Abstract
The canonical Notch signaling pathway mediated by Delta- and Jagged-like Notch ligands determines a variety of cell fates in metazoa. In Caenorhabditis elegans and sea urchins, canonical Notch signaling is essential for different cell fate specifications during early embryogenesis or the formation of endoderm, mesoderm, or ectoderm germ layers. Transcripts of Notch signaling pathway genes are present during mouse blastogenesis, suggesting that the canonical Notch signaling pathway may also function in early mammalian development. To test this directly, we used conditional deletion in oocytes carrying a ZP3Cre recombinase transgene to generate mouse embryos lacking both maternal and zygotic protein O-fucosyltransferase 1, a cell-autonomous and essential component of canonical Notch receptor signaling. Homozygous mutant embryos derived from eggs lacking Pofut1 gene transcripts developed indistinguishably from the wild type until approximately embryonic day 8.0, a postgastrulation stage after the formation of the three germ layers. Thus, in contrast to the case with C. elegans and sea urchins, canonical Notch signaling is not required in mammals for earliest cell fate specifications or for formation of the three germ layers. The use of canonical Notch signaling for early cell fate specifications by lower organisms may represent co-option of a regulatory pathway originally used later in development by all metazoa.
Identifying mechanisms responsible for early cell fate specifications and formation of the three germ layers is a fundamental issue in developmental and evolutionary biology. The canonical Notch signaling pathway stimulated by Delta- and Jagged-like ligands is functionally conserved among the metazoa (2, 29) and is required for early cell fate specifications or the formation of germ layers in Caenorhabditis elegans (12, 41) and sea urchins (1, 43-45, 52) and for endoderm patterning in zebrafish (3, 21). Drosophila melanogaster (5, 6) and C. elegans (28) need maternal contributions of certain Notch pathway components for early development, while sea urchins (25) and zebrafish (11, 35, 53) apparently do not. In mammals, there are four Notch receptors (Notch1 through Notch4) and five Notch ligands (Dll1, Dll3, Dll4, Jag1, and Jag2) that mediate the canonical Notch signaling pathway (4, 55). Drosophila and mammalian Notch receptors require protein O-fucosyltransferase 1 that transfers fucose to epidermal growth factor-like (EGF) repeats of their extracellular domain in order to signal through Delta and Jagged/Serrate ligands (24, 31-33, 40, 46). Inactivation of the mouse Pofut1 gene that encodes protein O-fucosyltransferase 1 leads to severe Notch signaling defects (46) similar to those of embryos lacking downstream effectors of Notch signaling through all four Notch receptors, such as RBP-Jκ (30), Psen1 and Psen2 (10, 17), and Mib1 (22). Protein O-fucosyltransferase 1 is therefore an essential, cell-autonomous component of the canonical Notch signaling pathway.
Gene expression studies at different stages of mouse blastogenesis have revealed a variety of Notch pathway gene transcripts, including Notch receptors, Notch ligands, downstream targets, and presenilins, leading to the proposal that canonical Notch signaling may be required for preimplantation development in mammals (8, 54). However, all mouse mutants defective in global Notch signaling survive to approximately embryonic day 9.5 (E9.5) exhibiting unimpeded development of the three germ layers (10, 17, 22, 30, 46), suggesting that canonical Notch signaling is not required before gastrulation. On the other hand, these Notch pathway mutant embryos may have been “rescued” by maternal transcripts that were present in the ovulated egg and obscured a Notch signaling requirement. To investigate whether canonical Notch signaling is essential for blastogenesis and early embryonic development, embryos lacking maternal and zygotic transcripts of a nonredundant gene whose action is essential and specific for signaling by the four mammalian Notch receptors are required. Ablation of the Pofut1 gene in oocytes would allow such embryos to be produced. The only gene in metazoan genomes related to Pofut1 is Pofut2, whose product transfers fucose to thrombospondin repeats but not to EGF repeats (27). Embryonic stem (ES) cells that lack Pofut1 but possess Pofut2 do not transfer fucose to EGF repeats but do transfer fucose to thrombospondin repeats (27).
In this study, we inactivated the Pofut1 gene specifically in oocytes by using a Cre recombinase transgene driven by the zona pellucida 3 (ZP3) promoter. Various ZP3 promoter constructs have been used to express Cre specifically in oocytes and thereby delete DNA flanked by loxP sites in early oogenesis (9, 20, 23, 42, 47). When eggs lacking maternal Pofut1 transcripts were fertilized by Pofut1Δ sperm, embryos with no maternal or zygotic Pofut1 transcripts developed through embryogenesis to approximately E8.0 in a manner indistinguishable from that of wild-type embryos. Thus, in contrast to the case with more primitive species, canonical Notch signaling is dispensable for early cell fate specifications in mammals.
MATERIALS AND METHODS
Oocyte-specific deletion of the Pofut1 gene.
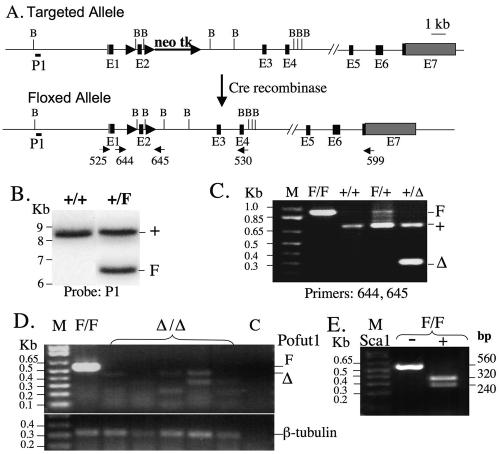
WW6 ES cells (18) were previously engineered to contain exon 2 of the Pofut1 gene flanked by two loxP sites and a selection cassette (neomycin-thymidine kinase) as described previously (46). Following Cre recombinase expression, ES cell lines with a Pofut1F allele and devoid of the selection cassette were derived and injected into C57BL/6 blastocysts. Germ line transmission was confirmed by PCR of genomic DNA (gDNA) with primers 644 and 645 (46) and by Southern analysis with the P1 probe (46) after digestion of gDNA with BamHI (Fig. 1A). Pofut1F/F females were mated with Pofut1F/+:ZP3Cre transgenic males (47) to obtain Pofut1F/F:ZP3Cre females.
