Abstract
Set2p, which mediates histone H3 lysine 36 dimethylation (H3K36me2) in Saccharomyces cerevisiae, has been shown to associate with RNA polymerase II (RNAP II) at individual loci. Here, chromatin immunoprecipitation-microarray experiments normalized to general nucleosome occupancy reveal that nucleosomes within open reading frames (ORFs) and downstream noncoding chromatin were highly dimethylated at H3K36 and that Set2p activity begins at a stereotypic distance from the initiation of transcription genome-wide. H3K36me2 is scarce in regions upstream of divergently transcribed genes, telomeres, silenced mating loci, and regions transcribed by RNA polymerase III, providing evidence that the enzymatic activity of Set2p is restricted to its association with RNAP II. The presence of H3K36me2 within ORFs correlated with the “on” or “off” state of transcription, but the degree of H3K36 dimethylation within ORFs did not correlate with transcription frequency. This provides evidence that H3K36me2 is established during the initial instances of gene transcription, with subsequent transcription having at most a maintenance role. Accordingly, newly activated genes acquire H3K36me2 in a manner that does not correlate with gene transcript levels. Finally, nucleosomes dimethylated at H3K36 appear to be refractory to loss from highly transcribed chromatin. Thus, H3K36me2, which is highly conserved throughout eukaryotic evolution, provides a stable molecular mechanism for establishing chromatin context throughout the genome by distinguishing potential regulatory regions from transcribed chromatin.
In eukaryotic cells, the accessibility of the DNA template is influenced by chromatin structure. For example, in Saccharomyces cerevisiae, transcription factors have been shown to bind to consensus sequences upstream of genes in preference to identical consensus sequences that occur within the coding sequences of transcribed genes (24, 31). Likewise, transposons preferentially insert into promoter regions (33), and the double-strand breaks required for meiotic recombination in S. cerevisiae occur preferentially in gene promoters rather than in the coding regions (12, 55). Chromatin context therefore is a major determinant of where on the genomic DNA template many biological phenomena will occur.
Regulation of accessibility to the DNA template is likely to be mediated in large part through differential regulation of nucleosome occupancy. Promoter regions of S. cerevisiae exhibit reduced nucleosome occupancy genome-wide (2, 26), and these differences in nucleosome occupancy are important for promoter accessibility (47). Furthermore, in S. cerevisiae, promoter and nonregulatory chromatin can be biochemically fractionated, indicating that those regions have distinct physical properties (36). Nucleosomes can be moved or displaced from specific genomic regions by several general mechanisms, including nucleosome-remodeling complexes like SWI/SNF and RSC (34), binding of activators to DNA (3, 4, 35), transcriptional elongation by RNA polymerase II (RNAP II) (20, 26, 46), and inherent properties of DNA sequence (47). Template accessibility and nucleosome occupancy can also be mediated by posttranslational modification of the N-terminal histone tails, most notably acetylation (27, 48). Although chromatin context may be defined in part by regional differences in histone modifications, no chromatin mark has been shown to correspond specifically to coding or regulatory regions throughout the genome. Here, we present evidence that dimethylation of histone H3 at lysine 36 (H3K36me2), which is mediated by the methyltransferase Set2p (25, 51), may provide such a mark.
Set2p interacts with the C-terminal domain (CTD) of RNAP II (22, 29, 56), and this interaction is regulated by the phosphorylation state of the CTD. Serine 5 (Ser5) of the CTD repeat is phosphorylated by Kin28p during initiation of transcription, while serine 2 (Ser2) and Ser5 are phosphorylated by Ctk1p during elongation (8, 17, 19, 30). Set2p associates preferentially with Ser2/Ser5 phosphorylated repeats of the RNAP II CTD, and deletion of CTK1 abolishes H3K36me2 (22, 56). Set2p-RNAP II interactions are also dependent on the Paf1 complex (Paf1p, Rtf1p, Cdc73p, Ctr9p, and Leo1p) (22), which also associates with RNAP II (21, 40, 49). This and other biochemical data suggest that Set2p associates with RNAP II specifically during transcription elongation (18, 22, 29, 45, 56). Chromatin immunoprecipitation (ChIP) assays followed by quantitative PCR on a few selected loci have supported this assertion, showing that H3K36me2 is generally restricted to the transcribed regions of RNAP II-regulated genes (1, 18, 22, 45).
While there is strong evidence that Set2p is associated with elongating polymerase, the physiological functions of Set2p and H3K36me2 are still unknown. Evidence suggesting a function for Set2p in transcriptional elongation comes from results showing either sensitivity or resistance of set2Δ strains to the elongation inhibitor 6-azauracil. These phenotypes are similar to those exhibited by strains defective for genes encoding elongation factors like Chd1p, Iswi1p, and Fkh1p (22, 28, 29, 45, 57). A role in transcriptional elongation is also supported by synthetic genetic interactions between set2Δ and deletions of all members of the Paf1 complex, the chromodomain factor Chd1p, a putative elongation factor Soh1p, and the Bre1p or Lge1p components of histone H2B ubiquitination complex (22). However, whatever role Set2p plays in elongation is either not essential or redundant, since set2Δ strains are viable and, in many backgrounds, exhibit very mild phenotypes.
To further elucidate the cellular function of H3K36me2, we determined its pattern of distribution throughout the S. cerevisiae genome. We performed additional experiments to determine how the pattern of H3K36me2 changes in response to a change in global transcriptional state and the relationship between the H3K36me2 mark and nucleosome stability. H3K36me2 demarcates the structurally distinct regulatory and nonregulatory regions of yeast genomic chromatin and may serve as an indicator of chromatin context.
MATERIALS AND METHODS
Nomenclature.
Throughout the paper, we use a recently proposed uniform histone modification nomenclature (53). Thus, for example, “H3K36” refers to histone H3 lysine at residue 36, and “H3K36me2” refers to dimethylation of that residue.
Strains and culture conditions.
For H3K36me2 and histone H3 ChIPs, strain AS4 (MATα trp1-1 arg4-17 tyr7-1 ade6 ura3) was used (50). For histone H4 ChIPs, a previously described myc-tagged H4 strain constructed in strain UCC1111 [MATα ade2::his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2 pRS412 (ADE2 CEN ARS)-HHF2-HHT2] was used (37, 38). Unless otherwise described, yeast was grown to an optical density of 0.8 to 1.0 at 600 nm with shaking at 30°C in 100 ml of yeast extract-peptone-dextrose media (1% yeast extract, 2% peptone, 2% dextrose).
Antibodies.
Antibodies against histone H3 lysine 36 dimethylation have been described previously (51) and were derived from Upstate (catalog no. 07-369). myc-tagged antibodies were also obtained from Upstate (catalog no. 05-419). The rabbit histone H3 antiserum was obtained from Abcam, Inc. (AB1791), and was raised in rabbits using a peptide corresponding to amino acids 124 to 135 (CGIQLARRIRGERA) of human histone H3.
Dot blot.
Peptides (KSAPSTGGVKKPHRYKPGTGK-BIOTIN) in which the residue corresponding to H3K36 (underlined) was either mono-, di-, or trimethylated were resuspended in double-distilled H2O (10 μg/μl) and serially diluted in Tris-buffered saline (TBS) (150 mM NaCl, 10 mM Tris, pH 7.6). Aliquots of 100-μl peptide-TBS solution were spotted onto polyvinylidene difluoride membranes by using a Bio-Rad dot blot apparatus. Membranes were washed in TBS and then blocked in a solution of 2.5% (wt/vol) Carnation nonfat dry milk in TBS-Tween 20 (0.1% Tween 20 in TBS) for 10 min prior to incubation with a 1:10,000 dilution of the specified antibody for 2 h at room temperature. Membranes were washed with TBS-Tween 20 for 10 min three times, incubated with anti-rabbit horseradish peroxidase-conjugated immunoglobulin G for 2 h at room temperature, and then washed again for 10 min three times prior to detection using ECL-Plus from Amersham.
