Abstract
We have previously shown that Saccharomyces cerevisiae Isw2 complex slides nucleosomes to remodel chromatin in vivo. Our data suggested a model in which Isw2 complex binds the histone octamer and DNA separately to generate the force necessary for nucleosome movement. Here we find that the histone H4 “basic patch” is the only portion of any amino-terminal histone tail required for both target-specific association of Isw2 complex with chromatin and chromatin remodeling in vivo, whereas it is dispensable for basal levels of chromatin binding. Similarly, we find that nonremodeled chromatin structure and integrity of Isw2 complex are required only for target-specific association of Isw2 with chromatin. These data demonstrate fundamental differences between the target-specific and basal modes of chromatin binding by Isw2 complex in vivo and suggest that only the former involves contributions from DNA, histone H4, and sequence-specific DNA binding proteins. We propose a model for target recognition and chromatin remodeling by Isw2 complex in vivo.
In eukaryotes, chromatin structure imposes a barrier to processes that require access to DNA, due to the fact that histone-DNA interactions block DNA accessibility and distort its double helical structure within the nucleosome (26). Consequently, chromatin structure must be dynamically regulated in vivo: whereas disruption of chromatin structure is necessary to facilitate transcription, DNA replication, DNA repair, and other functions, these processes can be repressed in specific chromosomal regions by the active formation of inaccessible chromatin structure.
Two main classes of enzymes have evolved to remodel chromatin structure: histone modifying enzymes and ATP-dependent chromatin remodeling factors. ATP-dependent chromatin remodeling factors function by altering histone-DNA contacts of nucleosomes in regions of chromatin to which they are targeted. Their activities include assembly or disruption of nucleosomes (1, 3, 11, 36), sliding of nucleosomes to new positions on DNA (29, 43), and exchange of histones within existing nucleosomes (4, 25, 27, 35). All known ATP-dependent chromatin-remodeling factors are related through homology of their ATPase domains and fall into several conserved families, including Swi/Snf, ISWI, Chd/Mi-2, Ino80, and Rad54.
Members of the ISWI class of ATP-dependent chromatin remodeling factors display a number of biochemical activities in vitro, including chromatin disruption, nucleosome sliding, nucleosome spacing, and chromatin assembly activities (28). However, different ISWI complexes catalyze distinct subsets of these activities. For example, while Drosophila NURF can disrupt chromatin structure in vitro (39), ACF and CHRAC (from Drosophila and humans) and RSF (from humans) catalyze chromatin assembly and nucleosome spacing in vitro but do not catalyze chromatin disruption (21, 30-32, 40). However, all ISWI complexes tested to date catalyze nucleosome sliding in vitro, suggesting that this process may be central to the functions of all ISWI complexes in vivo (9, 18, 21, 23, 30, 42).
ISWI complexes have been implicated in a variety of nuclear processes in vivo, such as regulation of transcription, DNA replication, and chromosome segregation. However, the mechanisms by which ISWI complexes remodel chromatin in vivo are not well understood. Recently, we showed that Saccharomyces cerevisiae Isw2 complex functions in vivo at two classes of chromosomal target genes by sliding nucleosomes into a compact configuration that represses transcription (9). At the POT1 promoter, Isw2 complex slides nucleosomes extending from the promoter into the coding region into more upstream positions, whereas at the REC104 promoter, Isw2 complex slides upstream nucleosomes to downstream positions. Previously, we showed that Isw2 complex was recruited to the promoters of early meiotic genes such as REC104 by direct interaction with the sequence-specific DNA binding protein Ume6p (14). Upon close inspection of our previous results, we found that Isw2 complex appears to slide an upstream nucleosome to a position immediately adjacent to or possibly overlapping with the Ume6p-binding site URS1 at the REC104 promoter (9). Therefore, Isw2 complex functions at this locus by sliding nucleosomes toward its own site of recruitment. These results suggest a model in which Isw2 complex slides nucleosomes by forcing linker DNA adjacent to its site of recruitment into the nearest nucleosome, as proposed based on in vitro experiments (3). One prediction of this model is that Isw2 complex should require separate binding sites for the histone octamer and DNA in order to generate the force necessary to push DNA into the nucleosome.
Several possible candidates for an Isw2 complex-binding site on the histone octamer have emerged in recent years. A basic portion of the histone H4 amino-terminal tail was previously found to be required for stimulation of the ATPase activity of Drosophila ISWI monomer and nucleosome sliding by NURF complex in vitro (5, 6, 17), and the H4 lysine 16 acetyltransferase MOF appears to inhibit the function of Drosophila ISWI in vivo (7). While the H4 basic patch was not required for chromatin binding by ISWI in vitro, it appeared to function as an allosteric activator of the ISWI ATPase activity. A human ISWI complex, RSF, exhibits a complete requirement for the histone H4 tail for chromatin assembly, as well as a partial requirement for the H2A and H2B tails (32). In addition, association of yeast Isw1p with chromatin at the MET16 gene in vivo requires methylation of lysine 4 on histone H3 by the histone methyltransferase Set1p (37). However, Isw1 complex does not directly interact with the lysine 4-methylated H3; how Isw1 complex interacts with chromatin at the MET16 gene in vivo remains to be determined (37). Similarly, colocalization of a human ISWI complex with the H3 lysine 4 methylation mark in vivo has been reported, although it is unclear in this case whether lysine 4 methylation is required for association of the complex with chromatin (16). A role for the H3 tail in the function of ISWI complexes was further supported by biochemical studies implicating the H3 tail in binding of nucleosomes by ISWI proteins in vitro (15, 22, 37). While these examples illustrate the requirements for histone tails for the functions of ISWI complexes, there is no evidence of direct binding of histone tails by ISWI complexes.
