, 8% Vul) or lin-1(n2515) (2.97 VPCs induced, 4% Vul) alone (Table 2). This enhancement of lin-1(n2515) is not seen with other mutations, such as loss-of-function alleles of eor-1 and eor-2 that positively regulate the transcriptional outcome of the Ras-MAPK pathway in the vulva (20), suggesting that lin-1(n2515) and cbp-1(ku258) might be functioning synergistically at the same point in the pathway to negatively regulate induction. In support of this, cbp-1(ku258) was able to increase the induction defects and Vul percentage of a sur-6(ku123) (38) mutation only in an additive manner (Table 2).
cbp-1(ku258) was unable to suppress the Muv phenotype of a loss-of-function lin-31(n301) mutant; however, there was an increase in the number of Vul and Egl animals in the lin-31(n301); cbp-1(ku258) strain (Table 2). Although LIN-1 and LIN-31 are thought to function together to negatively regulate induction, it has previously been shown that loss of lin-31 is epistatic to a lin-1 gain-of-function allele (22). The above data may suggest that cbp-1 is genetically functioning parallel to or downstream of the transcriptional component lin-1 but independently of lin-31. This placement in the pathway correlates well with the known transcriptional coactivator role of CBP/p300 family members.
cbp-1(ku258) does not appear to be altering vulval induction primarily through the Wnt or SynMuv pathway.
Several studies of both Drosophila and C. elegans have demonstrated a role for CBP/p300 in antagonizing Wnt signaling (13, 29, 45). In C. elegans, Wnt signaling is required to maintain the VPCs in an unfused and competent state as well as to promote induction of the vulval cell fate (9, 10, 14, 18). These two functions of Wnt signaling are accomplished in part by the upregulation of the Hox gene LIN-39 (10). We sought to determine whether cbp-1(ku258) was functioning to inhibit Wnt signaling in the VPCs.
The ajm-1::gfp reporter can be used to visualize adherens junctions of epithelial cells, such as the VPCs, and thus determine whether or not they have fused with the surrounding epidermal syncytium (10, 32). If Wnt signaling were compromised, ectopic fusion events can be seen in the VPCs which normally remain unfused (10). Using this marker, we did not detect any ectopic VPC fusion events in the cbp-1(ku258) versus wild-type control animals (data not shown). These data indicate that Wnt signaling is not overtly compromised in the VPCs of cbp-1(ku258) animals, and it is unlikely that cbp-1(ku258) suppression of activated let-60 ras is a secondary consequence of ectopic VPC fusion.
To further investigate the possibility that cbp-1(ku258) disrupts Wnt signaling, we made double-mutant strains with cbp-1(ku258) and mutations in two genes that lead to Wnt pathway hyperactivation and a Muv phenotype. The first Wnt pathway component we examined was the Axin-like protein PRY-1, a negative regulator of the Wnt pathway in C. elegans (27). Mutations in pry-1 result in overinduction of the VPCs and a Muv phenotype (14, 27). The loss of function of components of the Ras pathway is unable to dramatically suppress this Muv phenotype, whereas the loss of function of Wnt pathway components can suppress this Muv phenotype. cbp-1(ku258) was only weakly, yet significantly, able to suppress the pry-1(mu38) Muv phenotype (Table 2). We also examined the ability of cbp-1(ku258) to suppress the Muv phenotype caused by a gain-of-function mutation in bar-1 under the control of a heat shock promoter (14). cbp-1(ku258) was unable to suppress the Muv phenotype of this bar-1 mutation following heat shock (Table 2). These genetic data in conjunction with the absence of ectopic VPC fusion support a model in which CBP-1 functions primarily downstream of Ras signaling but either does not or only partially functions to regulate Wnt signaling in the VPCs.
The Synthetic Multivulva (SynMuv) phenotype caused by mutation in the dual class SynMuv gene lin-15 has been shown to be at least partially dependent on the level of Ras signaling (11). Interestingly, cbp-1(ku258) is unable to substantially suppress the SynMuv phenotype of a lin-15(n765 ts) mutation at 20°C (Table 2). However, this SynMuv phenotype is highly penetrant, and other genes capable of suppressing the Muv phenotype of let-60(n1046) to near wild-type levels, such as ksr-1, have also been shown to have a weak effect on the SynMuv phenotype of lin-15(n765 ts) animals at 20°C (42). cbp-1(ku258)/lin-15(n765 ts) animals did display a high degree of lethality and were in general much sicker than other cbp-1(ku258) double-mutant strains (44% larval lethal; n = 69).