ChIP assays.
ChIP assays were performed as described previously (23). Briefly, whole-cell extracts were prepared from 1% formaldehyde-fixed wild-type and set2Δ cells by using lysis buffer (50 mM HEPES-KOH, pH 7.5, 300 mM NaCl, 1 mM EDTA, 1% Triton X, and 0.1% sodium deoxycholate) and sonicated to shear the chromatin (0.25- to 1-kb range). Immunoprecipitation was performed with anti-H3K36me2, anti-myc, or anti-H3. After cross-link reversal at 65°C, DNA was extracted by using a QIAGEN PCR purification kit according to the manufacturer's instructions.
DNA amplification, labeling, array hybridization, and data processing.
ChIP-enriched DNA and reference DNA in all experiments were amplified as described previously (5). Briefly, two initial rounds of DNA synthesis with T7 DNA polymerase using primer 1 (5′-GTTTCCCAGTCACGATCNNNNNNNNN-3′) were followed by 25 cycles of PCR with primer 2 (5′-GTTTCCCAGTCACGATC-3′). Cy3-dUTP or Cy5-dUTP was then incorporated directly with an additional 25 cycles of PCR using primer 2. Microarray hybridizations were performed using standard procedures (16). The arrays were scanned with a GenePix 4000 scanner, and data were extracted with Genepix 5.0 software. Data were normalized such that the median log2 ratio value for all quality elements on each array equaled zero, and the median of pixel ratio values was retrieved for each spot. Only spots of high quality by visual inspection, with at least 50 pixels of quality data (regression R2 of >0.6) and for which intensity of the reference signal was strong (>350 U), were used for analysis. Arrayed elements that did not meet these criteria on at least half of the arrays were excluded from analysis. All data were log transformed before further analysis. For normalization with the nucleosome occupancy data, the median log2 ratio values of H4-myc ChIP were subtracted from the median H3K36me2-ChIP ratio values. Unless otherwise noted, all data presented are nucleosome occupancy normalized in this way. While many methods of bulk nucleosome normalization are possible, all must contend with the inherent difficulties of combining ChIP data sets produced with two different antibodies (6). The method used here is simplest and provides a more realistic representation of the modification pattern than do unnormalized data. We provide all raw data (see below) so that readers may apply their preferred normalization method.
DNA microarray preparation.
Open reading frames (ORFs) and intergenic regions from yeast (S288C) were PCR amplified and printed on polylysine-coated glass slides by using a robotic arrayer as described previously (16). ORFs were generally represented by PCR products that extended from start codon to stop codon. Elements representing intergenic regions generally included all DNA between annotated ORFs, with the fragments divided such that PCR products were no longer than 1.5 kb.
Locus-specific detection of ChIP enrichment.
The sequences of the primers used (see Fig. 2 and 5) are shown in Table 1.
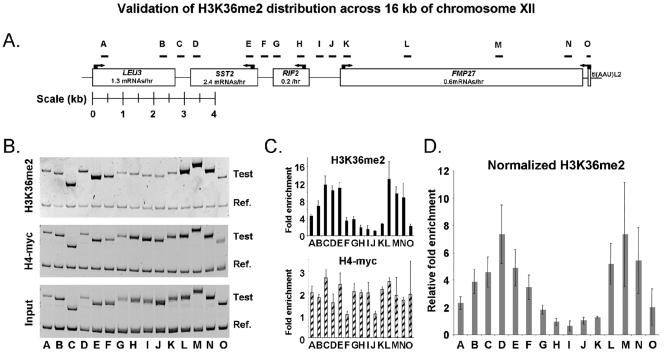
FIG. 2.
Validation and fine-scale mapping of H3K36me2 distribution across 16 kb of chromosome XII. (A) Schematic of chromosome XII coordinates 1,036,089 to 1,052,141, showing the locations of primers used to interrogate three of the independent H3K36me2 ChIP-chips shown in Fig. 1. Primer sets are listed in Table 1. (B) Polyacrylamide gel analysis of PCR products generated by the primer sets shown in panel A (Test). The reference product (Ref.) corresponds to 146 bp of a large noncoding region between YEL073C and YEL072W on chromosome V. (C) Quantitation of the gel shown in panel B. Graphs show the average enrichments detected by the PCR primer sets following H3K36me2 and histone H4-myc ChIPs. The value plotted for each fragment was calculated as follows: [(w/x)/(y/z)], where all values are the sum of pixel intensities for each band. w, ChIP fragment; x, ChIP reference; y, input test fragment; z, input reference. Numbers therefore reflect relative immunopreciptation efficiencies; values of less than 1 may be expected. Error bars illustrate the average deviations from the means. (D) H3K36me2 enrichment values after normalization with general nucleosome occupancy levels. The values plotted were calculated as follows: [(w/x)/ (y/z)]H3K36me2/[(w/x)/(y/z)]H4-myc.
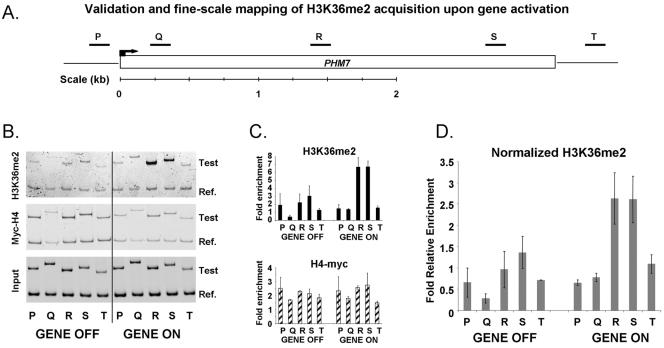
FIG. 5.
Validation and fine-scale mapping of H3K36me2 acquisition upon gene activation. (A) Schematic of the PHM7 locus, showing the locations of primers used to interrogate three of the independent H3K36me2 ChIP-chips shown in Fig. 4B, before and after heat shock. Primer sets are listed in Table 1. (B) Polyacrylamide gel analysis of PCR products generated by the primer sets shown in panel A (Test). The reference product (Ref.) is the same as that used for Fig. 2. (C) Quantitation of the gel shown in panel B. Graphs show the average enrichments detected by the PCR primer sets following H3K36me2 and histone H4-myc ChIPs, both before and after heat shock. The value plotted for each fragment was calculated as described in the legend for Fig. 2. (D) H3K36me2 enrichment values before and after heat shock after normalization with general nucleosome occupancy levels, calculated as described in the legend for Fig. 2. H3K36me2 levels increase after heat shock. Note that the increase is confined primarily to areas R, S, and T, which represent the center and 3′ end of the gene, but not areas P and Q, which represent, respectively, the promoter and coding sequences at the 5′ end.
