Abstract
Candida albicans possesses a plasma membrane-localized sensor of extracellular amino acids. Here, we show that in response to amino acids, this sensor induces the proteolytic processing of two latent transcription factors, Stp1 and Stp2. Processing removes negative regulatory motifs present in the N-terminal domains of these factors. Strikingly, Stp1 and Stp2 exhibit a clear dichotomy in the genes they transactivate. The shorter active form of Stp2 activates genes required for amino acid uptake. The processed form of Stp1 activates genes required for degradation of extracellular protein and uptake of peptides, and cells lacking Stp1 do not express the secreted aspartyl protease SAP2 or the oligopeptide transporter OPT1. Consequently, stp1 null mutants are unable to grow on media with protein as the sole nitrogen source. Cells expressing the STP1* allele that encodes a protein lacking the inhibitory N-terminal domain constitutively express SAP2 and OPT1 even in the absence of extracellular proteins or peptides. Also, we show that Stp1 levels, but not Stp2 levels, are downregulated in the presence of millimolar concentrations of extracellular amino acids. These results define the hierarchy of regulatory mechanisms that differentially control two discrete pathways for the assimilation of nitrogen.
Candida albicans is a commensal organism that lives as a benign member of the microflora of mammalian hosts. In response to changes in the host immune status or microflora, C. albicans ceases to be a commensal organism and infects a variety of host tissues (46). The capacity to shift from a commensal to a pathogenic state requires a coordinated metabolic response that triggers discrete developmental programs and induces the expression of virulence factors. Several virulence traits have been described for C. albicans, including adhesion, morphological and phenotypic switching, and the production of secreted hydrolytic enzymes (11). These virulence traits contribute to host tissue recognition, tissue invasion and colonization, and evasion of the host immune response and, importantly, influence the ability of C. albicans cells to take up required nutrients to survive and proliferate. In contrast to many microbial pathogens, C. albicans has a diverse metabolic repertoire and is able to colonize virtually any tissue and organ, each with a distinct nutritional content.
Although only limited information is available regarding what nutrient sources are actually utilized by C. albicans in situ within infected hosts, there are two obvious and abundant nitrogen sources, i.e., amino acids and host proteins. Amino acids are present at above millimolar concentrations in human blood (39). The Candida genome encodes a family of 22 amino acid permeases (AAPs) that facilitate amino acid uptake (9, 36, 49). C. albicans secretes a variety of hydrolytic enzymes, including secreted aspartyl proteases (SAPs) that are capable of digesting extracellular proteins. SAPs are encoded by a gene family of 10 related genes (SAP1 to SAP10) (38). Although the mechanisms regulating SAP gene expression have not been established, it has been shown that SAP genes are differentially regulated depending on growth conditions and are repressed in the presence of preferred nitrogen sources and high concentrations of amino acids (4, 25, 42, 52). The products of SAP activity, primarily oligopeptides, are transported into cells by a family of oligopeptide transporters (OPTs) that are encoded by a gene family comprised of eight members (OPT1 to OPT8) (35, 41).
The importance of nitrogen regulation is underscored by recent studies examining mutants lacking GAT1, a gene encoding a GATA factor that activates expression of nitrogen catabolic pathways in the absence of a preferred nitrogen source, e.g., ammonium (33). GAT1 null mutants are unable to derepress genes required for growth in the absence of preferred nitrogen sources and are highly attenuated in a murine model of systemic infections. These findings suggest that the capacity to utilize alternative nitrogen sources, i.e., certain amino acids and protein, is essential for virulent growth. Consistent with this notion, C. albicans strains lacking the ability to sense extracellular amino acids and to take up amino acids exhibit reduced virulence (36). Additionally, the importance of SAP production during virulent infections has been confirmed by several independent studies. Mutant strains with greatly reduced SAP activity are less virulent than parental wild-type (WT) strains (26, 43), and mice immunized with purified Sap2 exhibited dramatically reduced loads of C. albicans during systemic infections (51).
The yeast Saccharomyces cerevisiae is able to assess the availability of extracellular nutrients via sensors in the plasma membrane (for a review, see references 20 and 23). The capacity to sense amino acids was initially demonstrated by the observation that the expression of the dipeptide transporter (PTR2) and several AAP genes was derepressed by the presence of micromolar amounts of amino acids (16, 28). Induced expression of these nutrient uptake systems requires a plasma membrane-localized sensor complex, dubbed the SPS sensor (19). Cells lacking any one of the three components of this sensor, the SSY1, PTR3, and SSY5 gene products, are unable to respond to amino acid stimuli (8, 19, 29). Ssy1, the only integral membrane component of the SPS sensor, is a unique member of the AAP family that does not transport amino acids (17, 21, 27, 30). The SPS sensor functions as a ligand-activated receptor of external amino acids that controls nuclear localization of Stp1 and Stp2, two latently expressed transcription factors (2).
In response to the addition of amino acids, and in a strictly SPS sensor-dependent manner, Stp1 and Stp2 are endoproteolytically cleaved. This event liberates the DNA-binding and transactivation domains from an approximately 10-kDa N-terminal fragment that function to anchor unprocessed forms in the cytoplasm (1). The shorter forms of Stp1 and Stp2, lacking the negative regulatory domains, accumulate in the nucleus, where they function to transactivate SPS sensor-regulated genes. An additional component required for proper SPS sensor-induced Stp1 and Stp2 processing includes the integral endoplasmic reticulum (ER) membrane component Shr3 (30). Shr3 functions as a membrane-localized chaperone specifically required for AAPs, including the SPS sensor component Ssy1, to exit the ER (30, 31). Consequently, Shr3 is the most upstream component of the SPS-sensing pathway, and shr3Δ mutants are unable to both sense and take up amino acids.
Orthologs of the S. cerevisiae AAPs and the known SPS sensor pathway components are present in the C. albicans genome (36). Accumulating evidence indicates that C. albicans cells use the SPS sensor pathway to sense and respond to extracellular amino acids in a manner that is remarkably similar to that of yeast cells. Csy1, the Ssy1 ortholog, is required for amino acid-induced expression of AAP genes; consequently, csy1Δ cells exhibit decreased rates of amino acid uptake (9). Csh3, the ortholog of Shr3, is required for the proper localization of AAP and Csy1 to the plasma membrane (36). Consequently, csh3Δ mutants display all of the phenotypes of csy1Δ mutants; they have a greatly diminished capacity to take up amino acids and do not undergo morphological transitions in response to inducing amino acids (36). Importantly, the reduced virulence of csh3Δ mutants suggests that C. albicans cells require the capacity to take up amino acids for growth in mammalian hosts (36).
Clearly, the availability of csh3Δ and csy1Δ strains has provided novel insights regarding the influence of amino acid availability on C. albicans growth and virulence. Here, we extend our analysis of the SPS-sensing pathway and have focused on the ultimate downstream effector components, the transcription factors Stp1 and Stp2. Similar to signaling events in S. cerevisiae, extracellular amino acids induce the proteolytic processing of C. albicans Stp1 and Stp2. Strikingly, the Candida factors transactivate two distinct sets of genes. Processed Stp1 activates the expression of proteins required for the catabolic utilization of extracellular proteins, whereas processed Stp2 induces the expression of AAP genes. Also, we report that Stp1 levels, but not Stp2 levels, are downregulated in the presence of millimolar concentrations of extracellular amino acids. These results indicate that Candida cells use their capacity to sense extracellular amino acids to differentially control two discrete pathways for the assimilation of nitrogen for growth.
MATERIALS AND METHODS
Media.
Standard media, including yeast extract-peptone-dextrose (YPD) medium, ammonia-based synthetic minimal dextrose (SD) medium, and ammonia-based synthetic complex dextrose (SC) medium were prepared as described previously (44). Yeast carbon base (YCB)-bovine serum albumin (BSA) medium contains 23.4 g liter−1 yeast carbon base, 4 g liter−1 bovine serum albumin, and 25 mM Na citrate (pH 4.0). Ura− strains were grown in medium supplemented with uridine (25 μg ml−1). Individual amino acids were added to SD or YCB-BSA medium at the concentration indicated in each case. The ability to utilize different amino acids as the sole nitrogen source was examined on succinate-buffered yeast nitrogen base (YNB) (without amino acids and ammonium sulfate) (pH 6) containing 2% glucose and 50 μM histidine. The amino acids were added to a final concentration of 1 mM. Medium was made solid by the addition of 2% nitrogen-free agar. Where appropriate, 0.5 mg ml−1 5-fluoroortic acid, 10 μg ml−1 mycophenolic acid, 200 μg ml−1 nourseothricin (NAT), and 1.5 mg ml−1 sulfonylurea herbicide (MM) were added to the media.
Cloning of STP1 and STP2.
Plasmids used in this study are listed in Table 1, and the sequences of oligonucleotides are listed in the supplemental material (see Table S1 in the supplemental material). Here, we present a description of the cloning of STP1 and STP2. Details regarding the construction of derivative plasmids are provided in the supplemental material (see Table S1 in the supplemental material). The genomic region containing the STP1 open reading frame (ORF) was cloned using PCR with DNA from strain SC5314 as a template; primers 1F1 and 1R1 were used to amplify a 1.8-kb fragment initiating 479 bp upstream and ending at the stop codon of the STP1 ORF, and primers 1F2 and 1R2 were used to amplify a 480-bp fragment initiating at the stop codon and ending downstream of the ORF (Fig. 1B). The primers 1F1 and 1R2 introduced PstI and XbaI sites, respectively, and primers 1R1 and 1F2 introduced a BamHI site immediately preceding the stop codon. The 1F1-1R1 and 1F2-1R2 fragments were initially inserted into pCR-BluntII-TOPO and pCR2.1-TOPO (Invitrogen), respectively, and the PstI/BamHI 1F1-1R1 and BamHI/XbaI 1F2-1R2 fragments were subsequently cloned in PstI/XbaI-digested Bluescript KS(+), creating plasmid pPM67. The genomic region containing the STP2 ORF was cloned using PCR with DNA from strain SC5314 as a template; primers 2F1 and 2R1 were used to amplify a 2.6-kb fragment initiating 917 bp upstream and ending at the stop codon of the STP2 ORF, and primers 2F2 and 2R2 were used to amplify a 340-bp fragment initiating at the stop codon and ending downstream of the STP2 ORF (Fig. 1C). The primers 2R1 and 2F2 introduced a BamHI site immediately preceding the stop codon. These two products were recombined into HindIII/SmaI-restricted pRC2312 in S. cerevisiae to create plasmid pPM92.