CBP-1 localizes to the nuclei of the vulval precursor cells.
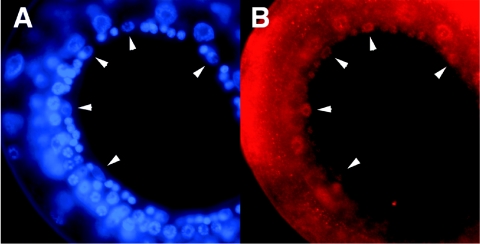
As one would expect for a transcriptional coactivator, it has previously been reported that CBP-1 localizes to the nuclei of embryonic cells (36). Using an antibody specific for CBP-1 (44), we stained C. elegans larvae to determine the expression pattern and subcellular localization of CBP-1 in larval-stage worms. CBP-1 was predominantly localized to the nuclei of most if not all somatic cells. Importantly, CBP-1 was found in the nuclei of the VPCs during the late L2 larval stage which corresponds to the time of let-60 Ras signaling and vulval induction (Fig. 3). These data are consistent with a model in which cbp-1(ku258) suppresses the Ras pathway at the transcriptional level.
FIG. 3.
Immunohistochemistry of CBP-1. Larval-stage worms were stained using a polyclonal rabbit antibody specific for CBP-1. Pictured is an L2 larval-stage worm which showed CBP-1 staining in the nuclei of the vulval precursor cells situated at the ventral surface of the animal (arrows point to VPC nuclei) (B). DAPI staining served as a positive control for the identification of nuclei (A).
CBP-1 (ku258) has increased histone acetyltransferase activity compared to wild-type CBP-1.
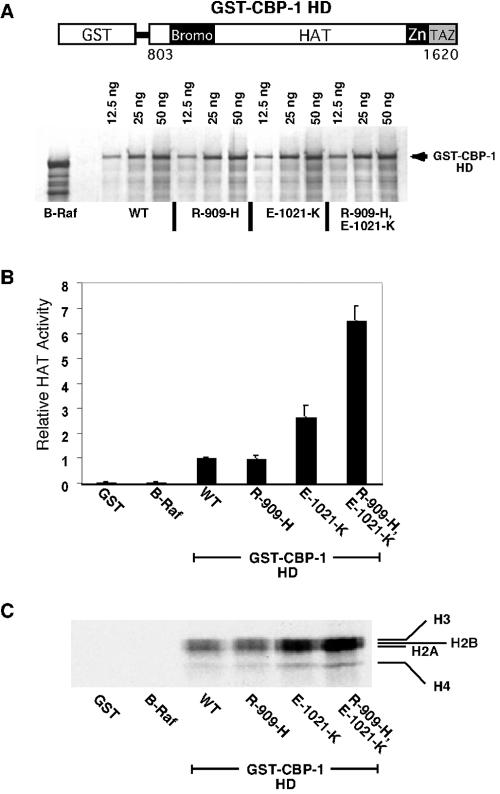
p300/CBP proteins are known histone acetyltransferases, and it has been reported that the C. elegans CBP-1 HAT domain can also acetylate histones in an in vitro assay (44). Since one of the two mutations in cbp-1(ku258) lies within the minimal HAT domain of CBP-1, we investigated the possibility of histone acetyltransferase activity being altered in the mutant protein. GST-CBP-1 fusion proteins containing the bromodomain and HAT domain region (amino acids 803 to 1620) (Fig. 4A) were expressed and purified from mammalian HEK 293 cells. The GST-CBP-1 fusion proteins were used in HAT assays after the levels of protein input were normalized by silver staining (Fig. 4A) and anti-GST Western blotting (data not shown). GST-CBP-1 containing the E-1021-K mutation alone and GST-CBP-1 containing both the R-909-H and E-1021-K mutations both displayed significantly higher HAT activity, 2.5-fold and 6.5-fold, respectively, than wild-type GST-CBP-1 did (Fig. 4B and C). Although the R-909-H mutation showed no increase in activity on its own, it appears that it is able to act synergistically with the E-1021-K mutation to produce a higher level of HAT activity than with E-1021-K alone. The HAT activity seen in this assay was specific for histones, as no detectable levels of autoacetylation or acetylation of a bovine serum albumin control protein were seen for any of the fusion proteins (data not shown). We propose that there are constitutively high levels of CBP-1 HAT activity in cbp-1(ku258) animals and it is this activity that is responsible for the suppression of let-60(n1046) ras as well as the other defects seen in these animals.