TABLE 1.
| Set | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| A | GGAGCCATGCACTAAGTGTG | CGTCTCAAAAGCACTCTTCCT |
| B | TAAGGCATCCAGAAAAGAAAGC | CAGAGGTTCATTGACAGCTTCAG |
| C | CTGCGGTGCTACAGGAGAGGTAA | CGTGAGCGCTTACGAATCTTC |
| D | CGTTAGGTGAACCACCTGAAGTG | CCTGAGGACCTTCAAAGTTGC |
| E | CACCTAAGGGAGTTGCTGTTTTAC | GGCAAGGTAATGGGTCATCTG |
| F | GAAGAAGCCATGAAGTATGCGAAG | GGACGATGAGAAGTAAAGACAGAG |
| G | CAGTCTTACAACTCGCTTGTCAC | GTGGAAGACGCTGGTTTTTC |
| H | GTGTTAGCAGCCCAAAGGTG | CTGAAACTGCACGGTCGATAC |
| I | CCACGTTCGTATTTATATCTATATTCG | CCGTTCTTGCTGACATGGTTTTC |
| J | CCTATTTTTTTGGCTTGTAACTCAAAC | CTGTCCTTTGTAATAGCTACCGTTC |
| K | CCGATTAATGTTCTACTGTACAAGTGG | CATTAGTAGATGATTTCTTGGAATCA |
| L | GGTGCTAGGTCGGTTTTTATG | CGGCAATACTGGACTTATGGTTAG |
| M | CGCAGATGTCTCACCATTATTC | GATGTCCACGTCCTGGCATAC |
| N | GCCGTTGCAAAGCTAAACTTTC | GTGGTCTCAACGTTTTTAACAACC |
| O | GTAGCATCATACATATCAATATGTTACCA | CATGGTCTCTTGGCCCAGTTG |
| P | CGTTAATCTCCCTGAAATACTTTAAATAC | CGCTGCTTTGAATATGATGTGC |
| Q | GGAGGAAGAGAGAACGGAACC | GTTGGTAACATTTGAAAATGATAGCAG |
| R | CTTCCTGATGGGTGTCATTACT | GTCAATAATCGAGTCAACGGTG |
| S | CGATGTTGAGGCATTGAGAAATAAG | CTCATCCTGACACTGTCGAAAG |
| T | CCGCATATCTGTGAGCTTGG | GATAGACACTCAACCTATAGGAACTAGTTTC |
Availability of data.
All raw microarray data and images are available to the public through the UNC microarray database (https://genome.unc.edu/). Data are also available in Table S2 of the supplemental material and through GEO (accession number GSE 2991).
RESULTS
Chromatin from ORFs and regions immediately downstream of ORFs is enriched for histone H3 lysine 36 dimethylation.
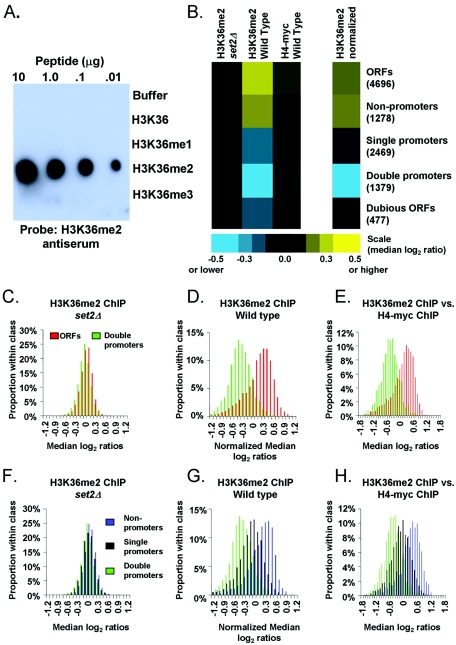
As the initial step in our goal to determine the genome-wide location of H3K36me2 in S. cerevisiae, we characterized a polyclonal antibody directed against H3K36me2. While the general specificity of this antibody for methylation at H3K36 had been previously verified (18, 57), its precise specificities to the different possible H3K36 methylation states (mono-, di-, and trimethylation) were unknown. To determine the specificity of this antibody for H3K36 methylation, we performed dot blots against peptides that were either mono-, di-, or tri-methylated at the residue corresponding to H3K36 (see Materials and Methods). As shown in Fig. 1A, the antiserum was specific to dimethylation of H3K36 and did not cross-react with any of the related modifications.
FIG. 1.
H3K36me2 is restricted to RNAP II-transcribed regions genome-wide. (A) The indicated amounts (top) of the specified peptides (right) were blotted onto a polyvinylidene difluoride membrane and probed with a 1:10,000 dilution of the H3K36me2 antiserum. To verify the presence of the H3K36me1 and H3K36me3 peptides, antibodies specific to each of those peptides were used in parallel to probe identical blots (see Fig. S1) in the supplemental material). (B) Colors (scale at bottom) represent the median of ratios [log2(H3K36me2 ChIP signal intensity/normalized reference signal intensity)] recorded from all arrayed elements in the indicated Saccharomyces Genome Database functional class (numbers of arrayed elements in parentheses). Data were derived from 12 independent wild-type and 8 set2Δ H3K36me2 ChIP experiments (biological replicates). Intergenic regions are organized into the following three categories: double, upstream of two divergent genes; single, upstream of one gene; non, upstream of zero genes. (C) A histogram showing the distribution of H3K36me2 ChIP median log2 ratios in a set2Δ strain. Ratios in panels C to H were normalized to bulk nucleosome occupancy by subtracting median H4-myc log2 ratios (eight independent experiments, wild type) or anti-H3 ChIPs (five independent experiments, set2Δ) from median H3K36me2 ChIP log2 ratios (same 12 ChIPs as mentioned for panel B). (D) Same as panel C, but for a wild-type strain. (E) Same as panel D, but instead of computational normalization, plotted ratios were derived from direct hybridization of H3K36me2 ChIP versus H4-myc ChIP (four independent ChIP sets). Note that for panels C to E, a positive correlation between the ratios reported at double promoters and the ratios reported at adjacent ORFs was observed (data not shown). Therefore, any high ratios reported at double promoters may, at least in part, be attributed to high ratios in adjacent ORFs. This is likely due to the limited resolution of the ChIP-chip procedure and our microarrays. (F) Same as panel C, but for noncoding regions. (G) Same as panel D, but for noncoding regions. (H) Same as panel E, but for noncoding regions.
Having verified the specificity of this antiserum, ChIP experiments were performed using extract from wild-type strains. To assess the relative abundance of genomic fragments enriched by the ChIP, samples were RNase treated and DNA was amplified and labeled fluorescently. In parallel, total genomic DNA was prepared from input extract, RNase treated, amplified, and labeled with a different fluorescent marker. The two samples were then analyzed by comparative hybridization to DNA microarrays. The microarrays used in this study cover the entire yeast genome, including the coding and noncoding regions, at approximately 1-kb resolution (see Materials and Methods). H3K36me2 ChIP-microarray (ChIP-chip) experiments were performed with a total of 12 independent wild-type yeast cultures (see Table S1 in the supplemental material for details). As a control, ChIP-chip experiments were performed from each of eight independent extracts in which H3K36me2 was eliminated by deletion of the SET2 gene. We found that H3K36me2 ChIPs enriched chromatin corresponding to ORFs relative to chromatin from genomic regions upstream of genes (Fig. 1B).
In S. cerevisiae, the noncoding regions downstream of two convergently transcribed genes are almost always completely transcribed, often on both strands, by the converging polymerases (15). We found that these regions, which correspond to 3′ untranslated regions (UTRs), were enriched by H3K36me2 ChIPs at a level equal to or greater than the enrichment observed at ORFs (Fig. 1B). To confirm that our ChIPs were reflections of H3K36me2 levels, we performed ChIP experiments using extracts from set2Δ strains. Very little DNA was recovered from these ChIPs, and analysis of the DNA that was recovered revealed none of the specific patterns described above (Fig. 1B). We therefore interpret the efficiency of DNA recovery at each locus after H3K36me2 ChIP to reflect relative H3K36me2 levels. The evidence presented thus far supports the hypothesis that regions of the genome transcribed by RNAP II are enriched for H3K36me2.
Chromatin upstream of ORFs is H3K36me2 deficient.