We recently found that Isw2 interacts with chromatin in two distinct modes, target-specific and basal levels of binding (12). However, the molecular basis for these distinct chromatin interactions is unknown. In this report we examine the features of chromatin necessary for binding and chromatin remodeling by Isw2 complex in vivo. Mutational analysis of each N-terminal histone tail revealed that the basic patch of histone H4 (R17 H18 R19), was solely required for chromatin remodeling by Isw2 complex in vivo. In addition, we found that the basic patch was essential for Isw2p to specifically associate with its target sites on chromatin and that this target-specific interaction required intact Isw2 complex. Furthermore, we found a partial requirement for nonremodeled chromatin structure for localization of Isw2 complex with its target sites in vivo. Unexpectedly, while the H4 basic patch, nonremodeled chromatin structure, the integrity of the complex, and sequence-specific DNA binding proteins were all required for target-specific chromatin interaction by Isw2, they were all dispensable for basal levels of chromatin binding. Our results demonstrate fundamental mechanistic differences between the two modes of chromatin interactions by Isw2 complex and suggest a model for how Isw2 complex identifies and remodels its chromatin targets in vivo.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
All S. cerevisiae strains were derived from W1588-4c, which is congenic to W303 (45). Strains expressing Isw2p with three tandem copies of the FLAG tag at its C terminus were constructed as described previously (13). The histone tail deletions and point mutations were constructed by site-directed mutagenesis, sequenced, and subcloned into pRS414. The deletion mutants of the H3 tail and the H4 tail were provided by Dan Gottschling and Jay Vary, respectively. For expression of H3-H4 basic patch mutant tetramer, the R17A R19A mutation was made by site-directed mutagenesis in a pET vector containing a version of yeast H4 in which codon usage was optimized for bacterial expression (13). This mutant H4 gene, along with wild-type yeast H3, were then cloned sequentially into the pET-DUET-1 vector.
Nuclease digestions and Southern hybridizations.
DNase I digestions and indirect end labeling experiments were performed as described previously (9), except cells were grown in YEPD (2% Bacto peptone 1% Bacto yeast extract, 2% dextrose) to and optical density at 600 nm of 0.7, at which time cells were harvested and processed. Zymolyase was used at 10 mg/ml.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (9), except that the RNase step was omitted and DNA was purified after reversal of cross-links and proteinase K digestion by one phenol extraction, one phenol-chloroform extraction, and ethanol precipitation. A detailed protocol is available at http://www.fhcrc.org/science/labs/tsukiyama/.
Assembly of NCPs and binding assays.
Wild-type nucleosome core particles containing recombinant yeast histones were assembled on the Xenopus borealis 5S nucleosome positioning sequence (19) by salt dialysis as described previously (13, 41), with a fraction of the DNA radioactively labeled for detection.
A soluble H3-H4 basic patch mutant tetramer was expressed in BL21-CodonPlus(DE3)-RIL cells. Cell extract was made in buffer H containing 100 mM KCl (buffer H0.1) (38). To purify soluble tetramer, the extract from a 2-liter culture was added to 1.5 ml of Q-Sepharose that had been washed twice with buffer H0.1 to “preclear” the extract. After binding at 4°C for 30 min on a rotating wheel, the beads were removed by centrifugation, and the unbound material was added to 1.5 ml of SP-Sepharose that had been washed twice with buffer H0.1. Extract was allowed to bind for 30 min at 4°C on a rotating wheel, and the beads were pelleted as above. Beads were washed twice with buffer H0.1 and transferred to a disposable column (Poly-Prep Chromatography Columns; Bio-Rad). The column was washed once with 5 ml of buffer H0.5 (500 mM KCl), and protein was eluted 5 times with 1.5 ml of buffer H1.0 (1 M KCl). NCPs containing the H4 basic patch mutant were assembled as above for wild-type H4, except that H3-H4 mutant tetramer and wild-type H2A-H2B dimer (obtained previously [13]) was substituted for wild-type histone octamer. Both the wild-type and mutant NCPs were heated at 55°C overnight to obtain a single correctly assembled and positioned species, and the resulting aggregate was filtered out using 0.45-μm-pore-size microcentrifuge filters (Ultrafree-MC; Microcon). Under the conditions used, no free DNA was detected in our preparations of wild-type or mutant NCPs. NCPs were stored at 4°C on ice.
Nucleosome binding assays were performed in 30-μl reaction mixtures containing buffer H0.05 (50 mM KCl). We incubated approximately 20 fmol NCPs for 30 min at room temperature with two different concentrations (approximately 16 and 40 fmol, as estimated by silver staining) of purified wild-type Isw2 or Isw2-K215R complex (13, 14). Approximately 2 μg of anti-Myc (9E10) or anti-FLAG (M2) antibodies was added for supershift experiments. Complexes were resolved on 5% native polyacrylamide gels, using 0.25× Tris-borate-EDTA as the running buffer.
Reconstitution of nucleosome arrays and ATPase assays.
Reconstitution of nucleosome arrays onto immobilized templates and assays of Isw2 ATPase activity were performed as described previously (13). ATPase assays were performed in triplicate on the indicated chromatin preparations.
Whole-genome expression analyses.
The whole-genome expression profile of the H4 R17A R19A basic patch double mutant was determined using spotted microarrays as described previously (8) using two sets of dye-swapped pairs (four total replicates). The data are available for downloading at http://www.fhcrc.org/labs/tsukiyama/supplemental_data/H4basicpatch/.