TABLE 1.
Plasmids used
| Plasmid | Description | Reference or source |
|---|---|---|
| pRC2312 | YRp CaARS LEU2 URA3 | 12 |
| pPM67 | STP1 in Bluescript KS(+) | This work |
| pPM72 | STP1 in pRC2312 (LEU2 URA3) | This work |
| pPM73 | STP1Δ62 in pRC2312 (LEU2 URA3) | This work |
| pPM74 | STP1-myc in pRC2312 (LEU2 URA3) | This work |
| pPM79 | STP1Δ62 in a YIp derivative of pRC2312 (URA3) | This work |
| pPM80 | STP1 in a YIp derivative of pRC2312 (URA3) | This work |
| pPM84 | stp1Δ1 in a YIp derivative of pRC2312 (URA3) | This work |
| pPM77 | PADH1-STP1-myc in YPB1 (URA3) | This work |
| pPM88 | PADH1-STP1-myc in a pPM77 derivative (RP10 URA3) | This work |
| pPM92 | STP2 in pRC2312 (LEU2 URA3) | This work |
| pPM94 | STP2-HA in pRC2312 (LEU2 URA3) | This work |
| pPM97 | STP2Δ100 in pRC2312 (LEU2 URA3) | This work |
| pPM108 | STP1-myc in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM109 | STP2 in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM110 | STP2Δ100 in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM111 | STP2-HA in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM114 | stp2Δ2::AgTEF1p-CaNAT1 in pRC2312 (LEU2 URA3) | This work |
| pPM121 | STP1 in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM122 | STP1Δ62 in a pRC2312 derivative (HIS1 URA3) | This work |
| pPM126 | STP2 in a YIp derivative of pRS315 (URA3) | This work |
| pPM129 | stp2Δ4::dp1200-URA3 in a YIp derivative of pRS315 (URA3) | This work |
| pPM132 | stp2Δ5::MPA in a YIp derivative of pRS315 (URA3) | This work |
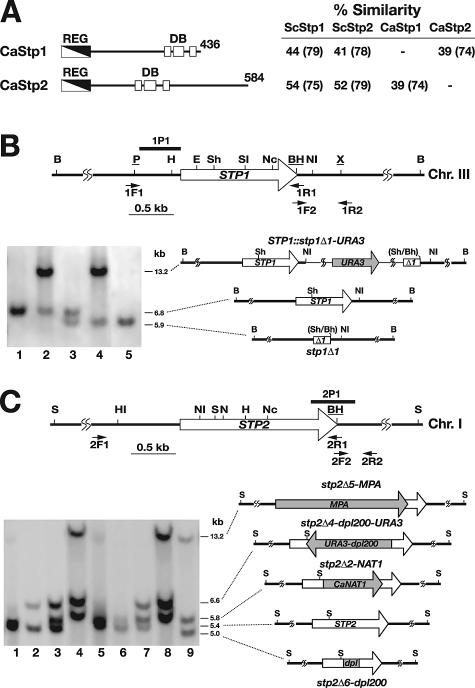
FIG. 1.
Schematic diagram of CaStp1 and CaStp2 and an illustration of the STP1 and STP2 chromosomal loci. (A) CaStp1 and CaStp2 have N-terminal regulatory domains (REG) (white/black diagonal boxes) and three DNA-binding domains (DB) (white boxes) with putative zinc fingers (aa 317 to 337, aa 345 to 378, and aa 396 to 419 in CaStp1 and aa 227 to 247, aa 255 to 293, and aa 312 to 335 in CaStp2) are marked. Percent similarities, calculated in accordance with the Lalign algorithm (http://www.ch.embnet.org) and the BLOSUM50 amino acid substitution matrix, to the S. cerevisiae ScStp1 and ScStp2 homologs and each other are indicated; values in parentheses indicate similarity within respective DNA-binding domains. (B) Diagrammatic presentation of stp1Δ and stp2Δ null mutations and the contruction of stp1Δ and stp2Δ mutant strains. The 6.8-kb BglII DNA fragment containing STP1 is shown. The positions of PCR primer pairs (1F1-1R1 and 1F2-1R2) used to clone STP1 and the oligonucleotide probe (1P1) used to verify correct integration of stp1Δ1-URA3 and subsequent URA3 excision that results in the unmarked stp1Δ1 allele are indicated. Southern analysis of BglII-digested genomic DNA of the parental strain CAI4 (STP1/STP1) and mutant derivatives PMRCA29 (STP1/STP1::stp1Δ1-URA3), PMRCA32 (STP1/stp1Δ1), PMRCA34 (stp1Δ1/STP1::stp1Δ1-URA3), and PMRCA35 (stp1Δ1/stp1Δ1) (lanes 1 to 5, respectively) is shown. The blot was hybridized with the labeled 1P1 probe. The BglII fragments containing STP1, STP1::stp1Δ1-URA3, and stp1Δ1 alleles are schematically depicted. (C) The approximate 5.4-kb SpeI DNA fragment on chromosome I containing STP2 is shown. The positions of PCR primer pairs (2F1-2R1 and 2F2-2R2) used to clone STP2 and the oligonucleotide probe (2P1) used to verify correct integration of the three stp2Δ deletion alleles are indicated. Southern analysis of SpeI-digested genomic DNA isolated from parental strain CAI4 (STP2/STP2/STP2) (lane 1) and stp2Δ mutant strains PMRCA54 (STP2/STP2/stp2Δ4::dpl200-URA3), PMRCA55 (STP2/stp2Δ4::dpl200-URA3/stp2Δ2::NAT1), and PMRCA57 (stp2Δ4::dpl200-URA3/stp2Δ2::NAT1/stp2Δ5::MPA) (lanes 2 to 4, respectively) is shown. The three stp2Δ alleles were individually introduced into the stp1Δ1/stp1Δ1 null mutant strain PMRCA35. Their correct insertion was monitored by Southern analysis using SpeI-digested genomic DNA from PMRCA35 (lane 5) and stp2Δ mutant strains PMRCA61 (STP2/STP2/stp2Δ2::NAT1), PMRCA62 (STP2/stp2Δ2::NAT1/stp2Δ4::dpl200-URA3), PMRCA88 (stp2Δ2::NAT1/stp2Δ4::dpl200-URA3/stp2Δ5::MPA), and PMRCA89 (stp2Δ2::NAT1/stp2Δ6::dpl200/stp2Δ5::MPA) (lanes 6 to 9, respectively). The blot was hybridized with the labeled 2P1 probe. The structure of the SpeI-flanked fragments containing the STP2, stp2Δ5::MPA, stp2Δ2::NAT1, stp2Δ4::dpl200-URA3, and stp2Δ6::dpl200 alleles are schematically depicted. Relevant restriction endonuclease sites are indicated as follows: B, BglII; BH, BamHI; E, EcoRV; H, HindIII; HI, HpaI; N, NheI; Nc, NcoI; NI, NsiI; P, PstI; S, SpeI; Sh, SphI; SI, SnabI; X, XbaI. The sites underlined were introduced in oligonucleotides used for PCR amplification, and sites in parentheses were inactivated during cloning.
Strain construction.
The C. albicans strains used in this study are listed in Table 2. Standard methods were used to construct two series of isogenic strains in two genetic backgrounds. Strain CAI4 (18) was used for the construction of strains carrying stp1 and stp2 null mutations, and strains derived from DAY286 (53) and CAEB4 (9) were used to examine Csy1-dependent processing of Stp1 and Stp2. PCR and Southern analysis were used to confirm each step of strain constructions (see Table S1 in the supplemental material for a detailed description of all strain constructions).
TABLE 2.