In further support of the hypothesis that H3K36me2 is restricted to transcribed regions, the lowest levels of H3K36me2 were found in chromatin upstream of two divergently transcribed genes (“double promoters”), which is not expected to be transcribed by RNAP II (Fig. 1B). On the other hand, “single promoters” are expected to be partially transcribed since they contain the 3′ UTR of the upstream gene. As predicted, single promoters exhibit a level of enrichment lower than that observed for ORFs and 3′ UTRs but higher than that observed for double promoters (Fig. 1B). These experiments provide evidence that dimethylation of histone H3 at lysine 36 is absent from regions of the genome that are not transcribed by RNAP II.
The genomic pattern of H3 lysine 36 dimethylation persists after normalization for general nucleosome occupancy.
Nucleosome occupancy is generally lower in noncoding regions upstream of genes than in ORFs (2-4, 26, 43, 52). Thus, we wondered if the pattern we observed with the H3K36me2-specific antiserum was a reflection, at least in part, of general nucleosome occupancy. To ensure that our results were specific to the H3K36me2 modification, we normalized our H3K36me2 distribution data to general nucleosome occupancy. We prepared extracts from yeast cells in which the only source of histone H4 was tagged with the myc epitope and performed ChIP assays using anti-myc antibodies. Histone H4 ChIPs were performed on five independent yeast cultures. Consistent with published data (26), results of the histone occupancy ChIPs revealed that nucleosomes were more enriched in the coding region of genes than in intergenic regions (Fig. 1B). Indistinguishable results were obtained with nucleosome ChIP-chips using an antibody specific to the C terminus of histone H3 (data not shown). In parallel, H3K36me2 ChIPs were performed using the same extracts. Even without normalization, the qualitative differences between the distribution of H3K36me2 and general nucleosome occupancy indicated that the H3K36me2 pattern was indeed distinct. Specifically, noncoding regions downstream of convergently transcribed genes were enriched by the H3K36me2 ChIP at a level nearly equal to the enrichment observed at ORFs, whereas in histone H3 or H4 ChIPs, ORFs were more strongly enriched than 3′ UTRs (Fig. 1B).
For further data analysis, we chose the simplest possible normalization routine by subtracting the median log2 ratio values of the H4-myc ChIP-chip data from the median ratio values of H3K36me2 ChIP-chip data (see Materials and Methods). After normalization, the clear enrichment of transcribed genomic regions and corresponding depletion of regulatory regions of the genome persisted (Fig. 1C, D, F, and G). As a test of the validity of this normalization approach, we performed direct comparative hybridizations between DNA enriched by H3K36me2 ChIP and DNA enriched with H4-myc ChIP. The data obtained from direct comparative hybridizations were essentially identical to the computationally normalized H3K36me2 data (Fig. 1E and H).
Higher resolution, locus-specific ChIPs confirm that H3K36me2 is concentrated in chromatin within coding regions and at the 3′ end of genes.
In H3K36me2 ChIPs, chromatin downstream of convergently transcribed genes was by far the most highly enriched class of noncoding chromatin (Fig. 1G). This suggested that Set2p is active throughout the entire transcript length, providing a possible mechanism for distinguishing nonregulatory intergenic regions from promoters. To validate this observation, we interrogated our ChIP results with PCR primers that represent regulatory and transcribed chromatin across a 16-kb region on chromosome XII (Fig. 2). Again we found that the chromatin heavily enriched by H3K36me2 ChIPs corresponded to coding regions and to regions lying downstream of two convergently transcribed genes. For example, SST2 and LEU3 are both transcribed under the conditions assayed. Their 3′ UTRs are each about 450 bp in length (15) and are represented by the primer sets B, C, and D in Fig. 2. Chromatin covered by these primer sets is among the most heavily enriched in the tested region.
Levels of histone H3K36 dimethylation do not correlate with transcription frequency.
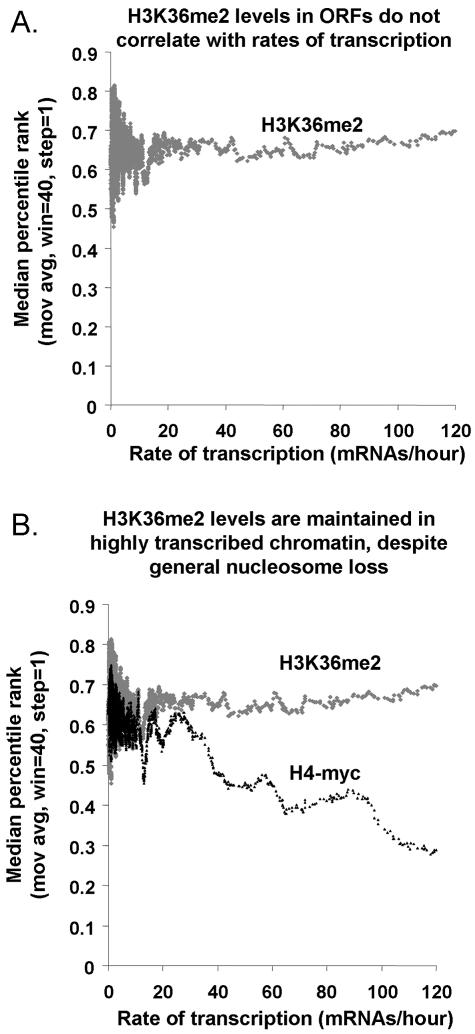
Localization of H3K36me2 to chromatin in the body of the RNAP II-transcribed genes is consistent with the earlier studies showing that Set2p associates with the elongating form of RNAP II. We wondered if the frequency of transcription correlated with the degree of H3K36 dimethylation. The transcription frequency (also called transcription rate) for each S. cerevisiae gene has been calculated based on measurements of steady-state RNA levels and RNA half-lives in exponentially growing yeast cells at 30°C (14). We compared these published transcription frequency values to the results of 12 independent H3K36me2 ChIP-chip experiments. We found that among genes with measurable transcription frequencies (>0 mRNA/hour), the level of H3K36me2 enrichment did not correlate with transcription frequency (Fig. 3A). Genes with transcription frequencies ranging from 1 to 120 mRNAs/hour were consistently enriched in the H3K36me2 ChIPs. For example, despite low rates of transcription, genes like BUD14 and TPK2 (both 1.8 mRNAs/hour) were enriched in H3K36me2 ChIPs (97th and 95th ChIP percentiles, respectively) as highly as were heavily transcribed genes like HXK2 (71 mRNAs/hour, 96th ChIP percentile). These results suggest that H3K36 dimethylation occurs chiefly in the initial instance or early instances of gene transcription, with subsequent transcription playing at most a maintenance role.
FIG. 3.
H3K36me2 levels in ORFs do not correlate with transcription frequencies. (A) Genes were sorted by their transcription rates during mitotic growth (x axis) (14), and a moving average (mov avg) (window [win] = 40, step = 1) of the degrees of their enrichment in H3K36me2 ChIPs was plotted (percentile rank, y axis). In this plot, H3K36me2 ChIP values are not normalized to overall nucleosome occupancy. (B) Same as panel A, except that both H3K36me2 ChIPs and H4-myc ChIPs are plotted.
H3K36me2 levels appear to compensate for transcription-coupled nucleosome loss.