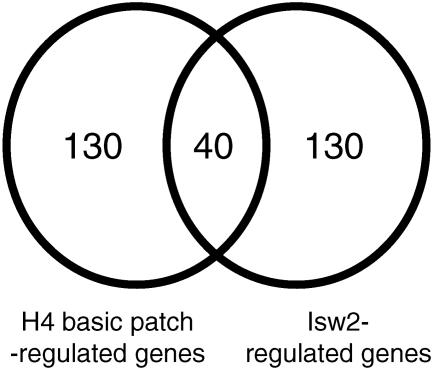
“Isw2-regulated genes” (see Fig. 4) were defined as those genes most upregulated in an isw2Δ rpd3Δ double mutant compared to an rpd3Δ single mutant (8). This allows for identification of Isw2 targets in a background sensitized to isw2 loss and reveals targets of Isw2-dependent repression not identified in analysis of an isw2 single mutant (12). To compare the genes regulated by Isw2 complex to those regulated by the H4 basic patch, we eliminated open reading frames (ORFs) missing in either data set; this step left 5,661 ORFs for further analysis. We then took the top 3% of each data set (170 ORFs) and characterized their degree of overlap in a Venn diagram (see Fig. 4). The significance of the overlap was calculated by hypergeometric distribution (P = 6.2 × 10−26)
FIG. 4.
The H4 basic patch mutant is broadly required for the function of Isw2 complex in vivo. The top 3% of genes upregulated in the H4 R17A R19A double mutant were compared to the top 3% of Isw2-regulated genes (170 genes each) as described in Materials and Methods.
RESULTS
Chromatin remodeling by Isw2 complex in vivo requires amino acids 15 to 19 of histone H4 but no other N-terminal histone tails.
We reasoned that histone mutations that prevented Isw2 complex from remodeling chromatin—due to an inability of Isw2 complex to bind histones or a failure of the mutant histones to support later steps in chromatin remodeling—would result in chromatin structure at Isw2 target genes similar or identical to that of an isw2 mutant. Therefore, we used chromatin structure at Isw2 target genes in vivo as a readout for Isw2 complex function. We constructed yeast strains in which the sole copy of the histone gene of interest was carried on a plasmid with a URA3 marker. We then replaced the wild-type histone genes with the indicated mutants by plasmid shuffling. Chromatin structure in each mutant was assayed by DNase I digestion of chromatin followed by indirect end-labeling.
Initially we focused on chromatin structure at the POT1 gene, which is repressed by Isw2-dependent chromatin remodeling in a parallel pathway with the Sin3/Rpd3 complex (8, 9). Using this assay, we found that the entire N-terminal tails of histones H3, H2A, and H2B were individually dispensable for Isw2-dependent chromatin remodeling (Fig. 1B, lanes 4, 10, and 11). The H4 tail was deleted in smaller segments because deletion of the entire tail causes severe growth defects (data not shown). As shown in Fig. 1B, two of four deletions in the histone H4 N-terminal tail resulted in chromatin structure that appeared identical to that of isw2 mutant cells (lanes 7 and 8). The smallest of these, deleting amino acids 15 to 19, removes most of a run of basic residues previously referred to as the basic patch of the histone H4 tail (Fig. 1A) (6). Importantly, a large deletion of amino acids 4 to 14, removing a significant portion of the H4 tail but leaving the basic patch intact, resulted in wild-type chromatin structure (lane 6). Therefore, the histone H4 basic patch is the only portion of any histone tail that is required for the function of Isw2 complex at this locus. Similar results were obtained analyzing the chromatin structure of a second Isw2 target gene, REC104 (data not shown).
FIG. 1.
A portion of the histone H4 basic patch is required for Isw2-dependent chromatin remodeling. (A) Diagram of the amino-terminal tails of the four core histones. The H4 basic patch is underlined. (B to D) Nucleosome positions at the POT1 gene were analyzed in wild type (WT) and the indicated histone tail mutants using DNase I as a probe of chromatin structure. Triangles and circles indicate DNase I-cut sites that resemble the wild-type and isw2 mutant patterns, respectively. Brackets indicate DNase I-cut sites that resemble intermediate states between the wild-type and isw2 mutant patterns. Size standards, numbered with respect to the POT1 initiation codon, are indicated to the left.
The requirement of Isw2 complex for the basic patch is limited to three residues: R17, H18, and R19.
The H4 basic patch was previously found to be necessary for activation of ATPase activity and facilitation of regular nucleosome spacing by Drosophila ISWI in vitro (5, 6). The authors found that nucleosomes lacking the first 15 or 16 amino acids of histone H4 were partially defective in stimulation of ISWI ATPase activity, while nucleosomes lacking the first 19 amino acids were incapable of stimulating ATPase activity beyond the level of DNA alone (6). To identify the H4 basic patch residues necessary for Isw2 function in vivo, we tested a series of amino acid substitutions within the H4 basic patch. Alanine substitutions in one or both of the lysines at positions 16 and 20 within the basic patch had no effect on Isw2-dependent chromatin remodeling, indicating that these residues were not required for Isw2 function (Fig. 1C, compare lanes 1 to 3 with lanes 4, 9, and 10). In addition, since acetylation of H4 lysine 16 has been implicated in the regulation of Drosophila ISWI activity (7), mutations of lysine 16 to glutamine (to mimic acetylation) or arginine (to conserve charge but prevent acetylation) were also tested. These mutations had no effect on Isw2-dependent chromatin structure (data not shown). While these data do not support a role for lysine 16 acetylation in regulation of Isw2 function through the basic patch, this possibility cannot be ruled out (see Discussion).
Previously, a double mutation in which both arginine 17 and arginine 19 residues were replaced by alanines was found to abolish ATPase stimulation of Drosophila ISWI in vitro (6). When we introduced this double mutation into yeast cells, we found that it resulted in chromatin structure identical to that of an isw2 mutant (Fig. 1C, lane 8). Single alanine substitutions of arginine 17 or histidine 18 (R17A and H18A) resulted in a partial defect in chromatin structure (lanes 5 and 6; note brackets). The single substitution of arginine 19 to alanine (R19A) had no noticeable phenotype (lane 7); however, since this mutation clearly enhances the phenotype of a single R17A substitution (compare lanes 5 and 8), we conclude that arginine 19 plays a role in facilitating chromatin remodeling by Isw2 complex in vivo.