FIG. 4.
CBP-1 histone acetyltransferase assay. (A) GST-CBP-1 HD fusion protein constructs were made using amino acid residues 803 to 1620 (this region contains the entire bromodomain and HAT domain). Combinations of mutations corresponding to those seen in cbp-1(ku258) animals were introduced into the constructs. Fusion proteins were purified from HEK 293 cells and quantified via silver staining. WT, wild type. (B) In histone acetyltransferase assays, the fusion proteins carrying the E-1021-K mutation in the HAT domain displayed significantly higher HAT activity than did the wild-type (WT) protein or the protein with the R-909-H mutation alone. GST-CBP-1 HD carrying both mutations present in cbp-1(ku258) displayed the highest level of HAT activity, which was nearly sevenfold higher than wild-type GST-CBP-1 HD. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of histone acetyltransferase reactions followed by autoradiography demonstrated that the GST-CBP-1 HD fusion proteins were specifically able to acetylate purified mammalian histones.
DISCUSSION
In this study, we analyzed the functions of the C. elegans CBP/p300 homolog by isolating and characterizing a semidominant suppressor of activated ras. Epistasis analysis suggests that cbp-1 likely acts in concert with the Ets family transcription factor gene lin-1 in regulating vulval induction. The suppression of the activated ras phenotype was caused by two closely linked point mutations in the C. elegans p300/CBP homolog, cbp-1. Furthermore, the cbp-1(ku258) mutation results in an increase in histone acetyltransferase activity over wild-type CBP-1 in an in vitro histone acetylation assay. Thus, we have identified a role for cbp-1 as a negative regulator of Ras signaling in the vulva.
The overwhelming majority of literature analyzing CBP/p300 family members has proposed that they possess a tumor suppressor-like function. Reminiscent of what is seen for p300/CBP in other systems, loss of cbp-1 in the embryo results in a hyperproliferation defect and failure of many embryonic tissues to differentiate (36). The resultant 100% embryonic lethality has made a comprehensive analysis of the roles of cbp-1 in various signaling pathways difficult to assess. Although we can't completely exclude the possibility of a neomorphic function introduced by the ku258 allele, our genetic data argue that we have isolated a novel gain-of-function allele of cbp-1 that has allowed us to investigate its role in a postembryonic ras signaling event. The antagonism of cbp-1(ku258) towards Ras signaling in the VPCs suggests that CBP-1 functions in a tumor suppressor-like capacity in the VPCs as well.
Our data suggest that CBP-1 is functioning with tissue-specific regulators downstream of the RTK-Ras-MAPK vulval induction pathway. It has been proposed that the tissue-specific LIN-1/LIN-31 heterodimer inhibits vulval induction and keeps VPCs out of the cell cycle until MAPK activation, which results in the disruption of this dimer through phosphorylation (43). We show in this report that CBP-1 is likely functioning at the transcriptional level downstream of ras, possibly in concert with the Ets family member LIN-1. This finding makes CBP-1 an attractive candidate to function as a transcriptional coactivator of the LIN-1/LIN-31 heterodimer. p300/CBP are involved in the regulation of cyclin-dependent kinase inhibitors, such as p21/Cip1/WAF1 (15). In the VPCs, the p21/Cip1/WAF1-like cyclin-dependent kinase inhibitor, cki-1, functions to keep the VPCs out of the cell cycle until induction (19). One possible model involves CBP-1 functioning as a transcriptional coactivator for the tissue-specific LIN-1/LIN-31 heterodimer to promote the expression of cell cycle regulators such as cki-1. Although there is evidence for CBP/p300 functioning as transcriptional coactivators for Ets proteins in other systems (48), we were unable to detect an interaction between CBP-1 and LIN-1 or LIN-31 using in vitro binding assays (data not shown). CBP/p300 can acetylate a number of transcription factors (reviewed in reference 15), and it is not implausible to think that LIN-1 or LIN-31 could be targets for CBP-1 acetylation as well. Additionally, p300/CBP have been shown to undergo phosphorylation by both cyclin-dependent kinases and MAPKs; therefore, CBP-1 may be a direct target of MPK-1 in the VPCs, or its activity may be regulated by Ras-MAPK activity indirectly (1, 2, 24). The ku258 mutant CBP-1 protein may be defective in its ability to sense and properly respond to the activation of Ras signaling in the VPCs.