Previous studies have shown that chromatin in very highly transcribed genes exhibits relatively low nucleosome occupancy, suggesting that nucleosomes are either removed or displaced temporarily on transcriptionally active chromatin (26, 46). However, we did not observe this phenomenon for nucleosomes that contain dimethylated H3K36. The levels of H3K36me2 appeared to remain constant on genes, irrespective of their rates of transcription (Fig. 3B). This suggests that the level of H3K36 dimethylation is maintained on highly transcribed chromatin either by preferential retention of nucleosomes containing H3K36me2 or by nonlinear increases in H3K36 methylation as a function of transcription rate. For the “nonlinear” hypothesis to be true, the H3K36me2 mark would have to be less than saturated at all genes, and increased enzymatic activity of Set2p would have to be linked to increased transcriptional activity for only the subset of very heavily transcribed genes at which bulk nucleosome loss has been observed. Since no correlation between transcription rate and H3K36me2 level is observed across a broad range of transcription rates (Fig. 3A), we favor the “preferential retention” hypothesis.
We then asked whether nucleosomes are less stable on highly transcribed chromatin in the absence of H3K36me2. We measured bulk nucleosome occupancy by performing histone H3 ChIPs in a set2Δ strain. However, we found that nucleosomes are lost from highly transcribed chromatin equally in set2Δ and wild-type strains, indicating that nucleosome occupancy is not directly affected by H3K36me2 (see Fig. S2 in the supplemental material). Therefore, additional mechanisms may work to stabilize H3K36me2 nucleosomes, or H3K36me2 may be an indicator, but not a cause, of transcription-stable nucleosomes.
H3K36 dimethylation correlates with “on” or “off” transcriptional state.
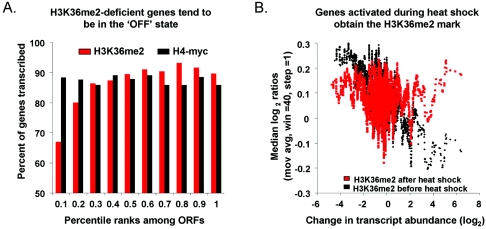
The hypothesis that H3K36 dimethylation is stable and occurs chiefly in the initial instance of gene transcription predicts no correlation between H3K36 dimethylation level and transcriptional rate (as we observed) but does predict a positive correlation between H3K36 dimethylation level and the on or off transcription state of a gene. We defined a gene as on if it had a measurable transcription rate (>0 mRNA/hour) (14) and off if it did not (0 mRNA/hour) (14). To test this prediction, we ranked the ORFs according to their enrichment levels in H3K36me2 ChIP experiments and divided them equally into 10 bins, such that the least enriched 10% of ORFs were in bin 1, the most enriched 10% were in bin 10, and so on. We then simply asked what proportion of genes in each bin was on (Fig. 4A). The results show that genes that were not enriched by H3K36me2 ChIPs were more likely than others to be off and that the likelihood of any given gene to be off decreased with increasing H3K36me2 ChIP enrichment. No such trend was observed with H4-myc ChIPs (Fig. 4A) or with H3K36me2 ChIPs performed with a set2Δ strain (data not shown). This result provides evidence that H3K36me2 is not linked with how often a gene is transcribed per se but rather with the occurrence of transcription.
FIG. 4.
H3K36me2 levels correlate with “on” or “off” transcriptional state. (A) ORFs were divided equally into 10 bins according to their degrees of enrichment in H3K36me2 ChIPs (normalized to H4-myc) or H4-myc ChIPs. The ORFs in each bin were then classified as either “on” (>0 mRNAs/hr) or “off” (0 mRNAs/hr). The percentage of genes in each bin that was classified as “on” is shown. We note that ORF size and transcription state are not independent variables, since shorter ORFs tend to be in the off state, even after removal of SGD-annotated dubious ORFs. However, analysis of ORFs of similar size rather than all ORFs reveals the same pattern shown here (Fig. S3 in the supplemental material). Note that ∼85% of genes are transcribed at detectable levels during mitotic growth (14, 15), so the uniform H4-myc data reflect a neutral relationship with transcription state. (B) Genes were sorted according to the degrees of their transcript level change upon heat shock (x axis) (10). A moving average (mov avg) (window [win] = 40, step = 1) of relative H3K36me2 ChIP enrichment (normalized to general histone H4 occupancy measured in parallel) at each ORF (y axis) is plotted before and after heat shock. Most genes are unchanged upon heat shock and are therefore clustered near the center of the graph. Points to the right represent ORFs that are activated by heat shock, and those to the left represent ORFs repressed by heat shock.
Dormant chromatin that becomes transcriptionally active acquires H3K36me2.
The hypothesis that H3K36 dimethylation is established by initial instances of gene transcription predicts that upon a switch from an inactive to an active state, chromatin will become dimethylated at H3K36. To test this prediction, we induced transcription at hundreds of genes simultaneously by subjecting yeast cells to transfer from 25°C to 37°C (7, 10). ORFs that become transcribed during heat shock acquire the H3K36me2 mark, while a relative decrease in H3K36me2 levels is observed in ORFs that are repressed (Fig. 4B). Of particular interest are the genes that were off during log phase but strongly induced after heat shock (measured expression increase of >log2 2). Of these genes, 75% (48/65) had increased levels of H3K36me2. In contrast, among genes that were off during log phase and remained off during heat shock (expression increase of <log2 1), only 47% (144 of 306) exhibited increased levels of H3K36me2. The difference between these groups was significant (χ2 test; P = 8.6 × 10−5).
We also observed a relative loss of H3K36me2 in the ORFs of repressed genes. Of genes that were on during log phase and that remained active (log2 expression ratios of >−1), only 46% (1,099/2,388) had increased H3K36me2 levels. In contrast, of genes that were on during log phase and repressed fourfold or more after heat shock, 65% (210/332) had decreased levels of H3K36me2 (χ2 test; P = 0.0014). By examining H3K36me2 ChIP data that were not normalized to nucleosome occupancy (data not shown), we found that this relative decrease is attributable in part to bulk nucleosome replenishment at repressed genes (26) rather than loss of H3K36me2 on existing nucleosomes.
To further confirm our ChIP-chip data, we performed quantitative ChIP analysis before and after heat shock along the length of PHM7 (Fig. 5A). PHM7 is repressed in logarithmically growing cell cultures (∼0 mRNA per hour) but is induced by a factor of 7 during heat shock (10). The results showed that this gene acquired the H3K36me2 mark after heat shock and only in the 3′ region of the ORF (primer sets R and S) (Fig. 5B to C). This result persisted after we normalized for bulk histone occupancy changes following heat shock (Fig. 5D).
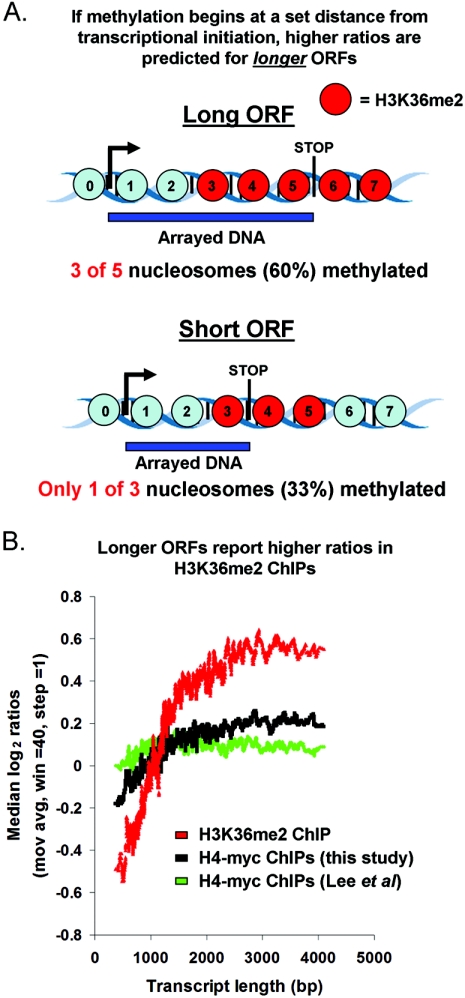
A positive correlation between gene length and measured H3K36me2 ChIP enrichment suggests that H3K36 dimethylation is initiated at a fixed distance from the start of transcription.