C. albicans strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Prototrophic wild type | 22 |
| SAP2MS4B | sap2Δ::FRT/sap2Δ::FRT | J. Morschhäusser |
| CAI4-derived strains | ||
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 18 |
| PMRCA10 | CAI4 csh3Δ3/csh3Δ3 | 36 |
| PMRCA12 | PMRCA10 ura3Δ::imm434/URA3 | 36 |
| PMRCA18 | CAI4 ura3Δ::imm434/URA3 | 36 |
| PMRCA24 | PMRCA10 STP1/STP1::STP1-URA3 | This work |
| PMRCA25 | PMRCA10 STP1/STP1::STP1Δ62-URA3 | This work |
| PMRCA35 | CAI4 stp1Δ1/stp1Δ1 | This work |
| PMRCA45 | PMRCA10 STP2/STP2/STP2-URA3 | This work |
| PMRCA46 | PMRCA10 STP2/STP2/STP2::STP2Δ100-URA3 | This work |
| PMRCA48 | CAI4 STP2/STP2/STP2::STP2-HA-URA3 | This work |
| PMRCA50 | PMRCA10 STP2/STP2/STP2::STP2-HA-URA3 | This work |
| PMRCA57 | CAI4 stp2Δ4::dpl200-URA3/stp2Δ2::CaNAT1/stp2Δ5::MPA | This work |
| PMRCA59 | ura3Δ::imm434/URA3 stp1Δ1/stp1Δ1 | This work |
| PMRCA60 | ura3Δ::imm434/URA3 stp1Δ1/STP1Δ62 | This work |
| PMRCA66 | CAI4 RP10/RP10/rp10::PADH1-STP1-myc-URA3 | This work |
| PMRCA68 | PMRCA10 RP10/RP10/rp10::PADH1-STP1-myc-URA3 | This work |
| PMRCA89 | PMRCA35 stp2Δ6::dpl200/stp2Δ2::CaNAT1/stp2Δ5::MPA | This work |
| PMRCA94 | ura3Δ::imm434/URA3 stp1Δ1/stp1Δ1 stp2Δ6::dpl200/stp2Δ2::CaNAT1/stp2Δ5::MPA | This work |
| PMRCA95 | PMRCA89 stp1Δ1/stp1Δ1::STP1-URA3 | This work |
| PMRCA96 | PMRCA89 stp2Δ/stp2Δ/stp2Δ::STP2-URA3 | This work |
| DAY286-derived strains | ||
| DAY286 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG-ARG4-URA3/arg4Δ::hisG | Aaron Mitchell |
| CAEB1 | ura3Δ::imm434/ura3Δ::imm434-his1Δ::hisG-HIS1/his1::hisG arg4Δ::hisG-ARG4-URA3/arg4::hisG | 9 |
| CAEB4 | ura3Δ::imm434/ura3Δ::imm434-his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG csy1Δ::ARG4/csy1Δ::URA3 | 9 |
| PMRCA74 | DAY286 STP2/STP2::STP2-HIS1 | This work |
| PMRCA75 | CAEB4 STP2/STP2::STP2-HIS1 | This work |
| PMRCA76 | DAY286 STP2/STP2::STP2Δ100-HIS1 | This work |
| PMRCA77 | CAEB4 STP2/STP2::STP2Δ::100-HIS1 | This work |
| PMRCA78 | CAEB4 STP1/STP1::STP1-myc-HIS1 | This work |
| PMRCA79 | CAEB4 STP2/STP2::STP2-HA-HIS1 | This work |
| PMRCA81 | DAY286 STP1/STP1::STP1-HIS1 | This work |
| PMRCA82 | DAY286 STP1/STP1::STP1Δ62-HIS1 | This work |
| PMRCA83 | CAEB4 STP1/STP1::STP1-HIS1 | This work |
| PMRCA84 | CAEB4 STP1/STP1::STP1Δ62-HIS1 | This work |
Secreted protease activity assays.
YPD-grown cells were inoculated in YCB-BSA with or without glutamine as indicated and incubated at 37°C. Aliquots of culture supernatants (500 μl) were diluted with an equal volume of water, and 250 μl of 0.5 M Na-citrate, pH 3.2, was added. Reactions were initiated by the addition of a 125-μl aliquot of 10% (wt/vol) bovine serum albumin and incubated at 30°C. Aliquots (300 μl) were taken immediately after the addition of BSA (t = 0) and at different time points in the linear range of the assay. Protein was precipitated by the addition of 250 μl cold 20% trichloroacetic acid, and samples were centrifuged for 10 min at 13,000 rpm. The levels of trichloroacetic acid-soluble peptides and free amino acids were spectrophotometrically determined at 280 nm. The t = 0 sample was used as the reference.
Immunoblot analysis.
Whole-cell extracts were prepared as described previously (45). Proteins within extracts resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 7.5% gels were analyzed by immunoblotting according to standard procedures. Immunoblots were probed with 1:1,000 dilutions of monoclonal antibodies recognizing the hemagglutinin (HA) (rat monoclonal anti-HA 3F10; Roche) or c-myc (mouse monoclonal anti-Myc horseradish peroxidase-conjugated 9E10; Roche) epitope. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura extended-duration substrate; Pierce) using the LAS1000 system (Fuji Photo Film Co., Ltd., Japan).
Semiquantitative reverse transcription (RT)-PCR.
Total RNA was isolated with RNeasy (QIAGEN) and treated with RNase-free DNase (QIAGEN). cDNA synthesis was performed in the presence of 2 μg of total RNA using Superscript II reverse transcriptase as recommended by the manufacturer (Invitrogen, Life Technologies). The cDNA-containing reactions were diluted 1:5, and 0.5 μl was used as a template in PCRs (25 μl). Samples were denatured at 94°C for 2 min, followed by 15 to 30 cycles (94°C for 45 s, 55°C for 45 s, and 72°C for 30 s). The levels of amplified products were determined at several cycle intervals to ensure that samples were analyzed during the exponential phase of amplification. Specific primers annealing to GAP1, GAP2, CAN1, PTR2, OPT1, OPT3, SAP2, and ACT1 were designed to yield products ranging from 250 to 300 bp (see Table S1 in the supplemental material). Specificity of each primer pair was empirically analyzed in a gradient PCR cycler using genomic DNA as a template, and reactions carried out in the absence of reverse transcriptase were used to control for the presence of contaminating DNA. The levels of the ACT1 fragment were used to control the levels of template cDNA.
RESULTS
Identification and cloning of CaSTP1 and CaSTP2.
Our previous work with csh3 null mutant strains provided strong evidence that C. albicans cells sense extracellular amino acids and use amino acids as nitrogen sources for growth in mammalian hosts (36). To obtain the necessary experimental tools to differentiate the relative importance of primary amino acid-sensing events and secondary effects due to diminished uptake, we identified the C. albicans orthologs of S. cerevisiae Stp1 and Stp2 (Fig. 1A). These proteins contain three zinc finger DNA-binding domains that share a pronounced degree of sequence conservation to each other (74% similarity) and with the DNA-binding domains in the S. cerevisiae orthologs (75 to 79% similarity). Based on the slight differences in the degree of similarity within the DNA-binding domains, we designated ORF pairs 19.5917 and 19.13338 CaStp1 and 19.4961 and 19.12426 CaStp2. Notably, CaStp1 (436 amino acids [aa]) is unique in two ways: it is significantly shorter than CaStp2 (584 aa), ScStp1 (520 aa), and ScStp2 (542 aa), and the DNA-binding domains of CaStp1 are located near the C terminus. Overall, CaStp1 and CaStp2 are more similar compared to ScStp1 and ScStp2 (44 to 54%) than with each other (39%). The low degree of sequence homology with each other raised the possibility that CaStp1 and CaStp2 may differentially activate gene expression. The genomic DNA fragments containing CaSTP1 and CaSTP2 (Fig. 1B and C, respectively) were cloned (see Materials and Methods) and sequenced; no discrepancies in our sequence and that in the C. albicans database were noted.
stp1Δ and stp2Δ null mutants exhibit a reduced capacity to take up amino acids.
To directly test whether CaStp1 and CaStp2 are effectors of the Candida SPS-sensing pathway, we constructed a set of isogenic mutant strains lacking STP1, STP2, or both (see Materials and Methods). Briefly, the two STP1 alleles were sequentially disrupted, creating an stp1Δ/stp1Δ (= stp1Δ) homozygous mutant strain with unmarked deletion alleles on chromosome III (Fig. 1B). STP2 is located on chromosome I that is present in three copies (14); consequently, three rounds of one-step gene replacements were carried out to obtain the stp2Δ stp2Δ stp2Δ (= stp2Δ) and stp1Δ stp2Δ null mutant strains (Fig. 1C). Each of the construction steps was confirmed by Southern analysis (Fig. 1B and C). Finally, the mutant strains (with the exception of the stp2Δ mutant) were made Ura3+ by integrating the URA3 gene into one of its endogenous loci. To control for possible transformation-induced mutations, the STP1 and STP2 wild-type alleles were independently reintroduced into the stp1Δ stp2Δ double mutant strain.
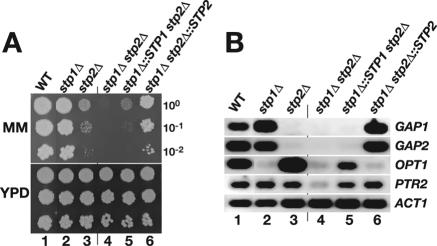
We compared the growth characteristics of wild-type (WT), isogenic null mutant, and complemented strains on YPD medium supplemented with the sulfonylurea herbicide MM. MM inhibits acetohydroxyacid synthase, resulting in a defect in the biosynthesis of branched-chain amino acids; consequently, cells must import branched-chain amino acids for growth (29). All strains grew equally well on YPD medium (Fig. 2A). In the presence of MM, the growth of the stp1Δ mutant appeared similar to that of the WT (Fig. 2A, compare dilution series 2 with 1), whereas the growth of the stp2Δ mutant was significantly impaired (compare dilution series 3 with 1). The residual growth of the stp2Δ mutant was dependent on STP1; the stp1Δ stp2Δ double mutant strain was unable to grow on MM (Fig. 2A, dilution series 4). Reintroduction of STP1 into the stp1Δ stp2Δ double mutant restored weak growth on MM, and the complemented strain exhibited an almost identical level of growth as the stp2Δ single mutant (Fig. 2A, compare dilution series 5 with 3). Reintroduction of STP2 into the stp1Δ stp2Δ double mutant restored robust growth on MM, almost to the level of the stp1Δ mutant (Fig. 2A, compare dilution series 6 with 2). These results indicate that the loss of Stp1 function is readily compensated by Stp2 but not vice versa. The subtle differences between the growth of homozygous single mutants and the heterozygous complemented strains are likely due to gene dosage effects.