Next we asked whether the basic patch needs only to be basic to promote Isw2-dependent chromatin remodeling or whether the specific sequence arginine-histidine-arginine is important for its function. To distinguish between these two possibilities, we tested the effect of a double mutation changing both arginines to lysines (R17K and R19K) on Isw2-dependent chromatin remodeling. This mutation resulted in an intermediate phenotype similar to that of the R17A and H18A single mutants (Fig. 1D). This indicates that the specific arginine-histidine-arginine sequence of the wild-type basic patch is required for full function in Isw2-dependent chromatin remodeling but that substitutions maintaining the positive charge are partially functional. Together, these results point to a central role for the three interior basic patch residues, arginine 17, histidine 18, and arginine 19, in chromatin remodeling by Isw2 complex in vivo.
An H4 basic patch mutation prevents specific association of Isw2 complex with target loci in vivo.
We next sought to determine the step at which the H4 basic patch is required for Isw2-dependent chromatin remodeling in vivo. Previously, it was shown that Drosophila ISWI was capable of binding chromatin templates in vitro that lacked the H4 basic patch (5, 6). However, basic patch mutant chromatin failed to stimulate the ATPase activity of ISWI, nor did it support ISWI-dependent chromatin remodeling. These findings suggested that the H4 basic patch functioned as an allosteric activator of ISWI at a step subsequent to chromatin binding.
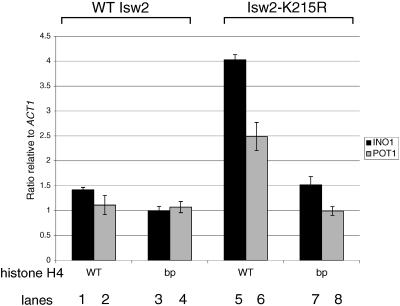
To test this model in vivo, we first determined whether the basic patch is required for Isw2p to interact with its known chromatin targets. We recently found that Isw2p interacts with chromatin by at least two distinct modes (12). The first is a basal level of interaction that is observed globally, which generally does not lead to chromatin remodeling. The second is a target-specific chromatin interaction that depends on DNA binding factors such as Ume6p and α2p and that correlates with sites of Isw2-dependent chromatin remodeling (12, 14). The latter interaction is detected only with a catalytically inactive mutant of Isw2p, Isw2p-K215R, using ChIP assays (12).
We therefore tested the ability of both wild-type Isw2p and Isw2p-K215R to associate with known target sites on chromatin in vivo, in the presence or absence of the R17A R19A H4 basic patch mutation (Fig. 2). Consistent with our previous results (12), the ChIP signals for wild-type Isw2p were similar at Isw2 target promoters POT1 and INO1 and a negative control promoter, ACT1. This level was essentially unchanged in an R17A R19A H4 basic patch mutant (Fig. 2, compare lanes 1 and 2 with 3 and 4). In contrast, we observed a substantial enrichment of Isw2p-K215R cross-linking at the POT1 and INO1 promoters relative to the ACT1 promoter (lanes 5 and 6). However, in the presence of the H4 basic patch mutation, the ChIP signals of Isw2p-K215R at POT1 and INO1 were reduced to levels similar to those of wild-type Isw2p (lanes 7 and 8). Our results show that the basal level of chromatin interaction detected by wild-type Isw2p is independent of the H4 basic patch. In contrast, target-specific chromatin interaction exhibited by Isw2p-K215R is completely dependent on the H4 basic patch. The signals at ACT1 were similar in all strains used (data not shown).
FIG. 2.
The basic patch is the only portion of any N-terminal histone tail required for specific association of Isw2p-K215R with its chromatin targets in vivo. Chromatin immunoprecipitation was performed as described in Materials and Methods. Radioactive duplex PCR was carried out for the POT1 or INO1 and ACT1 promoters, using a serial dilution of input chromatin (to ensure linearity of the PCR) and precipitated chromatin. The fraction of the input that immunoprecipitated was quantitated for each sample using a phosphorimager, and the ratios of the signals from Isw2 targets (POT1 or INO1) to those from a control locus (ACT1) were calculated. The averages and the data points from two experiments are shown by bars and vertical lines, respectively. The signals at ACT1 were similar in all strains used (data not shown). bp, R17A R19A basic patch mutant.
The H4 basic patch is required for binding of Isw2 complex to nucleosome core particles in vitro.
Our finding that the H4 basic patch is required for specific association of Isw2p with target genes in vivo suggests that Isw2 complex may directly bind to the basic patch. However, Drosophila ISWI was previously found to bind nucleosomes lacking the entire H4 N-terminal tail (5), leading to the proposal that the H4 tail was required at a step subsequent to nucleosome binding. However, these experiments employed mononucleosomes assembled with extensive linker DNA on each end, leaving the possibility that DNA might have provided a sufficient platform for ISWI to interact with mononucleosomes in an H4 tail-independent manner. Consistent with this, we have previously shown that both wild-type Isw2 and Isw2-K215R complexes bind nucleosome arrays and free DNA equally well (13). We therefore tested binding of Isw2 to NCPs. NCPs have only enough DNA to form 1.65 turns on the histone octamer, with no free DNA extending outside of the octamer to which Isw2 complex can bind (33). Isw2 complex was recently shown to be capable of interacting with NCPs, though less robustly than its interactions with mononucleosomes with extended linker DNA (22). Similar results were obtained for Drosophila ISWI (44). We reconstituted NCPs as described in Materials and Methods, containing either wild-type or the R17A R19A basic patch mutant histone H4 and assayed binding of Isw2 complex using native polyacrylamide gel electrophoresis.