A number of in vitro and cell culture studies as well as recent work using Drosophila and C. elegans have highlighted the requirement of HAT activity for the proper function of CBP/p300 (15, 29, 44). However, other studies have shown that this intrinsic HAT activity is dispensable for CBP/p300-mediated transcription at some promoters. Our data show that the cbp-1(ku258) mutation produces a mutant protein with a nearly sevenfold increase in intrinsic HAT activity over the activity of wild-type CBP-1. This increase in HAT activity is likely responsible for the suppression of activated ras seen in the VPCs and provides a biochemical explanation for the gain-of-function nature of the cbp-1(ku258) allele. It is unlikely that the suppression of activated ras in the VPCs is caused by nonspecific effects of acetylation by CBP-1, since cbp-1(ku258) was unable to significantly suppress the Muv phenotypes caused by mutations in lin-31 and the Wnt pathway component bar-1. Previous studies on CBP/p300 in vitro have suggested that phosphorylation of CBP/p300 or interactions with other factors can lead to changes in the protein that also result in an increase in intrinsic HAT activity (1, 2). Interestingly, the change in CBP/p300 HAT activity seen in these experiments is an approximately six- to sevenfold increase. The authors of these studies suggest that the phosphorylation-dependent increase in CBP/p300 HAT activity is the result of a conformational change in the protein. Thus, the cbp-1(ku258) mutations could be mimicking a physiologically relevant conformational state of CBP-1 possessing higher HAT activity and reveal a connection between a Ras signaling event and the regulation of CBP-1 HAT activity in vivo. Not only do high levels of CBP-1 HAT activity appear deleterious to Ras signaling in the vulva, but the pleiotropic defects seen in cbp-1(ku258) animals suggest that the failure to maintain an equilibrium in the acetylation state driven by CBP/p300 HAT activity can have deleterious repercussions on a number of signaling pathways and on a global transcriptional level.
Acknowledgments
We thank K. Grant and E. Desjardins for help with the initial mapping of cbp-1(ku258), Y. Shi for the anti-CBP-1 antibody and strains, D. Eisenmann for the bar-1(gf) strain, A. Coulson at the Sanger Centre for cosmids, and Y. Kohara for cDNA clones. Additionally, we thank K. L. Guan and W. Wood for helpful discussions. Some strains used in this paper were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
This work was supported by an RO1 grant from the NIH (GM47869) to M.H., who is an Investigator of the Howard Hughes Medical Institute
REFERENCES
- 1.Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet, P. Robin, B. Rudkin, A. Harel-Bellan, and D. Trouche. 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. [DOI] [PubMed] [Google Scholar]
- 3.Aurandt, J., H. G. Vikis, J. S. Gutkind, N. Ahn, and K. L. Guan. 2002. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc. Natl. Acad. Sci. USA 99:12085-12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 5.Beitel, G. J., S. Tuck, I. Greenwald, and H. R. Horvitz. 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9:3149-3162. [DOI] [PubMed] [Google Scholar]
- 6.Bettinger, J. C., K. Lee, and A. E. Rougvie. 1996. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122:2517-2527. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 9.Eisenmann, D. M., and S. K. Kim. 2000. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics 156:1097-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenmann, D. M., J. N. Maloof, J. S. Simske, C. Kenyon, and S. K. Kim. 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125:3667-3680. [DOI] [PubMed] [Google Scholar]
- 11.Fay, D. S., and M. Han. 2000. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26:279-284. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, E. L., and H. R. Horvitz. 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110:17-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gay, F., D. Calvo, M. C. Lo, J. Ceron, M. Maduro, R. Lin, and Y. Shi. 2003. Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev. 17:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gleason, J. E., H. C. Korswagen, and D. M. Eisenmann. 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16:1281-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 16.Han, M., A. Golden, Y. Han, and P. W. Sternberg. 1993. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature 363:133-140. [DOI] [PubMed] [Google Scholar]
