Abstract
Caspase-mediated proteolysis is a critical and central element of the apoptotic process, and caspase 3, one of the effector caspases, is proposed to play essential roles in the nuclear morphological changes of apoptotic cells. Although many substrates for caspase 3 localize in the nucleus and caspase 3 translocates from the cytoplasm to the nuclei after activation in apoptotic cells, the molecular mechanisms of nuclear translocation of active caspase 3 have been unclear. Recently, we suggested that a substrate-like protein(s) served as a carrier to transport caspase 3 from the cytoplasm into the nucleus. In the present study, we identified A-kinase-anchoring protein 95 (AKAP95) as a caspase 3-binding protein. Small interfering RNA-mediated depletion of AKAP95 reduced apoptotic nuclear morphological changes, suggesting that AKAP95 is involved in the process of apoptotic nuclear morphological changes. The association of AKAP95 with active caspase 3 was analogous to an enzyme-substrate interaction. Furthermore, overexpression of AKAP95 with nuclear localization sequence mutations inhibited nuclear morphological changes in apoptotic cells. These results indicate that AKAP95 is a potential carrier protein for active caspase 3 from the cytoplasm into the nuclei in apoptotic cells.
Apoptosis plays important roles in a variety of biological events, including morphogenesis, maintenance of tissue homeostasis, and removal of harmful cells. Apoptosis is morphologically characterized by chromatin condensation, nuclear fragmentation, and formation of membrane-enclosed vesicles called apoptotic bodies, which are phagocytosed by other cells. Caspases, a family of cysteine proteases, are required for apoptosis execution (2, 11, 12, 39). Caspase 3, one of the effector caspases, has been implicated as a key mediator of apoptosis in mammalian cells (11, 12, 39) and plays essential roles in the nuclear changes in apoptotic cells (25, 43, 44) despite the cytoplasmic localization of the precursor form of caspase 3 (28, 34). In addition, although many nuclear substrates for caspase 3 have been identified (11, 12, 18, 39), the precise localization of active caspase 3 in apoptotic cells had been unclear. Recently, we confirmed the nuclear localization of active caspase 3 in apoptotic cells by using antibodies specific for the large and small subunits of active caspase 3 (22). Furthermore, we showed that the nuclear translocation of caspase 3 required its proteolytic activation and substrate recognition, whereas caspase 7, another effector caspase, was not translocated into the nuclei (22). These results suggested that the nuclear translocation of active caspase 3 is not mediated by passive diffusion but requires an active transport system and that active caspase 3 may be translocated in association with a substrate-like protein(s) from the cytoplasm into the nucleus in apoptotic cells.
A-kinase-anchoring proteins (AKAPs) bind to the regulatory subunit of cyclic AMP-dependent protein kinase (PKA) to direct the kinase to discrete intracellular locations (10). A 95-kDa AKAP, designated AKAP95, has been identified from human (692 amino acids), mouse (687 amino acids), and rat (687 amino acids) sources (8, 14). AKAP95 proteins are highly conserved among species, with human AKAP95 showing 78% identity (85% similarity) with mouse and rat AKAP95. AKAP95 contains several characteristic sequences, including a nuclear matrix targeting site, overlapping putative bipartite nuclear localization sequences (NLSs), two zinc fingers, and a type II PKA regulatory subunit (RII) binding domain (see Fig. 5A), and is suggested to be localized to the nuclear matrix (1, 8, 14). Recently, it was reported that AKAP95 plays an essential role in chromatin condensation during mitosis through the anchoring of a cyclic AMP/PKA-signaling complex and the recruitment of components of the condensin complex onto chromatin (9, 13, 36).
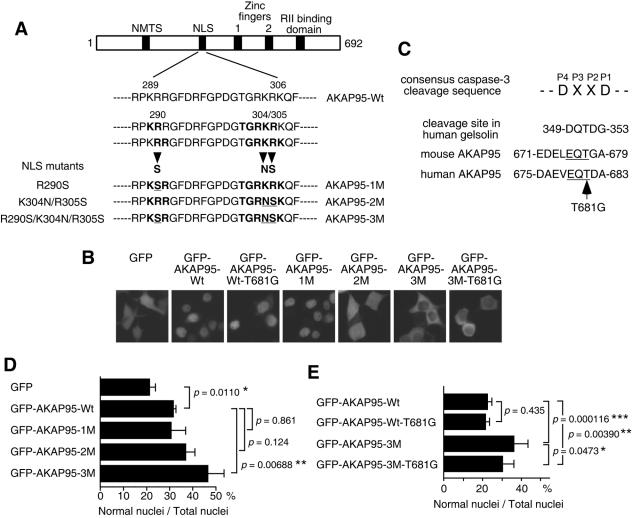
FIG. 5.
Function of AKAP95 in nuclear morphological changes of apoptotic cells. (A) Diagram showing the NLS mutations of AKAP95. Mutated amino acid residues are underlined. NMTS, nuclear matrix targeting site; RII, type II PKA regulatory subunit. (B) Localization of GFP-AKAP95 fusion proteins with or without mutations. 293T cells were transiently transfected with GFP or GFP-AKAP95 expression plasmids as indicated. After 18 h, cells were observed by fluorescence microscopy. (C) Amino acid sequences of active caspase 3 binding region in AKAP95. Conserved EQT sequence between human and mouse AKAP95 is underlined. (D and E) Overexpression of AKAP95 with NLS mutations inhibits nuclear morphological changes. HepG2 cells were transfected with GFP-AKAP95 expression plasmids as indicated. After 24 h, cells were treated with an agonistic anti-Fas antibody in the presence of actinomycin D for 12 h (D) or 15 h (E) and collected and fixed with 3.7% formaldehyde for 10 min. After staining with Hoechst 33342, the percentage of the cells showing normal nuclei to the total transfected cells was measured. At least 100 GFP-positive cells were counted for each measurement in all experiments. The data (mean ± the standard deviation) were obtained from at least three independent experiments. Significant test results (P values) are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To identify a substrate-like protein(s) that might serve as carrier proteins to transport active caspase 3 from the cytoplasm into nucleus in apoptotic cells, we used a cloning method for caspase substrates that uses the yeast two-hybrid system (21). In this manner, we identified AKAP95 as a caspase 3-binding protein and obtained evidence that AKAP95 functions as a carrier protein for the nuclear translocation of active caspase 3.
MATERIALS AND METHODS
Cell culture and apoptosis induction.
HepG2 and Jurkat cells were cultured in RPMI 1640 medium with 10% fetal bovine serum. HeLa (clone D98AH2) and 293T cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. For induction of apoptosis, HepG2 cells were treated with 1 μg of an agonistic anti-Fas antibody (CH-11; Kamiya Biomedical Company)/ml in the presence of 0.2 μg of actinomycin D/ml or with 200 μg of etoposide/ml. Transfection was performed using Lipofectamine (Life Technologies) for 293T cells and GenePORTER 2 (Gene Therapy Systems) for HepG2 cells according to the manufacturer's instructions.