Previous studies at selected genes, including ADH1, PYK1, PMA1, and SCC2, have shown that dimethylation of H3K36 is initiated after transcriptional initiation, concomitant with association of Set2p with the elongating polymerase (1, 18, 22). If this mechanism operates genome-wide, and if the interval between transcriptional initiation and Set2p association is constant regardless of gene length, longer genes will appear to be enriched by our H3K36me2 ChIPs to a greater extent than shorter genes. The reason for this predicted relationship is illustrated in Fig. 6A and the corresponding legend and is a consequence of the fact that the DNA on our microarrays covers each ORF from the start codon to the stop codon, regardless of length.
FIG. 6.
Evidence that H3K36dimethylation begins at a set distance from the initiation of transcription genome-wide. (A) The hypothesis that H3K36 dimethylation begins at a determined and consistent distance from transcriptional initiation predicts that higher ratios will be reported for longer ORFs. Note that “input chromatin” is used as a reference for these experiments, and the null expectation is that the raw signal intensities in both channels will be proportional to ORF length, resulting in neutral ratios for all ORFs. The prediction of higher ratios for longer ORFs is based entirely on the relative proportion of the arrayed element that is dimethylated, not the absolute length of the arrayed element or genomic feature per se. For example, if the entire ORF were modified, or if the distance at which the modification began from transcriptional initiation were proportional to ORF length, equal ratios would be obtained for long and short ORFs. Blue circles, nucleosomes not dimethylated at H3K36. (B) ORFs were sorted by length, and moving averages (mov avg) (window [win] = 40, step = 1) of their ratios of enrichment in H3K36me2 ChIPs (not normalized to H4-myc) and H4-myc ChIPs (from this study and reference 26) were plotted.
To test this prediction, we plotted enrichment in H3K36me2 ChIPs against ORF length (Fig. 6B). This analysis showed that longer ORFs appeared to be more efficiently enriched in H3K36me2 ChIPs, consistent with the prediction made by the hypothesis described above (Fig. 6B). The “leveling off” observed at ∼2,000 bp is also a predicted feature of a mark that begins at a set distance from the transcriptional start, since the proportion of the gene that is not modified becomes smaller with increasing length. No such relationship between ORF length and enrichment is observed with H3K36me2 ChIPs performed from set2Δ extracts (data not shown). We did observe a weak relationship between ORF length and apparent bulk nucleosome occupancy for the H4-myc ChIPs performed in this study (the effect was even less pronounced in reference 26). This may be due to nucleosome loss very near to the site of transcriptional initiation, which would be predicted to have this effect. In any case, the magnitude of the relationship between size and length was much stronger for the H3K36me2 ChIPs, suggesting a defined boundary for the initiation of H3K36me2 inside the ORFs genome-wide. This conclusion is further supported by the H3K36me2 ChIP profile across 16 kb of chromosome XII (Fig. 2D). For example, primer set A is situated ∼154 bp from the LEU3 transcription start site and does not detect significant enrichment, whereas chromatin represented by primer set E is situated ∼400 bp after the start site of SST2 and is highly enriched. Likewise, for the relatively long FMP27 gene, primer set K at the 5′ end of the coding region does not report enrichment, but the downstream primer sets L, M, and N show steady enrichment of the chromatin at the 3′ end of the coding region.
In the course of this analysis, we noted that short genes, on the whole, tend to be more frequently transcribed than long ones. Therefore, we wondered whether the correlations between length and ratio reported here confounded the conclusions presented in Fig. 3. To test this possibility, only ORFs greater than 1,000 bp in length, which do not show a relationship between length and transcription frequency, were used in the same analysis shown in Fig. 3. The resulting plot was indistinguishable from the one presented (data not shown).
Evidence that H3K36 dimethylation ends upon termination of transcription.
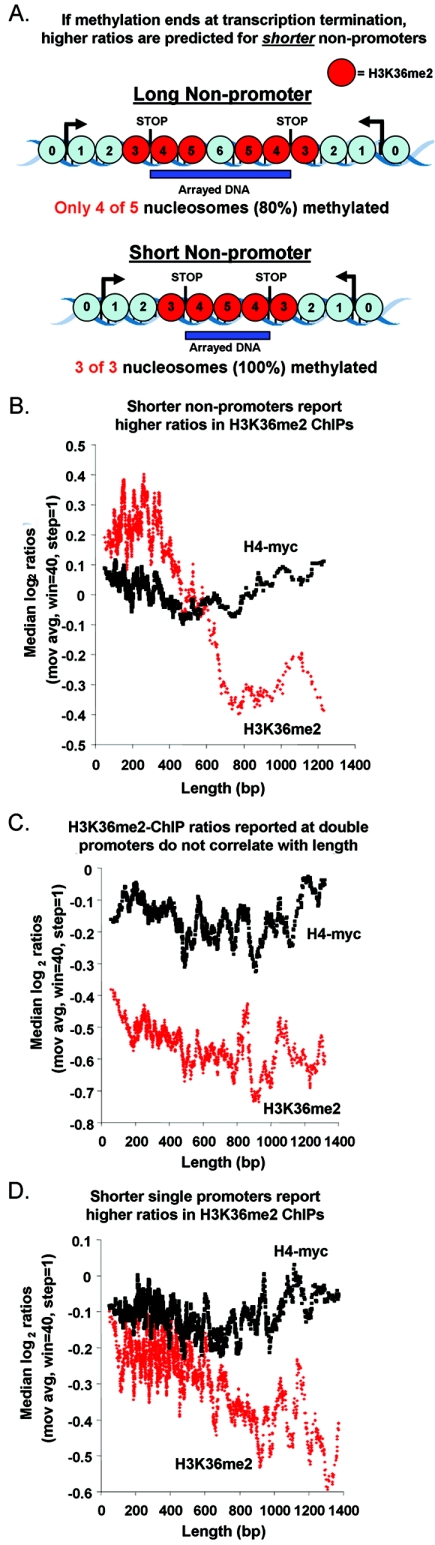
The results presented thus far provide evidence that dimethylation of H3K36 is restricted to transcribed genomic regions. A corollary to that hypothesis is that H3K36 dimethylation ends upon transcriptional termination. This hypothesis predicts that the smaller the interval between two convergently transcribed genes, the higher the measured ratio of enrichment in our H3K36me2 ChIPs. This is because these shorter regions are likely to be entirely transcribed, while as the distance between the two upstream genes grows, it becomes progressively less likely that the entire intergenic region will be transcribed. This would result in unmodified nucleosomes toward the center of the fragment, resulting in lower ratios (illustrated in Fig. 7A). Note that this prediction of the relationship between size and enrichment is the opposite of the previously described scenario for ORFs.
FIG. 7.
Evidence that H3K36 dimethylation ends at transcriptional termination genome-wide. (A) The hypothesis that H3K36 dimethylation ends upon transcriptional termination predicts that higher ratios will be reported for shorter nonpromoters (see the text for details). Blue circles, nucleosomes not dimethylated at H3K36. (B) Nonpromoters were sorted by length, and moving averages (mov avg) (window [win] = 40, step = 1) of their degrees of enrichment expressed as percentile ranks in H3K36me2 ChIPs (not normalized to H4-myc) and H4-myc ChIPs were plotted. (C) Same as panel B but for double promoters. (D) Same as panel B but for single promoters.