FIG. 2.
Stp1 and Stp2 have nonredundant functions. (A) Growth phenotypes of stp1Δ, stp2Δ, and stp1Δ stp2Δ null mutant strains. Serial dilutions of WT (PMRCA18), stp1Δ (PMRCA59), stp2Δ (PMRCA57), stp1Δ stp2Δ (PMRCA94), stp1Δ::STP1 stp2Δ (PMRCA95), and stp1Δ stp2Δ::STP2 (PMRCA96) strains were spotted on YPD medium and YPD medium containing MM. The plates were incubated for 2 days at 30°C and photographed. (B) Pattern of gene expression in stp1Δ, stp2Δ, and stp1Δ stp2Δ null mutant strains. Cultures of strains were pregrown in SD medium and induced with 5 mM glutamine for 1 hour. Total RNA was isolated, and GAP1, GAP2, OPT1, PTR2, and SAP2 expression was analyzed by RT-PCR. ACT1 was used as a positive control.
The mutant growth phenotypes were consistent with the prediction that Stp1 and Stp2 transactivate genes required for proper amino acid uptake. We examined this possibility directly by monitoring the expression levels of a select set of genes encoding amino acid permeases and peptide transporters. The transcript levels of two amino acid permease homologs (GAP1 and GAP2) (36), the dipeptide transporter (PTR2) (6), and the oligopeptide transporter (OPT1) (35) in cells grown in SD medium supplemented with glutamine were analyzed by RT-PCR (Fig. 2B). In wild-type cells, the transcripts of these genes were readily amplified (Fig. 2B, lane 1). In contrast, OPT1 transcripts were not detected in the stp1Δ mutant, indicating that its expression is dependent on Stp1 (Fig. 2B, lane 2). The deletion of STP1 did not reduce the levels of GAP1 and GAP2 transcripts. The inverse pattern of expression was observed in the stp2Δ null mutant; OPT1 transcripts were readily detected, but GAP1 and GAP2 transcripts were not (Fig. 2B, lane 3). PTR2 transcripts were detected in both stp1Δ and stp2Δ single mutants (Fig. 2B, lanes 2 and 3) but not in the stp1Δ stp2Δ double mutant (lane 4), indicating that either Stp1 or Stp2 can transactivate PTR2 expression. The reintroduction of STP1 and STP2 restored the respective wild-type pattern of gene expression (Fig. 2B, compare lane 6 with lane 2 and lane 5 with lane 3), demonstrating the recessive nature of the null alleles.
Extracellular amino acids induce the proteolytic processing of Stp1 and Stp2.
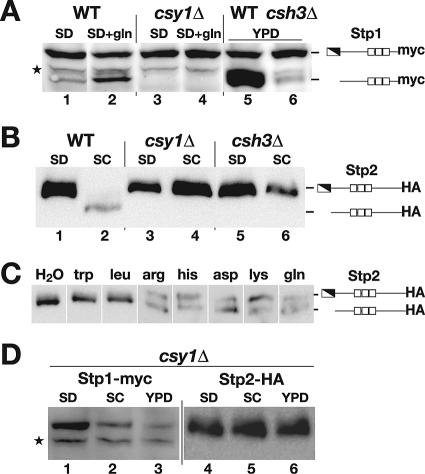
To assess whether Stp1 and Stp2 are regulated similarly to their orthologs in S. cerevisiae, we examined the electrophoretic behavior of Stp1 and Stp2 in cells grown in the presence and absence of amino acids (Fig. 3). To facilitate the analysis, STP1-myc and STP2-HA alleles encoding functional C-terminally-tagged proteins carrying three- and sixfold reiterated Myc and HA epitopes, respectively, were introduced into their endogenous loci in wild-type and mutant strains completely lacking (csy1Δ) (9) or with reduced (csh3Δ) SPS sensor function (36).
FIG. 3.
Extracellular amino acids induce the proteolytic processing of Stp1 and Stp2 in an SPS-sensor-dependent manner. (A) Characteristics of Stp1 processing and immunoblot analysis of whole-cell extracts from WT (PMRCA85, lanes 1 and 2) and csy1Δ (PMRCA78, lanes 3 and 4) strains expressing STP1-myc. Cells were grown in SD medium, and extracts were prepared 1 h after the addition of 5 mM glutamine or an equal aliquot of water as indicated. Immunoblotting of extracts from exponentially YPD-grown WT (PMRCA66, lane 5) and csh3Δ (PMRCA68, lane 6) strains constitutively expressing STP1-myc (RP10::PADH1-STP1-myc) is shown. (B) Characteristics of Stp2 processing and immunoblot analysis of extracts from WT (PMRCA48, lanes 1 and 2), csy1Δ (PMRCA79, lanes 3 and 4), and csh3Δ (PMRCA50, lanes 5 and 6) strains expressing STP2-HA. Cells were grown in SD or SC medium as indicated. (C) Stp2 processing is induced by a discrete subset of amino acids. Wild-type cells expressing STP2-HA (PMRCA48) were pregrown in SD medium, and subcultures were removed and incubated at 30°C for 1 h in the presence of amino acids (5 mM). Extracts were prepared and analyzed by immunoblotting. Immunoreactive forms of Stp1-Myc and Stp2-HA are schematically represented at their corresponding positions of migration. (D) Amino acid availability affects Stp1 but not Stp2 levels. Null mutant csy1Δ strains expressing STP1-myc (PMRCA78, lanes 1 to 3) or STP2-HA (PMRCA79, lanes 4 to 6) were pregrown in SD medium, washed, and resuspended in fresh SD, SC, or YPD medium and incubated at 30°C for 1 h. Extracts were prepared, and the levels of Stp1 and Stp2 were determined by immunoblotting. The star (A and D) marks the position of an unrelated antigen that cross-reacts with the anti-Myc antibody; this cross-reacting band fortuitously serves as a loading control.
In WT cells grown in the absence of external amino acids (SD medium), a major band corresponding to full-length Stp1-Myc (Fig. 3A, lane 1) was observed. One hour after 5 mM glutamine was added to cells, the full-length Stp1-Myc band decreased in intensity, and a faster-migrating band with an approximately 10-kDa-lower molecular weight was detected (Fig. 3A, lane 2). The faster-migrating band was not detected in csy1Δ cells (Fig. 3A, lanes 3 and 4). To detect Stp1-Myc in csh3 null mutant strains, we placed the STP1-myc allele under the control of the strong ADH1 promoter. The overexpressed Stp1-Myc was readily detected in both wild-type and csh3Δ mutant strains grown in amino acid-rich YPD medium (Fig. 3A, lanes 5 and 6, respectively). However, in wild-type cells, the bulk of Stp1-Myc migrated as a band corresponding to the shorter form, whereas in csh3Δ cells, the major portion of Stp1-Myc migrated as the full-length protein. A similar change in the electrophoretic mobility of Stp2-HA was observed in wild-type cells grown in the absence (SD medium) and presence (SC medium) of amino acids (Fig. 3B, lanes 1 and 2). A band corresponding to full-length Stp2-HA was readily detected in cells grown in SD medium, whereas in cells grown in SC medium, a faster-migrating band with an approximately 10-kDa-lower molecular weight was observed. No alterations in migration were observed in csy1Δ and csh3Δ mutants (Fig. 3B, lanes 3 to 6).
We tested the ability of all 20 common l-amino acids, ornithine, and citrulline to induce the proteolytic processing of Stp2 (Fig. 3C). Each of these potential Csy1 ligands was individually added to a final concentration of 5 mM to exponentially growing wild-type cells in SD medium. One hour after induction, cell extracts were prepared and the electrophoretic mobility of Stp2-HA was analyzed. When cells were challenged with arginine, histidine, lysine, aspartate, and glutamine, two distinct bands corresponding to the unprocessed and processed forms of Stp2 were detected. Limited processing was also detected when cells were induced with asparagine, serine, and ornithine. No processing was detected when cells were challenged with citrulline or the other amino acids tested (G, A, V, L, I, P, C, M, F, W, K, T, and Y) (data not shown). The amino acids that induce the cleavage of Stp2 are the same as those previously shown to stimulate amino acid uptake in C. albicans (9).
In summary, these results indicate that the appearance of the shorter forms of Stp1 and Stp2 is dependent upon the presence of extracellular amino acids, the amino acid sensor component Csy1, and the dedicated AAP-specific chaperone Csh3 (37). These findings are entirely consistent with our understanding of the SPS sensor pathway in S. cerevisiae (2, 20) and suggest that similar to their orthologs in S. cerevisiae, Stp1 and Stp2 are synthesized as latent inactive precursors with negative regulatory domains within their N termini.
Stp1 levels are regulated by amino acid availability.
In the course of examining the posttranscriptional processing of Stp1 and Stp2, we found that the steady-state levels of Stp1 were below detectable levels in SC medium-grown cells (data not shown). In contrast, Stp1 was readily detected in cells grown in SD medium, a medium lacking amino acids but containing high levels of ammonium (Fig. 3A). This observation raised the possibility that amino acid availability, and not the overall nitrogen status of the cell, affects the steady-state levels of Stp1. To prevent the possibility of processing-induced downregulation, we analyzed the levels of full-length Stp1 and Stp2 in SD medium-grown csy1Δ cells 1 h after they were placed in amino acid-rich SC and YPD media (Fig. 3D). In contrast to Stp2 levels that remained constant (Fig. 3D, lanes 4 to 6), the levels of Stp1 diminished in cells placed in SC (lane 2) and YPD (lane 3) media. These results indicate that C. albicans cells adjust levels of Stp1 in response to high concentrations of amino acids.