Antibodies and immunoprecipitation.
Preparation of anti-active caspase 3 polyclonal antibodies (antibody 2622) and anti-active caspase 3 monoclonal antibody (clone CS-1) was described elsewhere (22). Anti-caspase 3 monoclonal antibody (C31720) and anti-AKAP95 monoclonal antibody (A74220) were obtained from Transduction Laboratories; anti-caspase 3 polyclonal antibodies (sc-1224) and anti-lamin B1 polyclonal antibodies (sc-6217) were from Santa Cruz Biotechnology; anti-AKAP95 polyclonal antibodies (06-417) were from Upstate Biotechnology, Inc.; anti-caspase 3 polyclonal antibodies (antibody 9662) were from Cell Signaling Technology; anti-green fluorescent protein (anti-GFP) monoclonal antibody (antibody 8371) was from Clontech; anti-GFP polyclonal antibodies (A-6455) were from Molecular Probes; anti-Xpress monoclonal antibody (R910-25) was from Invitrogen; and anti-α-tubulin monoclonal antibody (T-5168) was from Sigma. The anti-AKAP95 rabbit antiserum used for immunoprecipitation experiments was kindly provided by J. D. Scott.
For immunoprecipitation experiments, cells were lysed in lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.4% Nonidet P-40, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 μg of pepstatin/ml, 100 μg of phenylmethylsulfonyl fluoride/ml). Lysates were incubated with anti-AKAP95 serum or normal rabbit serum for 1 h at 4°C with constant rotation and then with 5% (vol/vol) protein A-agarose for an additional 1 h. Cell lysates and immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with the indicated antibodies.
siRNA experiments.
Synthetic 21-nucleotide double-stranded RNAs were obtained from Dharmacon Research. The targeting sequence of human AKAP95 was AACTACAATTACTATGGCGCC, corresponding to coding nucleotides 100 to 120 relative to the first nucleotide of the start codon. HepG2 cells were transfected with Oligofectamine reagent (Invitrogen). One day before transfection, cells were seeded at a density of 9 × 105 cells per 10-cm dishes. In a first tube, 600 μl of Opti-MEM was mixed with 36 μl of 20 μM small interfering RNA (siRNA) duplex. In a second tube, 144 μl of Opti-MEM was incubated with 36 μl of Oligofectamine for 10 min at room temperature. The two mixtures above were combined, gently mixed, and incubated for 20 min at room temperature. After addition of 384 μl of Opti-MEM to the mixture, the entire mixture was added to the cells, followed by incubation for 4 days.
Yeast two-hybrid assays.
The yeast reporter strain L40 (MATa trp1 leu2 his3 ade2 LYS2::lexA-HIS3 URA3::lexA-lacZ) was used as the host, and cells positive for growth on selective medium (-His/-Leu/-Trp) were examined for β-galactosidase activity using a colony filter-lift assay.
Plasmid constructions.
Construction of pBTM-casp3-p12p17m and pBTM-casp3-p12 was described previously (21). A fragment encoding caspase 3-p17m was generated by PCR using caspase 3 cDNA bearing the C163S mutation as a template and was cloned into the EcoRI site of pBTM116 to generate pBTM-casp3-p17m. A fragment encoding caspase 3-p12m was generated by PCR using pcasp3-R207E-GFP plasmid (22) as a template and cloned into the EcoRI-BamHI site of pBTM-casp3-p12p17m lacking the caspase 3-p12 to generate pBTM-casp3-p12mp17m. pGAD-AKAP95 (clone 13 6-687) was originally identified as a possible substrate for caspase 3 by yeast two-hybrid screening (21) and contained residues 6 to 687 of mouse AKAP95. Various deletion mutants of AKAP95 for yeast two-hybrid assays were constructed by using suitable restriction enzyme sites or PCR. To substitute Glu675 for Gly and Thr677 for Gly, a PCR method using mutagenic oligonucleotide primers was used.
Construction of pCAG-casp3, pcasp3-Wt-GFP, pcasp7-Wt-GFP, pcasp3-C163S-GFP, pcasp3-D175A-GFP, pcasp3-R64E-GFP, and pcasp3-R207E-GFP was described elsewhere (22). The prodomain deletion mutant of caspase 3 was constructed by PCR and cloned into the EcoRI-BamHI site of pEGFP-C1 (Clontech) to generate pGFP-Δpro-casp3-Wt. The C-terminally hemagglutinin (HA)-tagged procaspase 3 cDNA fragment was cloned into the EcoRI site of pUC-CAGGS (29) to generate pCAG-casp3-HA. To construct caspase 3 expression plasmids as a fusion to the N terminus of DsRed (3), the fragment encoding caspase 3 was cloned into the EcoRI-BamHI site of pDsRed1-N1 (Clontech) to generate pcasp3-Wt-DsRed.
The human AKAP95 cDNA (kindly provided by K. Tasken) was cloned into the XhoI site of pUC-CAGGS, the BamHI site of pcDNA3.1/His A (Invitrogen) and the XhoI site of pEGFP-C2 (Clontech) to generate pCAG-AKAP95-Wt, pcDNA-AKAP95-Wt and pGFP-AKAP95-Wt. To substitute Arg290 for Ser, Lys304 for Asn, Arg305 for Ser, and Thr681 for Gly, a PCR method using mutagenic oligonucleotide primers was used. The human AKAP95 cDNAs containing mutations at R290S, K304N/R305S, R209S/K304N/R305S, T681G, and R209S/K304N/R305S/T681G were cloned into pEGFP-C2 to generate pGFP-AKAP95-1M, pGFP-AKAP95-2M, pGFP-AKAP95-3M, pGFP-AKAP95-Wt-T681G, and pGFP-AKAP95-3M-T681G.
RESULTS
Identification of caspase 3-binding proteins is valuable for understanding the molecular mechanisms of apoptotic execution. Therefore, we used the yeast two-hybrid method, which has been successfully used to identify caspase substrates (21, 30). In this manner, we identified several potential caspase 3 substrates including gelsolin (clones 1, 9, and 12 in reference 21), and a potential binding protein to caspase 3 (clone 13 in reference 21). Clone 13 contained residues 6 to 687 of mouse AKAP95, which has bipartite NLSs (see Fig. 5A) and is suggested to play an essential role in chromatin condensation during mitosis (9, 13, 36).
AKAP95 is a caspase 3-binding protein.