Upon mixture of Isw2 complex with NCPs, we observed the appearance of a slower-migrating species during native gel electrophoresis (Fig. 3A, open arrow). Furthermore, the intensity of the slower migrating band increased with a higher dose of Isw2 complex. This slower-migrating band was substantially more prominent for wild-type nucleosomes than for those with a mutated H4 basic patch (Fig. 3, compare lanes 2 and 3 with lanes 5 and 6). Upon quantitation of the NCPs shifted in each lane, we found that the fraction of wild-type nucleosomes shifted was 4.0- to 5.5-fold higher than that of the basic patch mutant. To confirm that the shifted species contained Isw2 complex, we added either a control antibody or an antibody directed against the FLAG epitopes fused to the C terminus of Isw2p to the binding reactions (Fig. 3B, lanes 8 and 9, respectively). The control antibody had no effect on either the unshifted NCPs or the shifted species, whereas the anti-FLAG antibody super-shifted the slower-migrating band (Fig. 3B, arrowhead). These data confirm that the slowly migrating species contains Isw2 complex. We conclude that optimal binding of Isw2p to NCPs is dependent upon the histone H4 basic patch. Neither ATP in the binding reactions nor the K215R mutation in Isw2p affected binding of Isw2 complex to NCPs in vitro (data not shown).
FIG. 3.
The H4 basic patch is required for binding of Isw2 complex to NCPs. (A) Electromobility shift assay measuring binding of Isw2 complex to wild-type or mutant NCPs. NCPs bound by Isw2 complex and unbound NCPs are indicated by white and black arrows, respectively. In this experiment we used purified catalytically inactive Isw2 complex, but essentially identical results were obtained with wild-type Isw2 complex (data not shown). There were no detectable effects of Mg-ATP on nucleosome binding by Isw2 complex (data not shown). (B) Wild-type NCPs were incubated with Isw2 complex and the indicated antibody. The supershifted species is marked with an arrowhead. M, anti-Myc; F, anti-FLAG. (C) Wild-type (WT) and mutant histone octamers containing the H4 R17A R19A basic patch double mutation (b.p.) were reconstituted into nucleosome arrays on immobilized templates and digested with micrococcal nuclease (MNase). Purified DNA was subjected to Southern blotting to analyze nucleosome density and verify chromatin assembly; “lo” and “hi” refer to lower and higher nucleosome densities in the assembled chromatin. (D) ATPase assays measuring the fraction of ATP hydrolyzed by Isw2 complex were carried out on samples identical to those described in panel C. The averages and standard deviations of three ATPase assays per sample are shown.
The dependence of Isw2 complex on the H4 basic patch for binding to nucleosome core particles suggests that this epitope should also be important for the biochemical activities of Isw2 complex. To test this possibility, we reconstituted nucleosome arrays onto immobilized templates using either wild-type histones or those with the R17A R19A double mutation within the H4 basic patch (Fig. 3C) and tested the chromatin samples for their ability to stimulate the ATPase activity of Isw2 complex (Fig. 3D). As expected, chromatin lacking a functional H4 basic patch failed to stimulate the ATPase activity of Isw2 complex, consistent with data obtained previously for Drosophila ISWI (6). Therefore, the inability of Isw2 complex to bind nucleosomes lacking a functional H4 basic patch prevents Isw2 function both in vivo and in vitro.
The H4 basic patch is required for Isw2-dependent transcriptional regulation in vivo.
To determine the extent to which the H4 basic patch is required for the functions of Isw2 complex in vivo, we analyzed the whole-genome expression profile of R17A R19A basic patch mutant cells and compared it with the expression profile of an isw2 mutant (8, 12) as described in Materials and Methods. As shown in Fig. 4, the genes most derepressed in the H4 basic patch mutant overlap significantly with those whose transcription is repressed most strongly by Isw2 complex (P = 6.2 × 10−26). Therefore, we conclude that the histone H4 basic patch functions in Isw2-dependent chromatin regulation at many loci in vivo.
Intact Isw2 complex is required for target-specific association of Isw2p-K215R with chromatin.
Nearly all Isw2p exists in one of two complexes in vivo: a four-subunit complex containing Isw2p, Itc1p, Dls1p, and Dpb4p and a smaller Isw2p-Itc1p heterodimeric complex (13, 20, 34, 38). The largest subunit of both complexes, Itc1p, is required for all known in vivo functions and biochemical activities of Isw2 complex (13), and in the absence of Itc1p, Isw2p exists as a monomer in vivo (13; A. D. McConnell and T. Tsukiyama, unpublished). However, the precise role of Itc1p in Isw2-dependent chromatin remodeling is not well understood.
To examine the role of Itc1p in Isw2 complex function in vivo, we used ChIP assays to measure the association of Isw2p and Isw2p-K215R with the Isw2 target genes, POT1 and tT(CGU)K, in the presence or absence of Itc1p. At both target genes, wild-type Isw2p association was similar to the negative control locus, PPA1, in both ITC1 and Δitc1 cells (Fig. 5, compare lanes 1 and 2 and lanes 5 and 6). As expected, Isw2p-K215R was enriched at both Isw2 target genes in ITC1 cells. However, in Δitc1 cells, Isw2p-K215R association at both target genes was reduced to the level of wild-type Isw2p (compare lanes 3 and 4 and lanes 7 and 8). These data indicate that Isw2p must form a complex with Itc1p for robust, target-specific association with chromatin, while Isw2p monomer is sufficient for basal levels of chromatin binding.
FIG. 5.
Itc1p is required for specific association of Isw2p-K215R with Isw2 targets in vivo. Chromatin immunoprecipitations of Isw2p and Isw2p-K215R were carried out in cells with wild type (ITC1) or null mutations (itc1) of the ITC1 gene. Each immunoprecipitation was carried out a total of three times using two independent preparations of chromatin for each strain. Shown are the average ratios and standard deviations of experimental loci relative to PPA1, calculated as follows: enrichment (n-fold) = experimental locus (immunoprecipitation as percentage of input)/PPA1 (immunoprecipitation as percentage of input).
Nonremodeled chromatin structure is required for robust association of Isw2 complex with most target loci in vivo.