Stp1 and Stp2 activate distinct patterns of gene expression reflecting divergent roles in nitrogen assimilation.
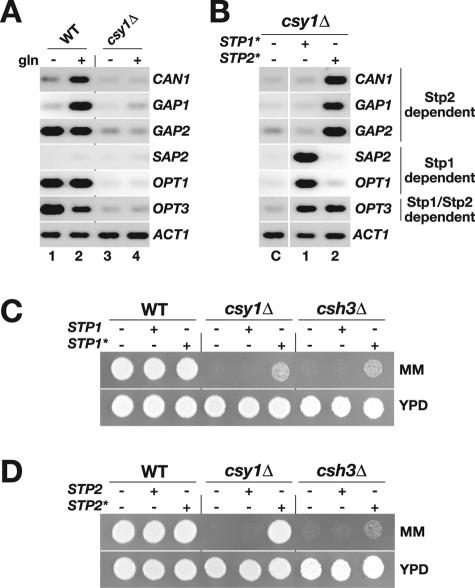
To address the in vivo role of Stp1 and Stp2, we determined the expression levels of several genes encoding proteins involved in nitrogen acquisition in wild-type and csy1Δ cells. First, we checked whether the addition of amino acids induced gene expression (Fig. 4A). Cells were grown in SD medium, and cultures were divided into two equal portions; one half received 5 mM glutamine, and the other half received a corresponding aliquot of water. Total RNA was isolated after 1 h, and RT-PCR was used to quantitate the relative levels of amino acid permease (CAN1, GAP1, and GAP2), oligopeptide transporter (OPT1 and OPT3), and secreted aspartyl protease (SAP2) gene expression. The addition of glutamine clearly induced the expression of CAN1 and GAP1 (Fig. 4A, lanes 1 and 2). Consistent with the lack of Stp1 and Stp2 processing, the expression of these genes was not induced in csy1Δ cells (Fig. 4A, lanes 3 and 4). Although GAP2, OPT1, and OPT3 transcripts were detected in uninduced wild-type cells, these genes were not expressed in csy1Δ cells, confirming the strict dependence on the SPS sensor in regulating their expression.
FIG. 4.
STP1* (STP1Δ62) and STP2* (STP2Δ100) alleles lacking N-terminal regulatory domain sequences encode constitutively active transcription factors. (A) Amino acid-induced processing of Stp1 and Stp2 is required for derepression of SPS sensor-regulated genes. Cultures of wild-type (CAEB1) and csy1D (CAEB4) strains were grown in SD medium, and total RNA was isolated 1 h after the addition of 5 mM glutamine or an equal aliquot of water. The levels of amino acid permease (CAN1, GAP1, and GAP2), oligopeptide transporter (OPT1 and OPT3), and secreted aspartyl protease (SAP2) transcripts were analyzed by RT-PCR. ACT1 was used as a positive control. (B) STP1* and STP2* exert differential effects on gene expression. Cultures of csy1Δ (CAEB4), csy1Δ plus STP1* (PMRCA84), and csy1Δ plus STP2* (PMRCA77) were grown in SD medium, and total RNA was isolated. The expression of the same set of genes as described above (A) were analyzed by RT-PCR. Phenotypic analysis of WT, csy1Δ, and csh3Δ strains carrying (C) an extra copy of STP1 or STP1* or (D) an extra copy of STP2 or STP2* is shown. Aliquots of cell suspensions were spotted on YPD medium and on YPD medium containing MM, and plates were incubated at 30°C for 2 days and photographed. Strains in C are as follows: WT (CAEB1), WT plus STP1 (PMRCA81), WT plus STP1* (PMRCA82), csy1Δ (CAEB5), csy1Δ plus STP1 (PMRCA83), csy1Δ plus STP1* (PMRCA84), csh3Δ (PMRCA12), csh3Δ plus STP1 (PMRCA24), and csh3Δ plus STP1* (PMRCA25). Strains in D are as follows: WT (CAEB1), WT plus STP2 (PMRCA74), WT plus STP2* (PMRCA76), csy1Δ (CAEB5), csy1Δ plus STP2 (PMRCA75), csy1Δ plus STP2* (PMRCA77), csh3Δ (PMRCA12), csh3Δ plus STP2 (PMRCA45), and csh3Δ plus STP2* (PMRCA46).
Since Stp1 and Stp2 are proteolytically processed in response to extracellular amino acids (Fig. 3), we postulated that the deletion of the N-terminal regions of these factors would result in constitutively active transcription factors. We created the STP1Δ61 (STP1*) and STP2Δ100 (STP2*) deletion alleles lacking codons 2 to 61 and codons 2 to 99, respectively. The breakpoints of these deletions were chosen based on sequence comparisons between ScStp1 and ScStp2 and CaStp1 and CaStp2; a conserved LFP motif in CaStp1 (aa 60 to 62) and a similar IFP motif in CaStp2 (aa 98 to 100) were identified. Analogous mutations in S. cerevisiae constitutively activate SPS sensor-regulated genes (1, 2). The STP1* and STP2* alleles were individually introduced into a csy1Δ strain, and we examined the pattern of gene expression in cells grown in SD medium in the absence of added amino acids (Fig. 4B). The results clearly show that STP1* and STP2* alleles encode constitutively active transcription factors, and consistent with our earlier findings (Fig. 2B), these factors regulate the expression of discrete sets of genes. The expression of SAP2 and OPT1, two genes encoding proteins required for the catabolic utilization of extracellular proteins, was exclusively induced in the strain carrying the STP1* allele. In contrast, amino acid permease-encoding genes CAN1, GAP1, and GAP2 were exclusively expressed in the strain carrying the STP2* allele. As previously found for PTR2 expression (Fig. 2B), OPT3 expression was induced in strains carrying either STP1* or STP2*.
STP2* induces amino acid uptake independent of SPS sensor function.
The specific pattern of gene expression induced by STP1* and STP2* alleles prompted us to examine the growth characteristics of strains carrying these constitutively active alleles. Based on our finding that STP2* but not STP1* induces AAP gene expression, we posited that only STP2* would bypass the requirement of a functional SPS sensor and restore amino acid uptake in csy1Δ cells. We tested this possibility by individually introducing STP1* and STP2* alleles and corresponding full-length alleles (STP1 and STP2) into their endogenous loci in wild-type, csy1Δ, and csh3Δ strains. The resulting strains were spotted on YPD medium and YPD medium containing MM (Fig. 4C and D, respectively). The wild-type strains grew well on either medium, indicating that the presence of constitutive alleles did not adversely affect growth. Due to impaired amino acid uptake capacity, csy1Δ mutants are unable to grow on MM medium. The introduction of extra copies of full-length alleles of STP1 or STP2 did not improve the growth of these strains. In contrast, the csy1Δ strain carrying the STP2* allele exhibited robust growth (Fig. 4D). The ability to complement csy1Δ mutations demonstrates that the deletion of the N-terminal regulatory domain of Stp2 mimics amino acid-induced activation and leads to induced amino acid uptake independent of SPS sensor function. As expected, the STP2* allele barely complemented the growth defect of the csh3Δ mutant, indicating that induced amino acid permease gene expression cannot efficiently bypass the specific secretory block imposed by csh3Δ mutations (36). Interestingly, the STP1* allele weakly complemented the growth defect of both csy1Δ and csh3Δ mutations (Fig. 4C). The ability to complement both csy1Δ and csh3Δ mutations suggested that the STP1* allele enabled cells to use alternative nitrogen sources present in limited amounts in YPD medium, e.g., proteins or oligopeptides. This notion is consistent with our finding that Stp1 transactivates SAP2, OPT1, OPT3, and PTR2 expression, genes that encode proteins that exit the ER and localize to the plasma membrane independently of Csh3 (Fig. 2B and 4B). The heterozygous csy1 and csh3 strains carrying reintroduced wild-type CSY1 and CSH3 alleles, respectively, grew as the wild-type strain on MM medium (data not shown).
stp2Δ null mutants are defective in the utilization of amino acids as the sole nitrogen source.
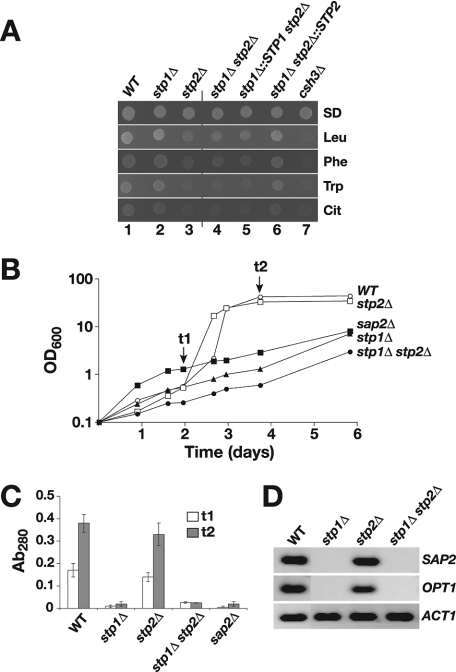
The finding that the STP2* allele constitutively induced the expression of amino acid permease-encoding genes CAN1, GAP1, and GAP2 (Fig. 4B) and complemented the amino acid uptake defects of the csy1Δ mutant (Fig. 4D) suggested that cells lacking STP2 would inefficiently use amino acids as sole nitrogen sources. To test this prediction, we analyzed the growth characteristics of wild-type, isogenic null mutant, and complemented strains on succinate-buffered YNB containing different amino acids as the sole nitrogen source (Fig. 5A). The medium was supplemented with 50 μM histidine to induce the SPS sensor and processing of Stp1 and Stp2 (Fig. 3C), and a csh3Δ mutant was used to control for nonspecific amino acid uptake. All strains grew equally well on plates containing ammonium (SD medium). The WT and the stp1Δ strains were equally able to utilize the amino acids tested (Fig. 5A, compare dilution series 2 with 1). In contrast, the growth of cells lacking STP2 was significantly impaired when forced to use either leucine, phenylalanine, tryptophan, or citrulline (Fig. 5A, compare dilution series 3 to 5 with 1). The growth of the stp2Δ mutants was similar to that exhibited by the csh3Δ mutant (Fig. 5A, compare dilution series 3 to 5 with 7). Reintroduction of STP2, but not STP1, into the stp1Δ stp2Δ double mutant restored growth to the level of the stp1Δ mutant (Fig. 5A, compare dilution series 5 and 6 with 2). These results indicate that Stp2 is required for proper amino acid uptake and that Stp1 is not.