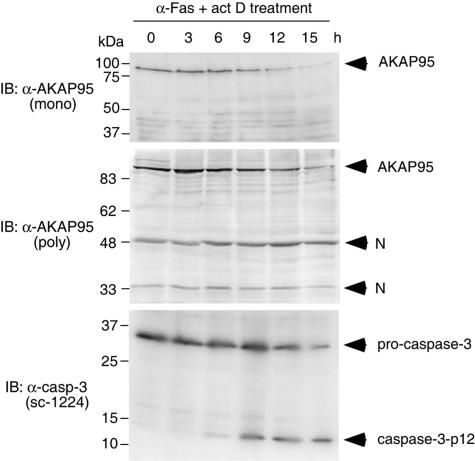
AKAP95 (clone 13 in reference 21) was not cleaved completely in in vitro cleavage assays even though it has three tetrapeptides DNSD90, DCRD175, and DLCD349, which fit the consensus caspase 3 cleavage sequence, whereas gelsolin (clones 1, 9, and 12 in reference 21) was cleaved completely. These results suggested that AKAP95 is not a good substrate for caspase 3. Therefore, we examined whether AKAP95 is a substrate for caspase 3 in vivo by using anti-AKAP95 monoclonal or polyclonal antibodies, which were prepared against C-terminal fragments of AKAP95. After treatment of HepG2 cells with an agonistic anti-Fas antibody, procaspase 3 was cleaved and activated (Fig. 1, lower panel). However, although the levels of AKAP95 protein gradually decreased, specific AKAP95 cleavage products were not detected by immunoblotting with an anti-AKAP95 monoclonal (Fig. 1, upper panel) or polyclonal (Fig. 1, middle panel) antibodies during apoptosis. These results suggested that AKAP95 is probably not a substrate for caspase 3 in vivo and lacks a cleavage site in the interaction domain.
FIG. 1.
AKAP95 is not a substrate for caspase 3 in vivo. HepG2 cells were treated with an agonistic anti-Fas antibody in the presence of actinomycin D for the indicated periods. Cell lysates were subjected to SDS-PAGE and immunoblotted with anti-AKAP95 monoclonal antibody, anti-AKAP95 polyclonal antibodies, or anti-caspase 3 polyclonal antibodies (sc-1224; Santa Cruz Biotechnology) that detect both procaspase 3 and caspase 3-p12 as indicated. N, nonspecific band.
AKAP95 plays a role in apoptotic nuclear morphological changes.
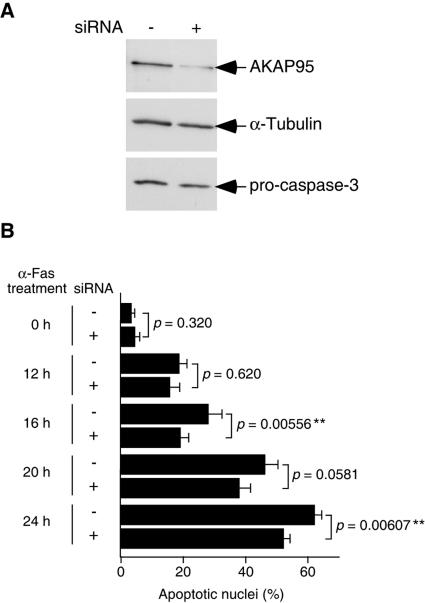
To investigate a possible role for AKAP95 in apoptotic execution, we examined the effects of AKAP95 overexpression in cells. However, we did not observe any differences between transfected and nontransfected cells (data not shown and see Fig. 5B). As an alternative means of determining whether AKAP95 plays a role in apoptosis, we used siRNA (15) to deplete AKAP95 protein in HepG2 cells (Fig. 2). At 4 days after transfection of siRNA for AKAP95, the level of AKAP95 was decreased significantly (to <15%), whereas the levels of α-tubulin and procaspase 3 were unaffected (Fig. 2A). Therefore, the same number of cells transfected with siRNA for AKAP95 were reseeded, and apoptosis was induced by treatment with anti-Fas antibody. HepG2 cells transfected with siRNA for AKAP95 exhibited delayed nuclear morphological changes compared to control cells (Fig. 2B), suggesting that AKAP95 plays a role(s) in the nuclear morphological changes of apoptotic cells.
FIG. 2.
Inhibition of nuclear morphological changes of apoptotic cells by siRNA for AKAP95. (A) Lysates from HepG2 cells transfected with or without siRNA for AKAP95 for 4 days were fractionated by SDS-PAGE and immunoblotted with anti-AKAP95 polyclonal antibodies, anti-α-tubulin monoclonal antibody, or anti-caspase 3 monoclonal antibody. (B) HepG2 cells transfected with or without siRNA for AKAP95 were treated with an agonistic anti-Fas antibody in the presence of actinomycin D for the indicated periods and collected and fixed with 3.7% formaldehyde for 10 min. After staining with Hoechst 33342, the percentage of the cells showing apoptotic nuclei to the total cells was measured. At least 100 cells were counted for each measurement in all experiments. The data (mean ± the standard deviation) were obtained from at least three independent experiments. Significant test results (P values) are shown. **, P < 0.01.
Association of caspase 3 with AKAP95 in yeast.
Recently, we proposed that active caspase 3 is translocated in association with a substrate-like protein(s) from the cytoplasm into the nucleus in apoptotic cells (22). A carrier protein for the nuclear translocation of active caspase 3 would not be expected to be a caspase substrate per se because a typical enzyme-substrate complex is not stable but rather should associate reasonably stably with caspase 3 and should have a functional NLS. AKAP95 has all of these properties and therefore is a candidate for a carrier protein of caspase 3. To investigate this possibility, we next defined the AKAP95-binding region in caspase 3 using direct yeast two-hybrid assays (Table 1). Cotransformation of pBTM-casp3-p12p17m, which expresses both p17 containing C163S mutation and p12 subunits of caspase 3, and pGAD-AKAP95 (clone 13 6-687) yielded His+ transformants that were also β-galactosidase positive. However, neither caspase 3-p12 nor caspase 3-p17m alone was able to bind to AKAP95, indicating that both subunits of caspase 3 are required for its binding to AKAP95. Furthermore, the R207E mutation, which prevents recognition of the P3 amino acid of substrates by caspase 3 (32, 42) and inhibited the nuclear translocation of active caspase 3 (22), abolished the association with AKAP95. These results suggest that the interaction of caspase 3 and AKAP95 may be analogous to an enzyme-substrate interaction.
TABLE 1.