- 17.Hanna-Rose, W., and M. Han. 1999. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development 126:169-179. [DOI] [PubMed] [Google Scholar]
- 18.Hoier, E. F., W. A. Mohler, S. K. Kim, and A. Hajnal. 2000. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 14:874-886. [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, Y., R. Roy, and V. Ambros. 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125:3585-3597. [DOI] [PubMed] [Google Scholar]
- 20.Howard, R. M., and M. V. Sundaram. 2002. C. elegans EOR-1/PLZF and EOR-2 positively regulate Ras and Wnt signaling and function redundantly with LIN-25 and the SUR-2 mediator component. Genes Dev. 16:1815-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu, V., C. L. Zobel, E. J. Lambie, T. Schedl, and K. Kornfeld. 2002. Caenorhabditis elegans lin-45 raf is essential for larval viability, fertility and the induction of vulval cell fates. Genetics 160:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, D., G. J. Beitel, S. G. Clark, H. R. Horvitz, and K. Kornfeld. 1998. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics 149:1809-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubowski, J., and K. Kornfeld. 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janknecht, R., and T. Hunter. 1996. Versatile molecular glue. Transcriptional control. Curr. Biol. 6:951-954. [DOI] [PubMed] [Google Scholar]
- 25.Koch, R., H. G. van Luenen, M. van der Horst, K. L. Thijssen, and R. H. Plasterk. 2000. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 10:1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornfeld, K. 1997. Vulval development in Caenorhabditis elegans. Trends Genet. 13:55-61. [DOI] [PubMed] [Google Scholar]
- 27.Korswagen, H. C., D. Y. Coudreuse, M. C. Betist, S. van de Water, D. Zivkovic, and H. C. Clevers. 2002. The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 16:1291-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 29.Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman, and S. M. Smolik. 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22:3832-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, L. M., M. E. Gallegos, B. A. Morisseau, and S. K. Kim. 1993. lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev. 7:933-947. [DOI] [PubMed] [Google Scholar]
- 31.Miller, R. W., and J. H. Rubinstein. 1995. Tumors in Rubinstein-Taybi syndrome. Am. J. Med. Genet. 56:112-115. [DOI] [PubMed] [Google Scholar]
- 32.Mohler, W. A., J. S. Simske, E. M. Williams-Masson, J. D. Hardin, and J. G. White. 1998. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 8:1087-1090. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 34.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riddle, D. L., T. Blumental, B. J. Meyer, and J. R. Priess. 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 36.Shi, Y., and C. Mello. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sieburth, D. S., Q. Sun, and M. Han. 1998. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell 94:119-130. [DOI] [PubMed] [Google Scholar]
- 38.Sieburth, D. S., M. Sundaram, R. M. Howard, and M. Han. 1999. A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 13:2562-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternberg, P. W., and M. Han. 1998. Genetics of RAS signaling in C. elegans. Trends Genet. 14:466-472. [DOI] [PubMed] [Google Scholar]
- 40.Sternberg, P. W., and H. R. Horvitz. 1986. Pattern formation during vulval development in C. elegans. Cell 44:761-772. [DOI] [PubMed] [Google Scholar]
- 41.Sulston, J. E., and H. R. Horvitz. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110-156. [DOI] [PubMed] [Google Scholar]
- 42.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 43.Tan, P. B., M. R. Lackner, and S. K. Kim. 1998. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93:569-580. [DOI] [PubMed] [Google Scholar]
- 44.Victor, M., Y. Bei, F. Gay, D. Calvo, C. Mello, and Y. Shi. 2002. HAT activity is essential for CBP-1-dependent transcription and differentiation in Caenorhabditis elegans. EMBO Rep. 3:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waltzer, L., and M. Bienz. 1998. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395:521-525. [DOI] [PubMed] [Google Scholar]
- 46.Wassarman, D. A., M. Therrien, and G. M. Rubin. 1995. The Ras signaling pathway in Drosophila. Curr. Opin. Genet. Dev. 5:44-50. [DOI] [PubMed] [Google Scholar]
- 47.Wu, Y., and M. Han. 1994. Suppression of activated Let-60 ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev. 8:147-159. [DOI] [PubMed] [Google Scholar]
- 48.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]