FIG. 5.
Stp1 and Stp2 differentially control two discrete pathways for assimilating nitrogen. (A) Stp2 is required for the proper uptake and utilization of amino acids. Aliquots of cells suspensions of WT (PMRCA18), stp1Δ (PMRCA59), stp2Δ (PMRCA57), stp1Δ stp2Δ (PMRCA94), stp1Δ::STP1 stp2Δ (PMRCA95), stp1Δ stp2Δ::STP2 (PMRCA96), and csh3Δ (PMRCA12) were spotted on succinate-buffered YNB containing 50 μM histidine and 1 mM of the indicated amino acid or citrulline (Cit). Cultures were incubated at 30°C. (B to D) Stp1 is required for the use of protein as the sole nitrogen source. Single colonies of WT (PMRCA18), stp1Δ (PMRCA59), stp2Δ (PMRCA57), stp1Δ stp2Δ (PMRCA94), and sap2Δ (SAP2MS4B) strains growing on solid YPD medium were inoculated into YCB-BSA medium, and growth (OD600) was monitored for 6 days at 37°C (B). Proteolytic activity present in culture supernatants was determined after 2 (t1) and 4 (t2) days (C). Total RNA isolated from cells at t1 and SAP2 and OPT1 expression were analyzed by RT-PCR (D). ACT1 was used as a positive control. Ab280, absorbance at 280 nm.
stp1Δ null mutants do not express SAP2 and are unable to utilize BSA as the sole nitrogen source.
The results presented so far demonstrate a remarkable degree of conservation of SPS sensor signaling pathways in C. albicans and S. cerevisiae with respect to regulating amino acid uptake. However, the finding that in C. albicans Stp1 does not appear to induce AAP gene expression and transactivates SAP2, a gene not present in the S. cerevisiae genome, is striking. This prompted us to examine whether STP1 is required when cells are forced to catabolize and utilize extracellular proteins. Strains lacking SAP2 grow poorly in medium containing protein as the sole nitrogen source (26). Thus, if Stp1 is indeed required for SAP2 expression, stp1Δ cells should also grow poorly in protein-based media. The growth of wild-type, stp1Δ, stp2Δ, and stp1Δ stp2Δ mutants in medium containing BSA as the sole nitrogen source (YCB-BSA medium) was assessed (Fig. 5B). For purposes of comparison, the growth of a sap2Δ mutant was monitored in parallel. Colonies of cells growing on YPD medium were resuspended in YCB-BSA medium to an optical density at 600 nm (OD600) of 0.1, the cultures were incubated at 37°C, and changes in cell densities were monitored spectrophotometrically (OD600) for a period of 6 days. During the first 2 days, all strains grew slowly. After 2 days, the wild-type and stp2Δ strains entered a phase of robust growth, and cultures reached a final OD600 of 44 and 35, respectively. In contrast, the strains lacking STP1 (stp1Δ and stp1Δ stp2Δ) continued to grow slowly, exhibiting identical rates of growth as the sap2Δ mutant; after 6 days, these cultures reached an OD600 of 7, 3, and 8, respectively. The lengthy lag phase experienced by wild-type and stp2Δ cells suggests that cells adapted for growth in media with high concentrations of amino acids must deplete intracellular nitrogen stores prior to expressing genes enabling them to catabolize extracellular proteins.
To test this possibility, we determined the protease activity present in the supernatants of YCB-BSA cultures at two time points (Fig. 5C). The first samples (t1) were analyzed at day 2, when all cultures had similar cell densities (OD600 ≈ 0.5); the second samples were analyzed when the wild-type and stp2Δ cultures had just entered stationary phase (t2) (Fig. 5B). Protease activity was readily detected and at similar levels in supernatants obtained from the wild-type and stp2Δ cultures. In these supernatants, the protease activity increased during growth; the activity at t2 was twofold higher than that at t1. Thus, the deletion of STP2 did not effect the expression of secreted proteases. In contrast, the supernatants from cultures of strains lacking STP1 (stp1Δ and stp1Δ stp2Δ) contained low levels of protease activity, as low as that present in supernatants from the sap2Δ mutant. These findings suggested that the level of protease activity present in culture supernatants was primarily dependent upon SAP2 expression. We analyzed the levels of SAP2 expression in cells at the t1 time point using RT-PCR (Fig. 5D). SAP2 was readily amplified using RNA extracted from wild-type and stp2Δ cells as a template but not from either of the stp1Δ strains. Similarly, the oligopeptide transporter gene OPT1 could not be amplified using RNA from strains lacking STP1. These results demonstrate that the inability of stp1Δ mutants to grow on YCB-BSA medium is due to their inability to express SAP2, and they are therefore unable to degrade extracellular proteins.
Micromolar concentrations of extracellular amino acids trigger the catabolic utilization of extracellular protein in an Stp1-dependent manner.
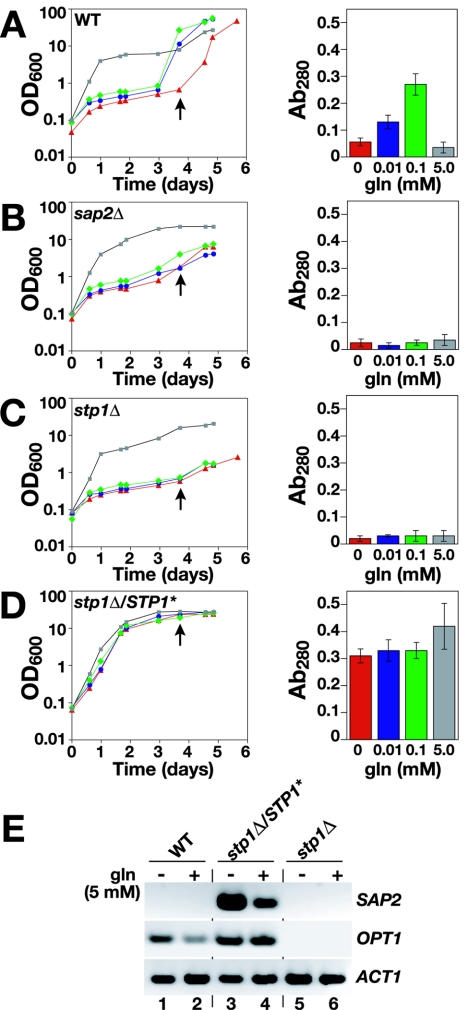
The results presented above have clearly shown that SAP2 expression depends on Stp1 (Fig. 4B and 5D). As Stp1 is synthesized as a latent factor that undergoes proteolytic processing in response to the presence of extracellular amino acids (Fig. 3A), we tested whether the addition of low concentrations of extracellular amino acids to YCB-BSA medium would induce SAP2 expression and reduce the time required for cells to adapt to using extracellular proteins as the sole nitrogen source. The cell density and the levels of secreted protease were determined in cultures of wild-type, sap2Δ, and stp1Δ strains shifted from YPD to YCB-BSA medium supplemented with 0, 0.01, 0.1, or 5 mM glutamine (Fig. 6). In the presence of 5 mM glutamine, all strains grew without a lag, and cultures reached similar high cell densities despite barely detectable levels of secreted protease. The observation that the sap2Δ and stp1Δ mutants grew as well as the wild type indicated that at high concentrations, glutamine provides cells with sufficient nitrogen for growth.
FIG. 6.
Extracellular amino acids induce the protein utilization response in an Stp1-dependent manner. (A to D) Cells with the indicated genotypes grown on solid YPD medium were inoculated in liquid YCB-BSA medium supplemented with 0 (red), 0.01 (blue), 0.1 (green), or 5 mM (gray) glutamine. The cultures were incubated at 37°C, and growth (OD600) was monitored for 5 to 6 days (left panels). The proteolytic activities present in culture supernatants at day 4 (arrow) were determined (right panels). (E) The dominant active STP1* allele constitutively derepresses SAP2 and OPT1 expression in the absence of protein and even in the presence of repressing concentrations of amino acids. Cultures of strains were pregrown in SD medium and induced with 5 mM glutamine for 1 hour as indicated, total RNA was extracted, and the level of SAP2 and OPT1 expression was determined by RT-PCR. ACT1 was used as a positive control. The strains analyzed were WT (PMRCA18), sap2Δ/Δ (SAP2MS4B), stp1Δ/Δ (PMRCA59), and stp1Δ/STP1* (PMRCA60). Ab280, absorbance at 280 nm.
In the absence of supplemental glutamine, wild-type cells exhibited a clear lag phase and entered exponential phase on day 4 (Fig. 6A, left). In media supplemented with 0.01 or 0.1 mM glutamine, the wild-type cells experienced a shorter lag phase, and exponential growth initiated on day 3; the reduced adaptation time correlated with higher levels of protease activity in culture supernatants (Fig. 6A, right). The sap2Δ mutant grew slowly in media without added glutamine (Fig. 6B). The presence of 0.01 or 0.1 mM glutamine did not improve growth, indicating that these concentrations of glutamine are too low to supply cells with nitrogen for growth. Similarly, the stp1Δ mutant also grew slowly, and culture supernatants contained low levels of protease activity (Fig. 6C).