Association of AKAP95 with active caspase 3 in yeast two-hybrid assaysa
| pGAD (Gal4 AD)b | pBTM (LexA BD)c | β-Galactosidase activityd |
|---|---|---|
| AKAP95 (#13 6-687) | casp3-p12p17m | + |
| AKAP95 (#13 6-687) | casp3-p12mp17m | − |
| AKAP95 (#13 6-687) | casp3-p17m | − |
| AKAP95 (#13 6-687) | casp3-p12 | − |
| AKAP95 (#13 6-47) | casp3-p12p17m | − |
| AKAP95 (#13 6-341) | casp3-p12p17m | − |
| AKAP95 (#13 6-542) | casp3-p12p17m | − |
| AKAP95 (#13 340-429) | casp3-p12p17m | − |
| AKAP95 (#13 428-687) | casp3-p12p17m | + |
| AKAP95 (#13 544-687) | casp3-p12p17m | + |
| AKAP95 (#13 544-670) | casp3-p12p17m | − |
| AKAP95 (#13 556-687) | casp3-p12p17m | + |
| AKAP95 (#13 556-679) | casp3-p12p17m | + |
| AKAP95 (#13 544-687 E675G) | casp3-p12p17m | − |
| AKAP95 (#13 544-687 T677G) | casp3-p12p17m | − |
Yeast L40 cells were cotransfected with expression plasmids for Gal4 activation domain (Gal4 AD) fusion proteins and for LexA DNA-binding domain (LexA BD) fusion proteins as indicated.
“#13” represents the clone number that was originally identified by yeast two-hybrid screening (21). The numbers (6-687, etc.) correspond to the encoded amino acids of AKAP95.
pBTM-casp3-p12p17m was used for expression of caspase 3-p12 and caspase 3-p17(C163S), pBTM-casp3-p12mp17m was used for expression of caspase 3-p12(R207E) and caspase 3-p17(C163S), pBTM-casp3-p17m was used for expression of caspase 3-p17(C163S), and pBTM-casp3-p12 was used for expression of caspase 3-p12.
Each transformation mixture was plated on a synthetic dropout plate lacking leucine, tryptophan, and histidine. Filter assays for β-galactosidase activity were performed to detect interactions between fusion proteins. +, development of blue color within 2 h; −, no growth of transformed yeast colonies.
Next we defined the active caspase 3-binding region in AKAP95 using yeast two-hybrid assays (Table 1). Various deletion mutants of AKAP95 were fused to the Gal4 activation domain and transformed into yeast with pBTM-casp3-p12p17m. Cotransformation of pGAD-AKAP95 (clone 13, 544-687) or pGAD-AKAP95 (clone 13, 556-679), but not pGAD-AKAP95 (clone 13, 544-670), with pBTM-casp3-p12p17m conferred the His+ phenotype and β-galactosidase activity, indicating that the binding site of AKAP95 for active caspase 3 is present in the C-terminal region (amino acids 556 to 679) of AKAP95 and that amino acids 671 to 679 are required for association with active caspase 3 in yeast. Although a consensus caspase 3 cleavage sequence (DXXD) was not found in this region, the tetrapeptide (E675QTG678), which is related to the caspase 3 cleavage site in gelsolin (DQTD) (21), is present (see Fig. 5C). To test whether this sequence is essential for binding to active caspase 3, we constructed E675G and T677G point mutants in a C-terminal fragment of mouse AKAP95 (amino acids 544 to 687). These mutations abolished the interaction of AKAP95 with active caspase 3 in yeast, suggesting that the E675QTG678 sequence in AKAP95 is required for binding to active caspase 3, possibly because this sequence functions as a noncleavable pseudosubstrate site for caspase 3. The fact that nuclear translocation of active caspase 3 did not require Arg64 or Cys163 (22), both of which are essential for recognition of, and cleavage after, Asp at P1 position (32, 42), is consistent with the absence of Asp at the position corresponding to P1 in the EQTG active caspase 3 binding site of mouse AKAP95.
Association of caspase 3 with AKAP95 in vivo.
Next we tested whether an in vivo association between active caspase 3 and AKAP95 could be detected by coimmunoprecipitation. GFP-tagged caspase 3 with or without mutations was transiently overexpressed in 293T cells together with human AKAP95. As a control for specificity, C-terminally GFP-tagged procaspase 7 was coexpressed with AKAP95. Even though the cells were not induced to undergo apoptosis, casp3-Wt-GFP, GFP-Δpro-casp3-Wt, and casp7-Wt-GFP were proteolytically activated, presumably as a result a stress, such as serum starvation or a toxic effect of liposomes during transfection, or the overexpression of the wild-type caspases (Fig. 3A, upper panel). After immunoprecipitation of AKAP95, coprecipitated GFP-fusion proteins were detected with anti-GFP antibody (Fig. 3A, middle panel). Proteolytically activated casp3-p12-GFP derived from casp3-Wt-GFP and GFP-casp3-p17 derived from GFP-Δpro-casp3-Wt, but not casp7-p12-GFP from casp7-Wt-GFP, were coprecipitated with AKAP95, suggesting that active caspase 3 interacts with AKAP95 in vivo and that the association of effector caspases with AKAP95 may be specific for caspase 3. These results are consistent with our findings that caspase 3, but not caspase 7, translocated from the cytoplasm to the nucleus in apoptotic cells (22). Interestingly, all of the unprocessed GFP-caspase fusion proteins were coprecipitated along with AKAP95, but the level of casp7-Wt-GFP precipitated was significantly less than that of casp3-Wt-GFP. Since the coprecipitation of N-terminally GFP-tagged caspase 3 with AKAP95 was less effective than that of C-terminally GFP tagged caspase 3, GFP fused to the N terminus of caspase 3-p17 may interfere with immunocomplex formation.
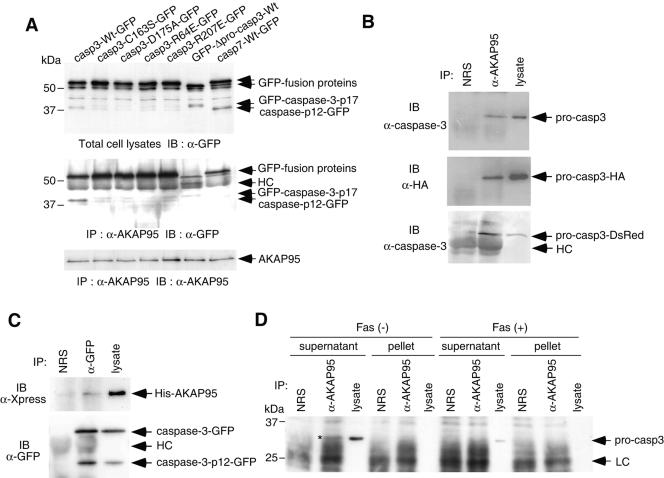
FIG. 3.