Why does a catalytically inactive form of Isw2p preferentially cross-link at targets in vivo? In the presence of wild-type Isw2 complex, chromatin structure at sites of Isw2 recruitment is remodeled by Isw2-dependent nucleosome sliding (9). If Isw2 complex has lower affinity for remodeled chromatin (the product of its enzymatic activity) than for nonremodeled chromatin (its substrate), it may dissociate from its chromatin targets after remodeling is completed, making its interactions with targets transient. In contrast, Isw2p-K215R is unable to remodel chromatin and, therefore, has constant access to the higher affinity nonremodeled substrate.
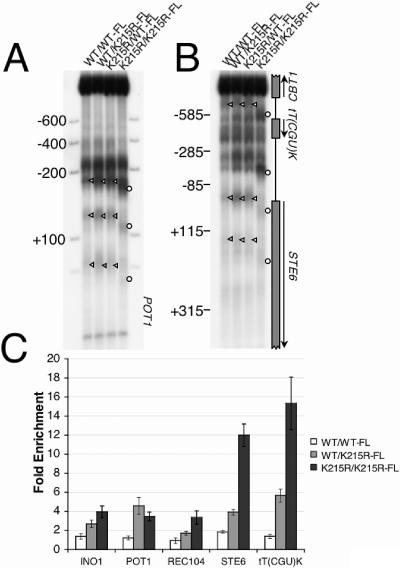
To test this possibility, we required a system in which target-specific chromatin binding of Isw2-K215R complex could be compared in the presence of either wild-type or isw2 mutant chromatin structure. Since isw2-K215R mutant cells have mutant (nonremodeled) chromatin structure, this experiment was not feasible in haploid cells. To solve this problem, we employed diploid yeast strains bearing epitope-tagged alleles of wild-type and catalytically inactive ISW2 expressed under endogenous promoters. As shown in Fig. 6A and B, strains heterozygous for wild-type and catalytically inactive ISW2 exhibit chromatin structure identical to the homozygous wild-type ISW2 strain at several Isw2-target loci: POT1, STE6, and the tT(CGU)K tRNA gene. Similar results were obtained at the REC104 gene (data not shown). In contrast, the homozygous catalytically inactive mutant exhibited chromatin structure identical to that observed previously in haploid isw2 deletion mutants. These data reveal that the isw2-K215R allele does not exhibit a noticeable dominant-negative phenotype in the heterozygote and is completely defective in remodeling chromatin in the homozygous mutant. Therefore, the localization of the Isw2-K215R complex can be compared in the presence of remodeled chromatin structure (in the heterozygote) and nonremodeled chromatin structure (in the homozygous mutant).
FIG. 6.
Isw2 complex requires nonremodeled chromatin structure for robust localization to target promoters in vivo. WT (wild type) and K215R refer to the genotype of each copy of the ISW2 gene. One allele of ISW2 was fused to three copies of the FLAG (FL) epitope. (A and B) The indicated diploid yeast strains were digested with DNase I, and their chromatin structure at Isw2 target genes POT1 (A), STE6 (B), and the nearby tRNA tT(CGU)K (B) was analyzed by indirect end-labeling analysis as above. (C) Chromatin immunoprecipitation of the diploid strains indicated in panel A. The relative enrichment (n-fold) for each ChIP was calculated as described in the legend of Fig. 5, except that ACT1 was used as the negative control in this experiment. We have found that neither ACT1 nor PPA1 is targeted by Isw2 complex, and therefore both can be used interchangeably as negative controls for Isw2 ChIP experiments (data not shown).
We performed ChIP assays to examine Isw2 localization in these diploid yeast strains. In each case, one copy of ISW2 was tagged with a triple-FLAG epitope. For the heterozygote, the isw2-K215R allele carried the FLAG tag. When the wild-type allele of the heterozygote was FLAG tagged and used in the ChIP assay, the wild-type Isw2p exhibited only basal levels of interaction with chromatin (data not shown). We tested Isw2 localization at five known Isw2 target loci, as well as a negative control promoter, ACT1 (Fig. 6C). The levels of cross-linking by wild-type Isw2p and Isw2p-K215R were similar at the ACT1 locus. In each case, association of wild-type Isw2 complex was near basal levels (Fig. 6C, white bars). At four of the five promoters, association of catalytically inactive Isw2p-K215R was significantly higher in the presence of mutant chromatin structure (Fig. 6C, dark gray bars) than wild-type chromatin structure (Fig. 6C, light gray bars) (P < 0.05, Student's t test). Isw2p-K215R association at the POT1 promoter was not significantly different between the wild-type Isw2p/Isw2p-K215R-FLAG (WT/K215R-FL) strain and the Isw2p-K215R/Isw2p-K215R-FLAG (K215R/K215R-FL) strain, although the WT/K215R-FL strain produced higher signals in all three replicate experiments. These data show that, in most cases examined, Isw2p-K215R associates more strongly with target loci that have not been remodeled. One common feature of chromatin structure at most Isw2 target genes is that nucleosomes are slid closer to Isw2 recruitment sites (8, 12, 14, 24, 34). Therefore, these results may be due to contributions to Isw2 binding from the increased lengths of linker DNA in nonremodeled chromatin, lack of steric constraints on Isw2 binding that result from tight nucleosome packing in remodeled chromatin, availability of the histone H4 tail, or a combination of these factors.
We cannot rule out the alternative possibility that a secondary consequence of nonremodeled chromatin, such as a defect in higher-order chromatin folding, accounts for the increased binding of Isw2-K215R complex to open chromatin. Previously, we found that binding of the Ume6 protein to its binding site within the REC104 promoter was unaffected by defects in nucleosome positioning and histone acetylation (14), suggesting that differences in transcription factor binding to open and closed chromatin do not account for the observed differences in Isw2 association. In addition, it is unclear why Isw2-K215R localization to the POT1 promoter is similar, or even slightly increased, in the presence of wild-type chromatin structure compared to isw2 mutant chromatin structure. One possibility is that there is sufficient linker DNA at the site of Isw2 recruitment for robust chromatin binding at this promoter in either case. Overall, however, our results are consistent with a model in which nonremodeled chromatin structure facilitates robust binding of Isw2 complex to chromatin target sites in vivo.