The critical role of Stp1 to induce SAP expression was further demonstrated by introducing the STP1* allele into the stp1Δ mutant strain (Fig. 6D). The STP1* allele constitutively complemented the growth defect imposed by the stp1Δ mutation; cells grew equally well in the absence or presence of glutamine, and the culture supernatants contained high levels of protease activity. Importantly, the cells did not experience a lag in growth, indicating that the constitutive expression of Sap2 enables cells to immediately hydrolyze BSA to obtain nitrogen for growth. These growth tests indicate that the presence of low concentrations of extracellular amino acids, not proteins, provides the initiating signals that ultimately regulate the functional expression of secreted proteases.
The amino acid-induced processing of Stp1 is the critical regulatory step that enables this factor to promote SAP2 expression. High concentrations of extracellular amino acids are known to prevent the production of SAPs (25); consistently, we found that supernatants from wild-type cells grown in the presence of 5 mM glutamine did not contain secreted proteases (Fig. 6A). Stp1 levels are low in cells grown in media containing high concentrations of amino acids (Fig. 3D), an observation that suggested a possible explanation for the lack of SAP activity. To examine this further, we determined the expression of SAP2 in SD medium-grown wild-type, stp1Δ/STP1*, and stp1Δ cells 1 h after the addition of 5 mM glutamine (Fig. 6E). In the presence of this high concentration of glutamine, wild-type cells did not express SAP2. The stp1Δ mutant strain carrying the dominant positive STP1* allele constitutively expressed SAP2 and OPT1 genes. As expected, neither SAP2 nor OPT1 transcripts were detected in cells lacking Stp1 (Fig. 6E, lanes 5 and 6). These results suggest that the growth lag experienced by wild-type cells that are shifted from YPD to YCB-BSA medium is dependent upon the nitrogen regulation of Stp1 expression. Accordingly, as cells utilize and deplete internal amino acid stores, Stp1 levels increase, leading to the expression of genes required for the catabolic utilization of proteins.
DISCUSSION
This study focused on the physiological role of the SPS-sensing pathway in C. albicans. We identified and characterized the effector components of this pathway and the mechanisms that regulate their activity. Our results show that the transcription factors Stp1 and Stp2 are synthesized as latent precursors, and in response to extracellular amino acids, they are proteolytically processed, a requisite event for their ability to efficiently induce gene expression. Strikingly, Stp1 and Stp2 exhibit a clear dichotomy in the genes they transactivate. Stp1 activates a set of genes that are required for the utilization of extracellular proteins (Fig. 4, 5, and 6), and Stp2 activates genes encoding AAPs (Fig. 2, 4, and 5). Additionally, the steady-state levels of Stp1 are controlled by amino acid availability (Fig. 3D). This latter result suggests that cells preferentially use extracellular amino acids when they are available and only induce secreted hydrolytic proteases when amino acids become limiting. This finding is consistent with previous studies that have shown that concentrations of amino acids sufficient to support nitrogen source requirements inhibit protease expression (4, 25) and that lower concentrations of amino acids (>3 mM) stimulate protease secretion even in the absence of extracellular proteins (24) and enhance di- and tripeptide transport (7). In summary, our results have illuminated the hierarchy of regulatory mechanisms in C. albicans that differentially control two discrete pathways for the assimilation of nitrogen.
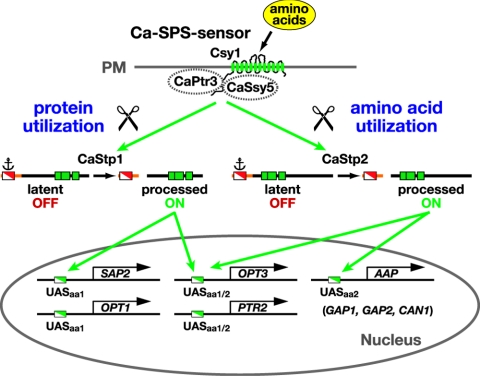
Our findings are consistent with a model schematically presented in Fig. 7. In analogy to S. cerevisiae (1, 2, 19), it is likely that CaPtr3 (orf19.4535) and CaSsy5 (orf19.6422) function together with Csy1 (9) at the plasma membrane forming a ligand-activated receptor complex (Ca-SPS sensor). The latent forms of Stp1 and Stp2 are proteolytically processed in response to extracellular amino acids and in a strictly SPS sensor-dependent manner. As is the case in S. cerevisiae (1), the full-length forms of Stp1 and Stp2 localize to the cytosol due to the presence of a cytoplasmic retention signal (anchor) in their N-terminal regulatory domains. These retention signals prevent the unprocessed full-length forms from efficiently entering the nucleus. In the presence of inducing amino acids, the SPS sensor is activated, leading to the proteolytic processing of Stp1 and Stp2 (scissor); the shorter activated forms of Stp1 and Stp2 lacking the inhibitory domains are targeted to the nucleus, where they induce transcription. The finding that Stp1 and Stp2 activate specific sets of genes is most easily explained if these factors bind to distinct upstream activating sequences (UASaa) in the promoters of amino acid-controlled genes. The processed form of Stp1 binds to UASaa1 present in the promoters of genes required for protein utilization (e.g., SAP2 and OPT1), whereas processed Stp2 binds to UASaa2 to induce the expression of amino acid permease genes (AAP). The transcription of PTR2 and OPT3 can be induced by both transcription factors, suggesting the presence of both UASaa1 and UASaa2 within their promoters.
FIG. 7.
Model of amino acid-induced activation of Stp1-dependent protein and Stp2-dependent amino acid utilization pathways. Csy1 functions together with CaPtr3 and CaSsy5 at the plasma membrane (PM) as a sensor of extracellular amino acids. The transcription factors Stp1 and Stp2 (DNA binding motifs, green boxes) are synthesized as inactive precursors that are restricted from entering the nucleus due to the presence of a cytoplasmic retention signal (anchor). In the presence of amino acids, the activated SPS sensor induces the proteolytic processing of Stp1 and Stp2 (scissor). The shorter activated forms of Stp1 and Stp2, lacking the inhibitory domains located in their N termini, are targeted to the nucleus, where they bind SPS sensor-regulated promoters (UASaa) and induce transcription. Processed Stp1 binds to UASaa1 in the promoters of genes required for protein utilization (e.g., SAP2 and OPT1), whereas processed Stp2 binds to UASaa2 in the promoters of amino acid permease genes (AAP). The expression of PTR2 and OPT3 can be induced by either factor, presumably due to the presence of both UASaa1 and UASaa2 within their promoters.
Experimental support for this model includes the following observations. First, mutants lacking Stp1 and Stp2 exhibit growth phenotypes consistent with the specific loss of gene expression; due to the inability to derepress SAP2 and OPT1 expression (Fig. 4 to 6), stp1Δ strains are unable to grow in media with protein as the sole nitrogen source (Fig. 5 and 6). Similarly, due to the inability to derepress AAP gene expression (Fig. 2 and 4), stp2Δ strains are unable to grow efficiently under conditions that require amino acid uptake (Fig. 2 and 5). Second, processing of Stp1 and Stp2, an event that liberates a 10-kDa N-terminal fragment, is induced by amino acids and is strictly dependent upon Csy1 and Csh3 (Fig. 3). Third, the STP1* and STP2* alleles lacking the 5′ region encode constitutively active transcription factors, clearly indicating that the N-terminal domains of Stp1 and Stp2 possess negative regulatory functions (Fig. 4). Strains carrying the STP1* allele constitutively express SAP2 and OPT1 and consequently grow without experiencing a lag phase when shifted from amino acid- to protein-based media (Fig. 6). The STP2* allele completely suppresses csy1Δ mutant growth phenotypes and induces AAP gene expression in amino acid-free medium (Fig. 4).
Compared to other characterized signaling pathways, the SPS-sensing pathway appears strikingly simple. However, a more complex interplay between specific and general factors undoubtedly exists. Depending upon growth conditions, cross talk between intersecting signaling pathways may affect the expression of particular SPS sensor-regulated genes. An interesting example of this is the dipeptide transporter (PTR2) in S. cerevisiae. Extracellular amino acids induce the expression of PTR2 in an SPS sensor-dependent manner (5); once induced, dipeptides entering cells stimulate Ubr1p-mediated ubiquitylation of Cup9p, a repressor that restricts the full level of PTR2 expression (10, 50). Additionally, in S. cerevisiae, several general factors that affect the expression of SPS sensor-modulated genes have been described, and conserved orthologs exist in Candida. These include Abf1, Leu3, Tup1, Ssn6, Uga35, ubiquitin, and the SCF-Grr1 E3 ubiquitin ligase complex (8, 15, 19, 27, 40). The rules governing how these factors operate to control gene expression remain to be elucidated.
Before this study, it was proposed that oligopeptides generated from the action of secreted proteases induce SAP2 expression (25, 32, 47). This hypothesis was supported by the observations that SAP2 expression was enhanced by the addition of early-log-phase YCB-BSA culture supernatants and was repressed by coaddition of proteinase inhibitor (25). These observations by no means contradict our conclusion that amino acids act as the primary inducers of SAP2 expression. Accordingly, when cells are shifted to media containing protein as the sole nitrogen source, amino acids generated by basal levels of secreted proteases could trigger the induced expression of SAP2. Indeed, it has been shown that aspartic proteinases can cleave di- and tripeptides, although quite inefficiently, to liberate free amino acids (3, 13). Although a parallel positive-feedback regulatory mechanism, similar to Cup9p regulation in S. cerevisiae, cannot be ruled out, the fact that the dominant allele STP1* induces SAP2 transcription in SD medium lacking peptides (Fig. 4) indicates a major role of the SPS sensor in the regulation of SAP2.