In vivo association of caspase 3 with AKAP95. (A) Active caspase 3, but not active caspase-7, binds to AKAP95. 293T cells were transfected with caspase-GFP expression plasmids as indicated, together with pCAG-AKAP95-Wt. Casp3-C163S-GFP contained the mutation at the catalytic Cys, casp3-D175A-GFP contained the cleavage site mutation between the p17 and p12 subunits, casp3-R64E-GFP and casp3-R207E-GFP contained the mutations of substrate recognition sites, and GFP-Δpro-casp3-Wt was a prodomain deletion mutant. After incubation for 24 h, lysates were immunoprecipitated with anti-AKAP95 serum. The input lysates (upper panel) and the immunoprecipitates (middle and lower panels) were fractionated by SDS-PAGE and immunoblotted with anti-GFP monoclonal antibody (upper and middle panels) or anti-AKAP95 monoclonal antibody (lower panel). HC, heavy chain. (B) Coprecipitation of procaspase 3 with AKAP95 in transiently overexpressed 293T cells. Lysates from 293T cells transfected with either pCAG-casp3, pCAG-casp3-HA, or pcasp3-Wt-DsRed, together with pCAG-AKAP95-Wt, were immunoprecipitated with normal rabbit serum (NRS) or anti-AKAP95 serum (α-AKAP95). The immunoprecipitates and the input lysates were fractionated by SDS-PAGE and immunoblotted with anti-caspase 3 monoclonal antibody (upper and lower panels) or anti-HA monoclonal antibody (12CA5) (middle panel). (C) Coprecipitation of AKAP95 with caspase 3 in transiently overexpressed 293T cells. Lysates from 293T cells transfected pcDNA-AKAP95-Wt together with pcasp3-Wt-GFP were immunoprecipitated with normal rabbit serum (NRS) or anti-GFP polyclonal antibodies (α-GFP). The immunoprecipitates and the input lysates were fractionated by SDS-PAGE and immunoblotted with anti-Xpress monoclonal antibody that recognizes the leader peptide from the pcDNA3.1/His vector between His tag and AKAP95 (upper panel) or anti-GFP monoclonal antibody (lower panel). (D) Procaspase 3 binds to AKAP95 at endogenous protein levels. HepG2 cells treated with or without an agonistic anti-Fas antibody in the presence of actinomycin D for 12 h were divided into supernatant or pellet fractions after lysis with digitonin and immunoprecipitated as described in panel B. The immunoprecipitates and the input lysates were fractionated by SDS-PAGE and immunoblotted with anti-caspase 3 monoclonal antibody. The asterisk indicates the procaspase 3 coprecipitated with AKAP95. LC, light chain.
To further analyze the interaction between procaspase 3 and AKAP95 in vivo, immunoprecipitations were carried out from lysates of 293T cells that were transiently transfected with AKAP95 and procaspase 3 expression plasmids (Fig. 3B). Procaspase 3 and C-terminally HA- or DsRed-tagged pro-caspase 3 was coprecipitated with AKAP95, suggesting that procaspase 3 also interacts with AKAP95 in 293T cells.
We also tested whether coprecipitation of AKAP95 with caspase 3 could be detected. For this purpose, N-terminally His-tagged AKAP95 was transiently overexpressed in 293T cells together with C-terminally GFP-tagged procaspase 3, and GFP-tagged caspase 3 was immunoprecipitated with anti-GFP antibodies, followed by detection of coprecipitated His-tagged AKAP95 by immunoblotting with anti-Xpress monoclonal antibody. As shown in Fig. 3C, His-tagged AKAP95 was coprecipitated with GFP-tagged caspase 3, although we could not determine whether His-tagged AKAP95 was coprecipitated with GFP-tagged procaspase 3, caspase 3-p12, or both.
To determine whether association between endogenous AKAP95 and caspase 3 proteins could be detected, lysates from HepG2 cells treated with or without anti-Fas antibody were separated into supernatant and pellet fractions after lysis with digitonin and immunoprecipitated with anti-AKAP95 serum, followed by immunoblotting with anti-caspase 3 antibodies. When we used anti-caspase 3 polyclonal antibodies for immunoblotting, which recognize both procaspase 3 and caspase 3-p12 subunit (Fig. 1), no active caspase 3 coprecipitating with AKAP95 was detected (data not shown). However, we detected a low level (<1%) of procaspase 3 coprecipitating with AKAP95 in the supernatant fraction of normal cells by using anti-caspase 3 monoclonal antibody in immunoblotting (Fig. 3D), suggesting that AKAP95 interacts with procaspase 3 endogenously in the cytoplasm of normal cells.
Colocalization of active caspase 3 and AKAP95.
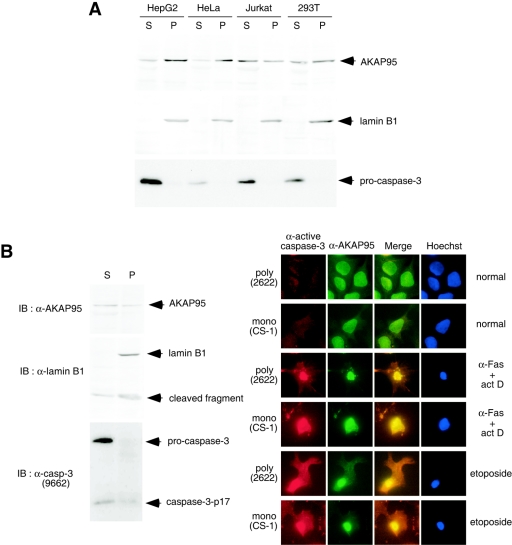
If AKAP95 functions as a carrier protein to transport active caspase 3 from the cytoplasm into nucleus, AKAP95 is expected to localize to the cytoplasm in normal cells and colocalize with active caspase 3 in apoptotic nuclei. To test this, various human cell lines were fractionated into supernatant and pellet fractions after lysis with digitonin, using lamin B1 as a nuclear marker. As shown in Fig. 4A, procaspase 3 was present only in the supernatant fraction, and AKAP95 was present in both the pellet and the supernatant fractions from normal cells. Next, apoptotic HepG2 cells were fractionated into supernatant and pellet fractions (Fig. 4B, left panel). Although procaspase 3 was present in the supernatant fraction, the caspase 3-p17 subunit and AKAP95 were present in both the pellet and supernatant fractions. Furthermore, HepG2 cells were stained with anti-AKAP95 and anti-active caspase 3 antibodies after treatment with anti-Fas antibody or etoposide (Fig. 4B, right panel). Although AKAP95 was detected in both nuclei and cytoplasm, active caspase 3 was not detected in cells before induction of apoptosis. However, AKAP95 and active caspase 3 were both highly enriched in the region around condensed nuclei in apoptotic cells, indicating colocalization of active caspase 3 and AKAP95 in apoptotic cells.
FIG. 4.