As predicted, we find that these shorter nonpromoter regions appear more highly enriched than longer nonpromoters in our H3K36me2 ChIPs (Fig. 7B). In contrast, the sizes of the regulatory regions upstream of two divergently transcribed genes, which themselves are not transcribed, show no relationship to the degrees of ChIP enrichment (Fig. 7C). Single promoters are expected to be partially transcribed, since they contain the 3′ UTR of an upstream gene. As predicted, single promoters exhibit an inverse relationship (weaker than that observed for nonpromoters) between size and reported H3K36me2 ratio value (Fig. 7D). No such relationship was observed when ChIPs were performed with histone H3 antibodies or in H4-myc strains or when H3K36me2 ChIPs were performed from set2Δ extracts (data not shown).
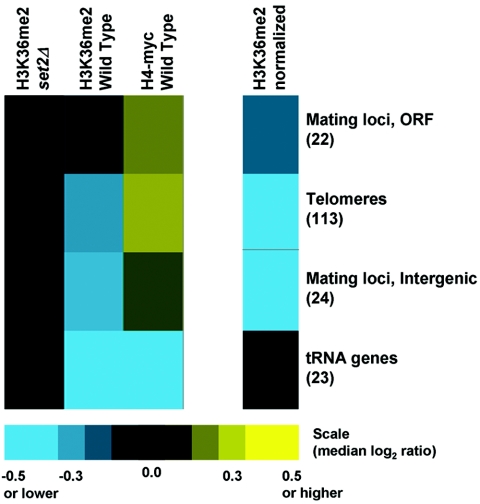
H3K36me2 is lacking in transcriptionally silent chromatin and in chromatin transcribed by RNA polymerase III.
To explore the possibility of other mechanisms of H3K36 dimethylation, we examined chromatin at telomeres and mating-type loci, two types of loci that are generally transcriptionally silent but serve specialized genomic functions. Both regions exhibit high nucleosome occupancy but are lacking in H3K36me2 (Fig. 8). We also asked whether H3K36 dimethylation was specific to chromatin transcribed by RNAP II or whether other polymerases might support cotranscriptional modification. Due to the repetitive nature of the RNAP I-transcribed rRNA genes, we were unable to make conclusions regarding H3K36me2 levels at these loci. However, we examined the RNAP III-transcribed tRNA loci and found that these regions were not enriched by our H3K36me2 ChIPs (Fig. 8). Although general nucleosome occupancy was also very low in regions transcribed by RNAP III, these results are consistent with the lack of evidence linking Set2p to RNA polymerase III. Therefore, Set2p's function in dimethylation of H3K36 appears to be mediated exclusively through its association with RNAP II.
FIG. 8.
H3K36me2 levels are very low or absent in transcriptionally silent and RNAP III-transcribed chromatin. Colors (scale at bottom) represent the medians of reported ChIP ratios, as described in the legend for Fig. 1B.
DISCUSSION
While previous studies had examined the distribution and behavior of S. cerevisiae H3K36me2 at a few loci, it was not known whether the reported characteristics were representative of other genes and genomic regions. The genome-wide analyses of H3K36me2 distribution and dynamics reported here allow us to unambiguously identify the core properties of this epigenetic mark in S. cerevisiae. The most important of these emergent properties are as follows. (i) H3K36me2 is scarce or absent in upstream gene regulatory regions, telomeres, mating loci, and regions transcribed by RNA polymerase III. This provides evidence that the enzymatic activity of Set2p is firmly restricted to its association with RNAP II. (ii) The degree of H3K36 dimethylation within ORFs correlates with the “on” or “off” state of transcription, but among genes that are measurably transcribed, the degree of modification does not correlate with the frequency of transcription. This provides evidence that H3K36 dimethylation occurs chiefly in the initial instance of gene transcription, with subsequent rounds playing at most a maintenance role. (iii) In support of the previous point, newly transcribed genes that had been transcriptionally dormant acquire the H3K36me2 mark but at levels that do not correlate with the degree of induction. (iv) Set2p enzymatic activity begins in ORFs at a fairly constant distance from the initiation of transcription, which does not vary with gene length. (v) Nucleosomes in noncoding regions immediately downstream of transcribed genes are as highly dimethylated at H3K36 as nucleosomes in ORFs. This result provides evidence that once Set2p is associated with RNAP II, Set2p continues to be active and remains associated with RNAP II throughout the rest of the transcript length. (vi) With increasing distance downstream of the stop codon, H3K36me2 levels decrease. This result provides evidence that Set2p histone methyltransferase activity is terminated along with the termination of transcription. (vii) Unlike the nucleosome depletion that is normally observed in the ORFs of highly transcribed chromatin, H3K36 dimethylated nucleosomes appear to be refractory to depletion, suggesting preferential retention of H3K36 methylated nucleosomes on DNA. However, nucleosome dynamics in a set2 deletion strain appeared to be normal, indicating that while H3K36me2 may be an indicator of stability, it is not likely to be a required factor. Although there may be some variation from locus to locus, these core properties shed light on important questions surrounding the function of the Set2p enzyme and H3K36me2 itself.
Challenges in determining the global distribution of chromatin modifications.
Before discussion of the biological function of H3K36me2, it is worth mentioning some of the challenges that are inherent to any experiment that aims to determine the distribution of a histone modification genome-wide. In this study, we used ChIP to specifically enrich for genomic regions that contain H3K36me2 nucleosomes and then interpreted the efficiency of DNA recovery at each locus to reflect the relative amount of H3K36 dimethylation at each locus. Using this approach, several factors could create nonbiological variation in results, including the effects of fixation, epitope accessibility, antibody specificity, microarray content, and underlying bulk nucleosome occupancy. These challenges have been discussed at length in recent reviews (6, 13, 32, 54).
This study includes important advances in addressing some of these issues. First, we thoroughly demonstrated the specificity of our H3K36me2 antibody by dot blot against H3K36me0, H3K36me1, and H3K36me3 peptides (Fig. 1A; see also Fig. S1 in the supplemental material), Western blots derived from whole-cell and nuclear extracts (data not shown), and control ChIPs in set2Δ strains (Fig. 1B). Second, we used DNA microarrays that cover the entire genome on a single slide. This represents a significant improvement over many published studies that used arrays representing only the ORFs or only noncoding intergenic regions or that split ChIP samples and hybridized them independently to separate arrays representing only the ORFs or the intergenic regions. Use of a whole-genome array was essential to most of the conclusions presented here (13). Third, the H3K36me2 data have been normalized to bulk nucleosome occupancy, using data from H3 or H4-myc ChIPs that were performed in parallel from the same extract. This is important because recent studies have shown that nucleosome occupancy throughout the yeast genome is heterogeneous (2, 26), and if left unaccounted for, misleading patterns could emerge. To our knowledge, this is the first instance of modified-nucleosome ChIP data being normalized to apparent bulk nucleosome occupancy genome-wide. Finally, we followed up each of our ChIP-chip experiments with high-resolution PCR-based detection at individual loci, which provided additional information and confirmed the conclusions drawn from the array results.
How is the intricate genomic pattern of histone H3K36 dimethylation specified?
The general mechanism of directing Set2p to specific genomic regions by piggybacking on elongating RNAP II through association with a doubly modified CTD (Ser2/Ser5) is entirely sufficient to explain the pattern of H3K36me2 we observed throughout the genome. This is an important conclusion because it indicates that Set2p modifies chromatin only when associated with RNAP II and not, for example, on soluble histone H3 prior to chromatin assembly. More specifically, the genome-wide analysis shows that H3K36me2 occurs at a determined distance from the initiation of transcription, regardless of ultimate transcript length (Fig. 6). This result is consistent with PCR-based ChIP assays performed on single genes (1, 18, 22, 45) and with locus-specific results presented here that imply H3K36 dimethylation begins approximately two nucleosomes downstream of the start codon. In addition, the data indicate that Set2p chromatin-modifying activity stops upon transcriptional termination (Fig. 7). We observed no relationship between the presence of introns and H3K36 dimethylation levels (data not shown).