DISCUSSION
We recently found that Isw2 complex exhibits basal levels of chromatin binding throughout the genome as well as target-specific chromatin interactions that can only be detected by a catalytically inactive Isw2-K215R mutant complex in vivo and that only the latter leads to chromatin remodeling (12). However, these experiments did not address the molecular basis for the two distinct chromatin interactions. Our previous finding that yeast Isw2 complex slides nucleosomes closer to its site of recruitment suggested that Isw2 complex should bind DNA and the histone octamer independently to generate the force necessary to disrupt histone-DNA contacts for nucleosome sliding. We tested this model to investigate the molecular requirements of each mode of chromatin interaction, as well as to further understand the mechanisms of Isw2-dependent chromatin remodeling in vivo.
We found that the histone H4 basic patch was the only region of any histone amino-terminal tail required for Isw2 complex to function in vivo. Furthermore, we found that the basic patch was required for specific association of Isw2p-K215R with Isw2 target sites in vivo. This was unexpected in light of previous data showing that Drosophila ISWI binds chromatin independently of the basic patch in vitro (5, 6). We also found that Isw2 complex binds NCPs in vitro in an H4 basic patch-dependent manner. These data strongly suggest that Isw2 complex interacts directly with the basic patch in the nucleosomal context. It should be noted that while these experiments demonstrate that the H4 basic patch is necessary for Isw2 interaction in vivo and in vitro, they do not demonstrate that this site is sufficient for Isw2 binding. Due to technical problems, we were unable to test whether Isw2 complex binds to the basic patch of free histone H4 tails in the absence of the nucleosome. It is therefore possible that the basic patch is only part of a more complex binding site that includes contributions from nucleosomal DNA, the core domain of histone H4, and/or the other histone core domains.
Since substitution mutations of lysines 16 and 20 of histone H4 had no effect on Isw2-dependent chromatin remodeling, we concluded that these residues are not required for the function of the basic patch. However, these results do not necessarily contradict previous results for Drosophila ISWI both in vitro and in vivo showing that acetylation of lysine 16 of histone H4 disrupts ISWI function. For example, an acetyl group on the ɛ-amino group of lysine 16 may directly block binding of ISWI complexes to the basic patch, while K16A, K16Q, and K16R mutations do not. Alternatively, if a protein recognizes and specifically binds to H4 tails that are acetylated at lysine 16, this protein may inhibit interaction of ISWI complexes with the basic patch and inhibit chromatin remodeling as a result. By replacing lysine 16 with alanine, glutamine or arginine, we likely would disrupt the binding site for this hypothetical ISWI inhibitor, rendering chromatin permissive for Isw2-dependent chromatin remodeling.
If the H4 basic patch is a binding site for Isw2 complex, how does Isw2 identify its specific target sites in vivo when it is found on every nucleosome? Presently, the only protein shown directly to recruit Isw2 complex to specific promoters is the dual repressor/activator protein Ume6p (14). However, other transcription factors likely function to recruit Isw2 complex to different classes of target genes. For example, chromatin remodeling and target-specific localization of Isw2 complex upstream of tRNA genes is dependent on Bdp1p (2).
In addition, we found that Isw2p requires nonremodeled chromatin structure and intact Isw2 complex for robust association with most target promoters in vivo. It is possible that some features of nonremodeled chromatin other than open chromatin structure are responsible for preferential binding of Isw2. Furthermore, Fitzgerald and colleagues recently proposed a model for a nucleotide-dependent reaction cycle of Isw2 (10). In this model, Isw2 stays bound to a nucleosome throughout the cycle, whereas its conformation and binding to linker DNA are regulated by nucleotides. Isw2 binds linker DNA in the absence of ATP. ATP binding causes compaction of Isw2 structure, generating force for nucleosome sliding. This is followed by ATP hydrolysis, which causes release of Isw2 from DNA as well as extension of Isw2 structure. Subsequent release of ADP from Isw2 allows rebinding of Isw2 to linker DNA. Therefore, we cannot rule out the possibilities that the K215R mutation prevents ADP-dependent release of Isw2 complex from chromatin in vivo or creates an intermediate of the chromatin remodeling reaction that permits better cross-linking to DNA. However, given that Isw2 complex interacts more strongly with linker DNA than nucleosomal DNA in vitro (10, 13, 22), we favor a model in which the longer lengths of linker DNA adjacent to Isw2 recruitment sites at nonremodeled chromatin targets play a role in recognition by Isw2 complex in vivo. The Itc1p subunit, on the other hand, may be required for direct recognition of target sites, structural integrity of the complex, or association of Dpb4p and Dls1p subunits with Isw2 complex, which may be necessary for interaction with target loci.