Localization of caspase 3 and AKAP95. (A) Localization of AKAP95 and procaspase 3 in normal cells. HepG2, HeLa, Jurkat, and 293T cells were harvested and fractionated into pellet (P) and supernatant (S) fractions after lysis with digitonin. Each fraction was subjected to SDS-PAGE and immunoblotted with anti-AKAP95 monoclonal antibody (upper panel), anti-lamin B1 polyclonal antibodies as a nuclear fraction marker (middle panel), or anti-caspase 3 monoclonal antibody (lower panel). (B) Colocalization of active caspase 3 and AKAP95 in apoptotic cells. HepG2 cells treated with an agonistic anti-Fas antibody in the presence of actinomycin D for 12 h were fractionated as described for panel A. Each fraction was subjected to SDS-PAGE and immunoblotted with anti-AKAP95 monoclonal antibody, anti-lamin B1 polyclonal antibodies, or anti-caspase 3 polyclonal antibodies (antibody 9662; Cell Signaling Technology) that detect both procaspase 3 and caspase 3-p17 as indicated (left panel). HepG2 cells were treated without or with an agonistic anti-Fas antibody in the presence of actinomycin D for 12 h or with etoposide for 40 h. After fixation and permeabilization, the cells were incubated with anti-active caspase 3 polyclonal (antibody 2622) or monoclonal (CS-1) antibodies, with anti-AKAP95 monoclonal or polyclonal antibodies, and with Texas Red (TXRD)- or fluorescein isothiocyanate-labeled secondary antibodies, followed by staining the nuclei with Hoechst 33342 (right panel).
Function of AKAP95 in apoptotic nuclear morphological changes.
If AKAP95 functions to carry caspase 3 from the cytoplasm into the nucleus, overexpression of AKAP95 with NLS-inactivating mutations should inhibit nuclear translocation of active caspase 3 by sequestration in the cytoplasm and thus prevent nuclear morphological changes in apoptotic cells. AKAP95 has potential overlapping bipartite NLSs (Fig. 5A) at amino acids 289 to 305 and amino acids 290 to 306. To identify amino acids necessary for the nuclear translocation of AKAP95, we constructed point mutants of basic amino acids in the putative NLS of human AKAP95 and fused these mutants to the C terminus of GFP. Mutation of Arg290 to Ser in AKAP95 (AKAP95-1M) had no effect on the nuclear localization of GFP-AKAP95 fusion protein (Fig. 5B), whereas a double mutation of Lys304 to Asn and Arg305 to Ser (AKAP95-2M) dramatically impaired nuclear accumulation of the fusion protein. Furthermore, combined mutation of R290S/K304N/R305S (AKAP95-3M) completely prevented nuclear localization of the AKAP95-GFP fusion protein (Fig. 5B). These results indicated that the overlapping bipartite NLSs located at amino acids 289 to 306 are essential for the nuclear import of AKAP95. Furthermore, introduction of the T681G point mutation (T677G in mouse AKAP95) (Table 1 and Fig. 5C) into AKAP95-Wt and AKAP95-3M did not affect the localization of GFP-AKAP95-Wt and GFP-AKAP95-3M, respectively (Fig. 5B).
To test whether overexpression of AKAP95 with NLS mutations inhibits nuclear morphological changes in apoptotic cells, GFP-AKAP95 with or without NLS mutations was transiently expressed in HepG2 cells, which were subsequently treated with anti-Fas antibody. GFP-AKAP95-2M and -3M had the greatest ability to prevent apoptotic nuclear morphological changes (Fig. 5D). Similar effects were also observed in HeLa cells (data not shown). Expression of GFP-AKAP95-Wt-T681G, which should not bind active caspase 3, did not decrease the percentage of cells with normal nuclear morphology compared to GFP-AKAP95-Wt (Fig. 5E), indicating that AKAP95 does not simply function as a competitive inhibitor of active caspase 3. Moreover, the T681G mutation partially impaired the protective effect of GFP-AKAP95-3M on apoptotic nuclear morphological changes in HepG2 cells (Fig. 5E). We could not determine whether this effect is a result of its inability to bind to active caspase 3 because the expression level of GFP-AKAP95-3M-T681G was lower than that of GFP-AKAP95-3M (<30% [data not shown]). However, this result clearly indicates that the protective effect of GFP-AKAP95-3M on apoptotic nuclear morphological changes depends on the expression of this protein. Collectively, our results showed that AKAP95 is a possible candidate of carrier proteins for the nuclear translocation of active caspase 3.
DISCUSSION
Mechanisms of nuclear translocation of active caspase 3.
We have identified here AKAP95 as a caspase 3-binding protein that functions as a potential cytoplasm to nucleus carrier protein for caspase 3 in the process of apoptosis execution.
Although we detected an interaction of procaspase 3 with AKAP95 in normal cells, we did not observe nuclear accumulation of procaspase 3, despite the fact that most AKAP95 was nuclear. However, since the population of procaspase 3 that binds to AKAP95 is very small, nuclearly localized procaspase 3 may be hard to detect, even if procaspase 3 is imported into nuclei in association with AKAP95. Alternatively, it is possible that procaspase 3 binding to AKAP95 prevents nuclear translocation of the bound AKAP95 by masking the AKAP95 NLS. The binding of procaspase 3 to AKAP95 may also play a role in placing procaspase 3 close to AKAP95, thereby allowing caspase 3 to have easy access to the binding site in the C-terminal region of AKAP95 once the associated caspase 3 molecule is activated.
We detected an association of endogenous procaspase 3 with AKAP95 in normal cells, but we were unable to detect an interaction of active caspase 3 with AKAP95 at endogenous protein levels. The failure to detect binding of active caspase 3 to AKAP95 at endogenous protein levels may be explained by a relatively small population of apoptotic cells, in which active caspase 3 binds to AKAP95, because apoptotic execution proceeds so fast (19, 31, 38, 41), or because the association between AKAP95 and activated caspase 3 is a weak and transient one, making it difficult to detect, especially because of the low sensitivity of anti-caspase 3 polyclonal antibodies for immunoblotting. In addition, degradation of active caspase 3 by the ubiquitin-proteasome pathway in apoptotic cells at early times reduces the amount of active caspase 3 (6, 20, 37).
Unexpectedly, we detected a weak interaction of procaspase 7 with AKAP95 by coimmunoprecipitation, but neither procaspase 7 nor activated caspase 7 was found to accumulate in nuclei (22). Although the prodomains of procaspase 3 and procaspase 7 are not conserved, caspase 3 and caspase 7 are highly conserved (54% identity) and have similar substrate specificities (40), and therefore procaspase 7 might be able to bind to the region containing the active caspase 3 binding site of AKAP95. Determination of the precise binding site of AKAP95 to procaspase 3 as well as procaspase 7 will be needed to resolve this issue.