SAPs are known to have multiple roles in promoting virulent growth. The SAP-catalyzed hydrolysis of host cell membrane proteins facilitates adhesion and tissue invasion, and SAPs have also been implicated in neutralizing cells and molecules of the host defense system, thereby enabling C. albicans cells to avoid or resist antimicrobial attack (reviewed in reference 38). Sap2 has been extensively studied, and Candida cells lacking this protein exhibit a reduced capacity to infect mammalian hosts (26, 43, 51). However, the regulation of individual SAP genes by host signals in vivo has not been defined (47, 48). Furthermore, it has recently been shown that the expression of genes encoding several SAPs, oligopeptide transporters, and AAPs is highly upregulated upon phagocytosis by macrophages, suggesting that proteolysis and uptake of peptides and amino acids are important processes in surviving phagocytosis (34). Although we have yet to examine whether stp1Δ, stp2Δ, and stp1Δ stp2Δ mutants are less virulent than wild-type strains, based on our previous findings that strains carrying csh3Δ mutations are less virulent (36), we predict that reduced virulence will be observed.
Finally, although several nutrient-regulated signal transduction pathways are known in Candida, including mitogen-activated protein kinase and cyclic AMP cascades, strikingly little is known about the primary sensing mechanisms that enable C. albicans cells to perceive changes in their growth environment and the precise nature of the downstream signaling pathways that ultimately affect patterns of gene expression. Our identification that the transcription factors Stp1 and Stp2 are the direct downstream components of the SPS sensor signaling pathway is intriguing, and it is noteworthy that these factors regulate not only AAP expression but also peptide transporters and SAP2 expression. The SPS sensor signaling pathway has apparently adapted in the course of evolution to enable Candida to effectively use both amino acids and proteins as nitrogen sources. Furthermore, these results illustrate how a rather mundane physiological process, i.e., nitrogen acquisition, can become an “accidental” but major virulence trait of an opportunistic human pathogen. Clearly, a more comprehensive understanding of the nutrients used by Candida and how this fungus senses and responds to the nutrient content within infected hosts will facilitate an understanding of the aggressive nature of this pathogen. Since the SPS sensor components are conserved in several fungal pathogens, the knowledge gained from analyzing this novel sensing system in Candida may provide the means to better understand similar mechanisms controlling virulent growth in other fungal pathogens.
Supplementary Material
Acknowledgments
We thank the members of the Ljungdahl laboratory for constructive comments throughout the course of this work and Claes Andréasson, Mirta Boban, and Stijn Heessen for critical comments on the manuscript. We also thank C. Ben Mamoun, G. R. Fink, A. Brown, W. A. Fonzi, J. Morschhäuser, J. Köhler, and A. Mitchell for strains and plasmids. Finally, we acknowledge the Stanford Sequence Database for making the C. albicans sequence available.
This research was supported by the Ludwig Institute for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andréasson, C., and P. O. Ljungdahl. 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24:7503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balbaa, M., A. Cunningham, and T. Hofmann. 1993. Secondary substrate binding in aspartic proteinases: contributions of subsites S3 and S′2 to kcat. Arch. Biochem. Biophys. 306:297-303. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, A., K. Ganesan, and A. Datta. 1991. Induction of secretory acid proteinase in Candida albicans. J. Gen. Microbiol. 137:2455-2461. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, D., W. Lai, M. Breslav, F. Naider, and J. M. Becker. 1998. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 29:297-310. [DOI] [PubMed] [Google Scholar]
- 6.Basrai, M. A., M. A. Lubkowitz, J. R. Perry, D. Miller, E. Krainer, F. Naider, and J. M. Becker. 1995. Cloning of a Candida albicans peptide transport gene. Microbiology 141:1147-1156. [DOI] [PubMed] [Google Scholar]
- 7.Basrai, M. A., H. L. Zhang, D. Miller, F. Naider, and J. M. Becker. 1992. Toxicity of oxalysine and oxalysine-containing peptides against Candida albicans: regulation of peptide transport by amino acids. J. Gen. Microbiol. 138:2353-2362. [DOI] [PubMed] [Google Scholar]
- 8.Bernard, F., and B. André. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41:489-502. [DOI] [PubMed] [Google Scholar]
- 9.Brega, E., R. Zufferey, and C. B. Mamoun. 2004. Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd, C., G. C. Turner, and A. Varshavsky. 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 12.Cannon, R. D., H. F. Jenkinson, and M. G. Shepherd. 1992. Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol. Gen. Genet. 235:453-457. [DOI] [PubMed] [Google Scholar]
- 13.Cao, Q. N., M. Stubbs, K. Q. Ngo, M. Ward, A. Cunningham, E. F. Pai, G. C. Tu, and T. Hofmann. 2000. Penicillopepsin-JT2, a recombinant enzyme from Penicillium janthinellum and the contribution of a hydrogen bond in subsite S3 to k(cat). Protein Sci. 9:991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, X., B. B. Magee, D. Dawson, P. T. Magee, and C. A. Kumamoto. 2004. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol. Microbiol. 51:551-565. [DOI] [PubMed] [Google Scholar]
- 15.de Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen, and R. J. Planta. 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28:974-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Didion, T., M. Grausland, C. Kielland-Brandt, and H. A. Andersen. 1996. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178:2025-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didion, T., B. Regenberg, M. U. Jørgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 18.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 21.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 23.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van de Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29:556-564. [DOI] [PubMed] [Google Scholar]
- 24.Homma, M., H. Chibana, and K. Tanaka. 1993. Induction of extracellular proteinase in Candida albicans. J. Gen. Microbiol. 139:1187-1193. [DOI] [PubMed] [Google Scholar]
- 25.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 26.Hube, B., D. Sanglard, F. C. Odds, D. Hess, M. Monod, W. Schafer, A. J. Brown, and N. A. Gow. 1997. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Island, M. D., F. Naider, and J. M. Becker. 1987. Regulation of dipeptide transport in Saccharomyces cerevisiae by micromolar amino acid concentrations. J. Bacteriol. 169:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 30.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kota, J., and P. O. Ljungdahl. 2005. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 168:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lerner, C. G., and R. C. Goldman. 1993. Stimuli that induce production of Candida albicans extracellular aspartyl proteinase. J. Gen. Microbiol. 139:1643-1651. [DOI] [PubMed] [Google Scholar]
- 33.Limjindaporn, T., R. A. Khalaf, and W. A. Fonzi. 2003. Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50:993-1004. [DOI] [PubMed] [Google Scholar]
- 34.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubkowitz, M. A., L. Hauser, M. Breslav, F. Naider, and J. M. Becker. 1997. An oligopeptide transport gene from Candida albicans. Microbiology 143:387-396. [DOI] [PubMed] [Google Scholar]
- 36.Martínez, P., and P. O. Ljungdahl. 2004. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol. Microbiol. 51:371-384. [DOI] [PubMed] [Google Scholar]
- 37.Martínez, P., and P. O. Ljungdahl. 2000. The SHR3 homologue from S. pombe demonstrates a conserved function of ER packaging chaperones. J. Cell Sci. 113:4351-4362. [DOI] [PubMed] [Google Scholar]
- 38.Naglik, J. R., S. J. Challacombe, and B. Hube. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson, J. F., and M. A. Pesce. 2000. Reference ranges for laboratory tests and procedures, p. 2181-2229. In R. E. Behrman, R. M. Kliegman, and H. B. Jenson (ed.), Nelson textbook of pediatrics, 16th ed. W. B. Saunders Company, Philadelphia, Pa.
- 40.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 41.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 42.Ross, I. K., F. De Bernardis, G. W. Emerson, A. Cassone, and P. A. Sullivan. 1990. The secreted aspartate proteinase of Candida albicans: physiology of secretion and virulence of a proteinase-deficient mutant. J Gen. Microbiol. 136:687-694. [DOI] [PubMed] [Google Scholar]
- 43.Sanglard, D., B. Hube, M. Monod, F. C. Odds, and N. A. Gow. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman, F. 1991. Getting started with yeast, p. 3-21. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 45.Silve, S., C. Volland, C. Garnier, R. Jund, M. R. Chevallier, and R. Haguenauer-Tsapis. 1991. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell Biol. 11:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soll, D. R. 2002. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 81:101-110. [DOI] [PubMed] [Google Scholar]
- 47.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 48.Staib, P., M. Kretschmar, T. Nichterlein, G. Kohler, S. Michel, H. Hof, J. Hacker, and J. Morschhauser. 1999. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol. Microbiol. 32:533-546. [DOI] [PubMed] [Google Scholar]
- 49.Sychrova, H., and J. L. Souciet. 1994. CAN1, a gene encoding a permease for basic amino acids in Candida albicans. Yeast 10:1647-1651. [DOI] [PubMed] [Google Scholar]
- 50.Turner, G. C., F. Du, and A. Varshavsky. 2000. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405:579-583. [DOI] [PubMed] [Google Scholar]
- 51.Vilanova, M., L. Teixeira, I. Caramalho, E. Torrado, A. Marques, P. Madureira, A. Ribeiro, P. Ferreira, M. Gama, and J. Demengeot. 2004. Protection against systemic candidiasis in mice immunized with secreted aspartic proteinase 2. Immunology 111:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, T. C., and N. Agabian. 1995. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J. Bacteriol. 177:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.