What is the biologically relevant function of H3K36 dimethylation?
Our results indicate that levels of H3K36me2 are not correlated with the frequency of transcription but rather with the occurrence of transcription per se (Fig. 3 and Fig. 4). This result suggests that H3K36 methylation does not generally act as a “rheostat” for gene transcription. In S. cerevisiae, set2Δ strains are viable and, in many backgrounds, exhibit only mild phenotypes. So, what does H3K36 methylation do?
At individual loci, Set2p has been shown to act as a transcriptional repressor (25, 51). In one of these studies, Set2p caused repression of GAL4 but not of other examined genes, and in the other, Set2p was artificially tethered to a promoter, which resulted in transcriptional repression. So while Set2p may act as a transcriptional repressor at individual genes or have the capacity to repress transcription if inappropriately tethered at promoters, a general role for repression of transcription at gene promoters is not consistent with the genomic pattern reported here.
Given Set2p's established interaction with elongating polymerase and the genomic pattern of H3K36me2 reported here, it is easy to envision a role for Set2p and H3K36me2 in transcriptional elongation. Several lines of evidence suggest that this is the case. Perhaps the most convincing is synthetic genetic array analysis, which revealed growth defects when a set2Δ mutant was combined with deletions of any of the five components of the Paf1p complex or of the transcription elongation factors Chd1p or Soh1p (22). It has also been shown that deletion of genes encoding either of two components of the Paf1 complex, Rtf1p or Cdc73p, resulted in a decrease in the recruitment of Set2p across the PMA1 gene and abolished H3K36 dimethylation at that locus (22). In addition to these findings, studies involving 6-azauracil help to confirm a role for Set2 as an elongation factor (18, 22, 28). However, it is still not clear exactly how Set2p or H3K36me2 might participate in the process of elongation itself. It remains possible that Set2p's association with elongation is solely a mechanism to control the distribution of H3K36 dimethylation, rather than an indication of any direct participation in the transcription elongation process. In this case, the defects in elongation observed in the absence of H3K36me2 could be indirect consequences of a failure to recruit chromatin-modifying enzymes or other factors important for transcriptional elongation to ORFs.
An epigenetic mark to distinguish regulatory and nonregulatory chromatin genome-wide.
A mark such as H3K36me2 could also function as a “molecular memory” of transcription patterns that are specified at only one point during development or the life cycle but must be maintained afterwards. This concept of transcriptional memory is similar to what has been proposed for S. cerevisiae H3K4me3, which remains stable on chromatin long after the transcription of chromatin at that locus has ceased (39). The putative “memory” role of H3K36 methylation, which appears to be similarly stable, could be accomplished by the ability of this mark to physically affect the chromatin fiber or, more likely, through the recruitment of other remodeling factors that alter chromatin structure (41, 44). For example, it has been recently observed that histone H3 and H4 acetylation is generally lower in coding regions than in promoters and that this global acetylation pattern is regulated by the protein Eaf3p (42). Eaf3 is a subunit of both the NuA4 histone acetylase complex and the Rpd3 histone deacetylase complex (9, 11). Reid et al. proposed that “Eaf3 might recognize some feature of chromatin (e.g., nucleosome conformation or nonhistone protein) that is distinct between promoters and coding regions” (42). H3K36 methylation could be just such a distinguishing feature of coding and noncoding chromatin.
One intriguing possibility along these lines is that Set2p mediates H3K36 dimethylation to create a stable epigenetic mark that generally serves to distinguish regulatory and nonregulatory chromatin genome-wide. What function might such a distinction serve? As described in the introduction, coding and noncoding chromatin exhibit several biologically important differences whose underlying physical basis remains unexplained. Higher nucleosome occupancy in the body of genes may serve to prevent nonproductive transcription factor-DNA interactions by occluding binding sites that occur in coding regions. Conversely, nucleosomes in promoter regions may be more prone to low occupancy or disassembly, thereby exposing binding sites and directing transcription factors to appropriate targets (47). These two tendencies, acting in concert, would have the effect of reducing the “sequence space” that must be searched by any given factor before an appropriate target is found.
In S. cerevisiae, about 85% of genes are transcribed at detectable levels during mitotic growth (14, 15), meaning that H3K36me2 distinguishes regulatory and nonregulatory chromatin throughout most of the genome. In actively transcribed chromatin domains, upstream regulatory sequences are clearly distinguished by their lack of the H3K36me2 mark. Therefore, the H3K36me2 mark, which is conserved throughout eukaryotic evolution, represents the first physical mark that has been shown to distinguish regulatory sequences from coding and nonregulatory intergenic sequences genome-wide. A transcription-coupled mark that does not correlate with transcription rate and is correlated with stabilized nucleosomes on transcribed chromatin, both properties of H3K36me2 described here, may be a general feature of eukaryotic chromatin that contributes to the mechanism of context-dependent targeting of DNA-associated proteins.
Supplementary Material
Acknowledgments
We thank Ronald N. Laribee for providing Fig. 1A and Fig. S1 in the supplemental material. We thank Michael Buck and Paul Giresi for help with data analysis and Arkady Khodursky for careful reading of the manuscript prior to submission.
This work was supported by NIH grants to B.D.S. (R01GM68088) and J.D.L. (K22HG002577). B.D.S. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di-and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280:17732-17736. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 5.Bohlander, S. K., R. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83:349-360. [DOI] [PubMed] [Google Scholar]
- 7.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckert, J., and K. Struhl. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 12.Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown, and T. D. Petes. 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanlon, S. E., and J. D. Lieb. 2004. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr. Opin. Genet. Dev. 14:697-705. [DOI] [PubMed] [Google Scholar]
- 14.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 15.Hurowitz, E. H., and P. O. Brown. 2003. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 5:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 17.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279:24957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf, and B. D. Strahl. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25:3305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 25.Landry, J., A. Sutton, T. Hesman, J. Min, R.-M. Xu, M. Johnston, and R. Sternglanz. 2003. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:5972-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 27.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 28.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 30.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 31.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 32.Loden, M., and B. van Steensel. 2005. Whole-genome views of chromatin structure. Chromosome Res. 13:289-298. [DOI] [PubMed] [Google Scholar]
- 33.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 35.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, P. L., M. L. Cleary, P. O. Brown, and J. D. Lieb. 2003. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. USA 100:6364-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 40.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 41.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 42.Reid, J. L., Z. Moqtaderi, and K. Struhl. 2004. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 44.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 45.Schaft, D., A. Roguev, K. M. Kotovic, A. Shevchenko, M. Sarov, K. M. Neugebauer, and A. F. Stewart. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735-748. [DOI] [PubMed] [Google Scholar]
- 48.Sewack, G. F., T. W. Ellis, and U. Hansen. 2001. Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol. 21:1404-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stapleton, A., and T. D. Petes. 1991. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svaren, J., and W. Horz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 53.Turner, B. M. 2005. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 12:110-112. [DOI] [PubMed] [Google Scholar]
- 54.van Steensel, B., and S. Henikoff. 2003. Epigenomic profiling using microarrays. BioTechniques 35:346-357. [DOI] [PubMed] [Google Scholar]
- 55.Wu, T. C., and M. Lichten. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263:515-518. [DOI] [PubMed] [Google Scholar]
- 56.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, T., C.-F. Kao, N. J. Krogan, Z.-W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.