The caspase substrate-binding groove is shaped by four surrounding loops, L1, L2, L3, and L4, whose sequences are highly conserved between caspase 3 and caspase 7 (35). However, the L2′ loop sequences, which correspond to the N-terminal region of the small subunits and are essential for substrate recognition (4, 5), are not conserved between caspase 3 and caspase 7, and this may explain why active caspase 3, but not active caspase 7, bound to AKAP95 and why only active caspase 3 accumulated in the nucleus.
From the results presented here, we can envision a molecular mechanism for the nuclear translocation of active caspase 3. In normal cells, a fraction of procaspase 3 molecules binds to AKAP95 in the cytoplasm. In response to apoptotic signals, procaspase 3 that is not bound to AKAP95 is activated and then cleaves cytoplasmic substrates, leading to apoptotic cytoplasmic changes. Procaspase 3 bound to AKAP95 is also activated, and the activated caspase 3 can remain bound to the C-terminal region of AKAP95. Since the population of procaspase 3 bound to AKAP95 in normal cells was very low, activated caspase 3 generated from procaspase 3 that was not bound to AKAP95 might also bind to AKAP95 after activation and be translocated into nucleus where it then cleaves nuclear substrates, leading to apoptotic nuclear morphological changes. Since neither the overexpression of AKAP95 NLS mutants nor siRNA-mediated depletion of AKAP95 completely abolished active caspase 3 nuclear entry, other active caspase 3 carriers might exist. Alternatively, the residual levels of AKAP95 might be sufficient for translocation of a small pool of active caspase 3 into the nucleus, where it acts on the nuclear pore from the inside and thereby allows larger proteins to diffuse in. It is reported that caspase-dependent disassembly of nuclear pores and disruption of the nucleocytoplasmic barrier precede nuclear entry of caspase 3 and DNA fragmentation mediated by caspase 3-dependent cleavage of ICAD/DFF45 (17, 23, 24), suggesting that dismantling of nuclear pores is essential for the early step of apoptotic nuclear changes. The nuclear pore membrane protein POM121, which is believed to play essential roles in formation of nuclear pores by anchoring other nucleoporins to the nuclear membrane, is cleaved in a caspase 3-dependent manner before nucleosomal DNA degradation during apoptosis (23, 24). Therefore, it seems possible that POM121 is a potential substrate for caspase 3 in nuclear pores at the early step of apoptotic nuclear morphological changes.
Regulation of nuclear morphological changes in apoptotic cells.
Caspase 3 plays essential roles in apoptotic execution, especially in the nuclear changes in apoptotic cells, as demonstrated by studies of caspase 3 knockout cells (43, 44). Although caspase-activated DNase (CAD)/DNA fragmentation factor (DFF) 40 and apoptotic chromatin condensation inducer in the nucleus (Acinus) were identified in the cytoplasmic fraction of apoptotic cells (16, 27, 33), CAD/DFF40 and Acinus are suggested to be localized in the nuclei even before apoptosis induction (26, 27, 33). Furthermore, many nuclear substrates for caspase 3 have been identified (11, 12, 18, 39). Recently, Cheung et al. (7) reported that apoptotic chromatin condensation is dependent on phosphorylation of histone H2B that is mediated by caspase 3-cleaved Mst1 kinase. Therefore, nuclearly translocated active caspase 3 may cleave and activate nuclearly localized caspase substrates such as DFF, Acinus, lamins, or Mst1 kinase, leading to apoptotic nuclear morphological changes. In addition, AKAP95 plays essential roles in mitotic chromatin condensation by recruiting PKA and the condensin complex onto chromatin (9, 13, 36). Therefore, these components might be substrates for caspase 3 during apoptotic execution and play a role in apoptotic chromatin condensation.
Acknowledgments
We thank Joel D. Leverson and Han-Kuei Huang for critical reading of the manuscript, Vishva M. Dixit for the pcDNA3/Yama plasmid, Kjetil Tasken for the human AKAP95 cDNA, John D. Scott for the anti-AKAP95 serum, and Jun-ichi Miyazaki for the pUC-CAGGS plasmid.
This study was supported in part by Grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (to S.K.) and by Public Health Service grants CA82683 and CA14195 from the National Cancer Institute (to T.H.). T.H. is a Frank and Else Schilling American Cancer Society Professor.
REFERENCES
- 1.Akileswaran, L., J. W. Taraska, J. A. Sayer, J. M. Gettemy, and V. M. Coghlan. 2001. A-kinase-anchoring protein AKAP95 is targeted to the nuclear matrix and associates with p68 RNA helicase. J. Biol. Chem. 276:17448-17454. [DOI] [PubMed] [Google Scholar]
- 2.Alnemri, E. S., D. J. Livingston, D. W. Nicholson, G. Salvesen, N. A. Thornberry, W. W. Wong, and J. Yuan. 1996. Human ICE/CED-3 protease nomenclature. Cell 87:171. [DOI] [PubMed] [Google Scholar]
- 3.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai, J., E. Shiozaki, S. M. Srinivasula, Q. Wu, P. Dataa, E. S. Alnemri, and Y. Shi. 2001. Structural basis of caspase-7 inhibition by XIAP. Cell 104: 769-780. [DOI] [PubMed] [Google Scholar]
- 5.Chai, J., Q. Wu, E. Shiozaki, S. M. Srinivasula, E. S. Alnemri, and Y. Shi. 2001. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell 107:399-407. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., L. Smith, Z. Wang, and J. B. Smith. 2003. Preservation of caspase 3 subunits from degradation contributes to apoptosis evoked by lactacystin: any single lysine or lysine pair of the small subunit is sufficient for ubiquitination. Mol. Pharmacol. 64:334-345. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, W. L., K. Ajiro, K. Samejima, M. Kloc, P. Cheung, C. A. Mizzen, A. Beeser, L. D. Etkin, J. Chernoff, W. C. Earnshaw, and C. D. Allis. 2003. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113:507-517. [DOI] [PubMed] [Google Scholar]
- 8.Coghlan, V. M., L. K. Langeberg, A. Fernandez, N. J. Lamb, and J. D. Scott. 1994. Cloning and characterization of AKAP 95, a nuclear protein that associates with the regulatory subunit of type II cAMP-dependent protein kinase. J. Biol. Chem. 269:7658-7665. [PubMed] [Google Scholar]
- 9.Collas, P., K. Le Guellec, and K. Tasken. 1999. The A-kinase-anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. J. Cell Biol. 147:1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colledge, M., and J. D. Scott. 1999. AKAPs: from structure to function. Trends Cell Biol. 9:216-221. [DOI] [PubMed] [Google Scholar]