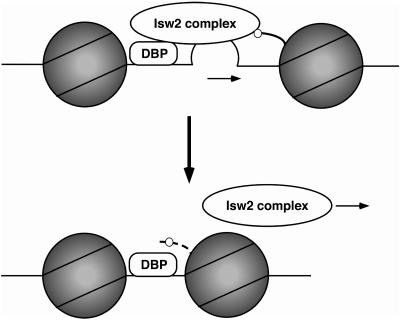
Together, these data suggest that at least three independent factors are required for target-specific chromatin binding by Isw2 complex in vivo: the presence of a sequence-specific DNA binding protein that binds to and recruits Isw2 complex, a functional H4 basic patch, and nonremodeled chromatin structure (Fig. 7). This is the first demonstration that target-specific chromatin interaction by an ATP-dependent chromatin remodeling complex in vivo requires contributions from these three separate factors, in contrast to the general DNA and chromatin binding activities of Isw2 complex in vitro. Based on these results, we propose that Isw2 complex recruited to specific sites by DNA binding proteins uses its interactions with linker DNA and the H4 tail basic patch to create the force necessary to push DNA into nucleosomes adjacent to its site of recruitment (Fig. 7, top). As a result, nucleosomes are slid closer to the site of Isw2 recruitment as previously observed in vivo (9). This remodeling reaction would destroy the high-affinity interaction sites for Isw2, resulting in release of the complex from its targets (Fig. 7, bottom). The reduced affinity of Isw2 for remodeled chromatin may be due to shorter linker DNA, reduced availability of the H4 tail basic patch, or a combination of both. This model is consistent with previous reports that Isw2 complex preferentially interacts with the linker DNA (10, 13, 22) and that a gap in linker DNA immediately adjacent to nucleosomes blocks Isw2-dependent nucleosome sliding in vitro (22).
FIG. 7.
A model for Isw2-dependent chromatin remodeling in vivo. (Top) Isw2 is targeted to specific loci via interactions with sequence-specific DNA binding proteins (DBP), linker DNA (bulge), and the basic patch (small circle) of the histone H4 tail (thick curved line). These interactions allow Isw2 to generate the necessary force to push DNA into nucleosomes (arrow). (Bottom) After nucleosome sliding, Isw2 is released from its targets as shown by an arrow. This may be due to the lack of sufficiently long linker DNA, reduced availability of histone H4 tail (dashed line), or a combination of both after remodeling.
In contrast to target-specific chromatin binding, the basal mode of chromatin binding by Isw2 complex is independent of DNA binding proteins, the histone H4 tail, integrity of the complex, and nonremodeled chromatin structure. These results demonstrate fundamental differences between the mechanisms underlying the target-specific and basal modes of chromatin binding by Isw2 complex in vivo. The basal levels of chromatin binding by Isw2 complex are observed globally throughout the yeast genome (12) and are independent of Itc1p, a subunit essential for all known Isw2 functions in vivo. Given that monomer Isw2p and intact Isw2 complex exhibit the same basal levels of chromatin binding, it is likely that active processes are not required for this mode of chromatin interaction. It is not clear at this time what, if any, functions of Isw2 complex are mediated by basal chromatin binding, but even passive chromatin interaction by Isw2 may be sufficient for the complex to identify its targets within the genome.
Acknowledgments
We thank members of the Tsukiyama, Biggins, and Gottschling laboratories for helpful discussions and members of the Tsukiyama laboratory for comments on the manuscript. We are also very grateful to Jeff Delrow for his kind help in statistical analyses of DNA microarray results.
This work was supported by National Institutes of Health grant GM58465 to T.T. T.G.F. and M.E.G are supported by predoctoral fellowships from HHMI. T.T. is a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does “chromatin remodeling” mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, N., M. E. Gelbart, T. Tsukiyama, and J. D. Boeke. 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 4.Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. Owen-Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 12:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clapier, C. R., G. Langst, D. F. Corona, P. B. Becker, and K. P. Nightingale. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapier, C. R., K. P. Nightingale, and P. B. Becker. 2002. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corona, D. F., C. R. Clapier, P. B. Becker, and J. W. Tamkun. 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, D. J., C. DeLuca, I. Berger, H. Gaillard, R. Sigrist, K. Schimmele, and T. J. Richmond. 2004. Reaction cycle of the yeast Isw2 chromatin remodeling complex. EMBO J. 23:3836-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 12.Gelbart, M. E., N. Bachman, J. Delrow, J. D. Boeke, and T. Tsukiyama. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19:942-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 15.Grune, T., J. Brzeski, A. Eberharter, C. R. Clapier, D. F. Corona, P. B. Becker, and C. W. Muller. 2003. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12:449-460. [DOI] [PubMed] [Google Scholar]
- 16.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 17.Hamiche, A., J. G. Kang, C. Dennis, H. Xiao, and C. Wu. 2001. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. USA 98:14316-14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, J. J., T. D. Tullius, and A. P. Wolffe. 1990. The structure of DNA in a nucleosome. Proc. Natl. Acad. Sci. USA 87:7405-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iida, T., and H. Araki. 2004. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 22.Kagalwala, M. N., B. J. Glaus, W. Dang, M. Zofall, and B. Bartholomew. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23:2092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassabov, S. R., N. M. Henry, M. Zofall, T. Tsukiyama, and B. Bartholomew. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22:7524-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-Related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 28.Langst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 29.Langst, G., and P. B. Becker. 2004. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta 1677:58-63. [DOI] [PubMed] [Google Scholar]
- 30.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 31.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 32.Loyola, A., G. LeRoy, Y. H. Wang, and D. Reinberg. 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 15:2837-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 34.McConnell, A. D., M. E. Gelbart, and T. Tsukiyama. 2004. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L. 2002. Chromatin remodeling: nucleosomes bulging at the seams. Curr. Biol. 12:R245-R247. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 38.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83:1011-1020. [DOI] [PubMed] [Google Scholar]
- 40.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 41.Vary, J. C., Jr., T. G. Fazzio, and T. Tsukiyama. 2004. Assembly of yeast chromatin using ISWI complexes. Methods Enzymol. 375:88-102. [DOI] [PubMed] [Google Scholar]
- 42.Vary, J. C., Jr., V. K. Gangaraju, J. Qin, C. C. Landel, C. Kooperberg, B. Bartholomew, and T. Tsukiyama. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehouse, I., A. Flaus, K. Havas, and T. Owen-Hughes. 2000. Mechanisms for ATP-dependent chromatin remodelling. Biochem. Soc. Trans. 28:376-379. [PubMed] [Google Scholar]
- 44.Whitehouse, I., C. Stockdale, A. Flaus, M. D. Szczelkun, and T. Owen-Hughes. 2003. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 23:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]