- 11.Cryns, V., and J. Yuan. 1998. Proteases to die for. Genes Dev. 12:1551-1570. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 13.Eide, T., C. Carlson, K. A. Tasken, T. Hirano, K. Tasken, and P. Collas. 2002. Distinct but overlapping domains of AKAP95 are implicated in chromosome condensation and condensin targeting. EMBO Rep. 3:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eide, T., V. Coghlan, S. Orstavik, C. Holsve, R. Solberg, B. S. Skalhegg, N. J. Lamb, L. Langeberg, A. Fernandez, J. D. Scott, T. Jahnsen, and K. Tasken. 1998. Molecular cloning, chromosomal localization, and cell cycle-dependent subcellular distribution of the A-kinase anchoring protein, AKAP95. Exp. Cell Res. 238:305-316. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 16.Enari, M., H. Sakahira, H. Yokoyama, K. Okawa, A. Iwamatsu, and S. Nagata. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391:43-50. [DOI] [PubMed] [Google Scholar]
- 17.Faleiro, L., and Y. Lazebnik. 2000. Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol. 151:951-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, U., R. U. Janicke, and K. Schulze-Osthoff. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10:76-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, J. C., N. J. Waterhouse, P. Juin, G. I. Evan, and D. R. Green. 2000. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2:156-162. [DOI] [PubMed] [Google Scholar]
- 20.Huang, H., C. A. Joazeiro, E. Bonfoco, S. Kamada, J. D. Leverson, and T. Hunter. 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275:26661-26664. [DOI] [PubMed] [Google Scholar]
- 21.Kamada, S., H. Kusano, H. Fujita, M. Ohtsu, R. C. Koya, N. Kuzumaki, and Y. Tsujimoto. 1998. A cloning method for caspase substrates that uses the yeast two-hybrid system: cloning of the antiapoptotic gene gelsolin. Proc. Natl. Acad. Sci. USA 95:8532-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada, S., U. Kikkawa, Y. Tsujimoto, and T. Hunter. 2005. Nuclear translocation of caspase 3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s). J. Biol. Chem. 280:857-860. [DOI] [PubMed] [Google Scholar]
- 23.Kihlmark, M., C. Rustum, C. Eriksson, M. Beckman, K. Iverfeldt, and E. Hallberg. 2004. Correlation between nucleocytoplasmic transport and caspase-3-dependent dismantling of nuclear pores during apoptosis. Exp. Cell Res. 293:346-356. [DOI] [PubMed] [Google Scholar]
- 24.Kihlmark, M., G. Imreh, and E. Hallberg. 2001. Sequential degradation of proteins from the nuclear envelope during apoptosis. J. Cell Sci. 114: 3643-3653. [DOI] [PubMed] [Google Scholar]
- 25.Kuida, K., T. S. Zheng, S. Na, C. Kuan, D. Yang, H. Karasuyama, P. Rakic, and R. A. Flavell. 1996. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384:368-372. [DOI] [PubMed] [Google Scholar]
- 26.Lechardeur, D., L. Drzymala, M. Sharma, D. Zylka, R. Kinach, J. Pacia, C. Hicks, N. Usmani, J. M. Rommens, and G. L. Lukacs. 2000. Determinants of the nuclear localization of the heterodimeric DNA fragmentation factor (ICAD/CAD). J. Cell Biol. 150:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, X., P. Li, P. Widlak, H. Zou, X. Luo, W. T. Garrard, and X. Wang. 1998. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA 95:8461-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancini, M., D. W. Nicholson, S. Roy, N. A. Thornberry, E. P. Peterson, L. A. Casciola-Rosen, and A. Rosen. 1998. The caspase 3 precursor has a cytosolic and mitochondrial distribution: implications for apoptotic signaling. J. Cell Biol. 140:1485-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193-199. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo, T., S. Kamada, T. Mikami, H. Murakami, and Y. Tsujimoto. 1999. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. 6:865-872. [DOI] [PubMed] [Google Scholar]
- 31.Rehm, M., H. DuBmann, R. U. Janicke, J. M. Tavare, D. Kogel, J. H. M. Prehn. 2002. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. J. Biol. Chem. 277:24506-24514. [DOI] [PubMed] [Google Scholar]
- 32.Rotonda, J., D. W. Nicholson, K. M. Fazil, M. Gallant, Y. Gareau, M. Labelle, E. P. Peterson, D. M. Rasper, R. Ruel, J. P. Vaillancourt, N. A. Thornberry, and J. W. Becker. 1996. The three-dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nat. Struct. Biol. 3:619-625. [DOI] [PubMed] [Google Scholar]
- 33.Sahara, S., M. Aoto, Y. Eguchi, N. Imamoto, Y. Yoneda, and Y. Tsujimoto. 1999. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401:168-173. [DOI] [PubMed] [Google Scholar]
- 34.Samali, A., B. Zhivotovsky, D. P. Jones, and S. Orrenius. 1998. Detection of pro-caspase 3 in cytosol and mitochondria of various tissues. FEBS Lett. 431:167-169. [DOI] [PubMed] [Google Scholar]
- 35.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 36.Steen, R. L., F. Cubizolles, K. Le Guellec, and P. Collas. 2000. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J. Cell Biol. 149:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, Y., Y. Nakabayashi, and R. Takahashi. 2001. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase 3 and enhances its antiapoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA 98:8662-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemoto, K., T. Nagai, A. Miyawaki, and M. Miura. 2003. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 40.Thornberry, N. A., T. A. Rano, E. P. Peterson, D. M. Rasper, T. Timkey, M. Garcia-Calvo, V. M. Houtzager, P. A. Nordstrom, S. Roy, J. P. Vaillancourt, K. T. Chapman, and D. W. Nicholson. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. J. Biol. Chem. 272:17907-17911. [DOI] [PubMed] [Google Scholar]
- 41.Tyas, L., V. A. Brophy, A. Pope, A. J. Rivett, and J. M. Tavare. 2000. Rapid caspase 3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 1:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei, Y., T. Fox, S. P. Chambers, J. Sintchak, J. T. Coll, J. M. C. Golec, L. Swenson, K. P. Wilson, and P. S. Charifson. 2000. The structure of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem. Biol. 7:423-432. [DOI] [PubMed] [Google Scholar]
- 43.Woo, M., R. Hakem, M. S. Soengas, G. S. Duncan, A. Shahinian, D. Kagi, A. Hakem, M. McCurrach, W. Khoo, S. A. Kaufman, G. Senaldi, T. Howard, S. W. Lowe, and T. W. Mak. 1998. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 12:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, T. S., S. F. Schlosser, T. Dao, R. Hingorani, I. N. Crispe, J. L. Boyer, and R. A. Flavell. 1998. Caspase 3 controls both cytoplasmic and nuclear events associated with Fas-mediated apoptosis in vivo. Proc. Natl. Acad. Sci. USA 95:13618-13623. [DOI] [PMC free article] [PubMed] [Google Scholar]