FIG. 1.
The conditional Pofut1 allele and phenotyping of Pofut1Δ/Δ eggs. (A) Diagram of the targeted Pofut1 locus before and after Cre recombinase treatment to give exon 2 flanked by loxP sites (solid arrows) and showing the positions of primers, probe P1 and BamHI (B) restriction sites. Black boxes represent exons (E) and gray boxes represent UTRs. neo tk, neomycin-thymidine kinase cassette. (B) Southern analysis after digestion of genomic DNA with BamHI probed with P1. (C) PCR of genomic DNA using primers 644 and 645. +, wild-type Pofut1 allele; F, floxed allele; Δ, deleted allele; M, molecular weight markers. (D) A single-egg equivalent of cDNA was prepared from eggs of Pofut1F/F and Pofut1F/F:ZP3Cre females and subjected to PCR with primers 525 and 530 and primers for β-tubulin. No RT control (C). (E) Pofut1 cDNA from Pofut1F/F eggs was digested with Sca1 and gave the expected Pofut1 fragments.
Reverse transcriptase (RT) PCR phenotyping of ovulated eggs.
Pofut1F/F:ZP3Cre females were injected with 5 IU of pregnant mare's serum gonadotropins (Calbiochem), followed after 44 to 46 h by 5 IU of human chorionic gonadotropin (Sigma). Sixteen hours later, eggs were collected, cumulus cells were removed by hyaluronidase treatment, and 10 eggs were transferred to 10 μl lysis buffer (Cells-to-cDNA kit; Ambion). The mixture was heated at 75°C for 10 min, and 1 μl (a single-egg equivalent) was taken for RT-PCR using the SuperScript III one-step RT-PCR system with platinum Taq polymerase (Invitrogen) and primers 525 (5′-ACTTGGATCCGCACTCTGGGGCTCTGCCGTCGACAT-3′) and 530 (5′-CGCTGAAGGAAACGCCTGTGAACAGTTCTGACTT-3′) that spanned three introns (Fig. 1A). β-Tubulin primers were 5′-TCACTGTGCCTGGAACTTACC-3′ (forward) and 5′-GGAACATAGCCGTAAACT-3′ (reverse), used in a parallel RT-PCR. Conditions for reverse transcription were 50°C for 20 min and 94°C for 2 min for predenaturing, followed by 45 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 1 min. The RT-PCR products were fractionated on a 1% agarose gel and confirmed as Pofut1 gene products by analysis after Sca1 digestion.
Notch coculture signaling assay and Pofut1 cDNA correction.
ES cell lines that were Pofut1+/+ or Pofut1Δ/Δ were derived from blastocyst outgrowths obtained from mating Pofut1+/Δ heterozygotes (46) and cultured on feeder-free gelatinized plates with ES cell culture medium (alpha-minimal essential medium [GIBCO], 10% ES-qualified fetal bovine serum, 1,000 U/ml leukemia inhibitory factor [Chemicon], ampicillin and streptomycin [Invitrogen], 0.0004% beta-mercaptoethanol [Sigma]). Primers 644 and 645 were used to genotype from gDNA (Fig. 1A). RT-PCR was performed to determine phenotype from cDNA using exon-spanning primers 525 and 530 (Fig. 1A). Coculture assays were performed essentially as described previously (7). Duplicate cultures were plated at 2 × 105 ES cells (clone 8-8 Pofut1+/+ and clone 5-6-3 Pofut1Δ/Δ) per well of a six-well dish in ES cell culture medium and, after ∼24 h, were cotransfected with a total of 0.2 μg of a plasmid carrying eight copies of an RBP-Jκ DNA binding sequence driving a firefly luciferase reporter gene termed the TP1-luciferase gene (50) and 0.05 μg of a plasmid with a Renilla luciferase reporter gene driven by the thymidine kinase promoter (pRL-TK; Promega) and with 1.8 μg of a mouse Pofut1 cDNA in pCDNA3.1/Zeo (Invitrogen) or vector alone using Lipofectamine 2000 (Invitrogen). The Pofut1 cDNA was generated from RT-PCR products obtained from total RNA prepared from WW6 ES cells and amplified with primer 525 in the 5′ untranslated region (UTR) and primer 741 in the 3′ UTR (5′-ATCAGGATCCTGGGAGGTGGGGGCTTCAGA-3′). At 24 h posttransfection, 106 rat Jagged1-expressing L cells that had been presorted for high Jagged1 expression by using a goat anti-rat Jagged1 antibody (AF599 R & D Systems) (7) or Delta1-expressing L cells that were presorted for high Delta1 expression by using a goat anti-human DLL1 antibody (AF 1818; R & D Systems) or parental L cells presorted for low expression of Jagged1 (7), which also had no detectable expression of Delta1, were overlaid. At 48 h after transfection, firefly and renilla luciferase activities were quantitated in cell lysates by using a dual luciferase assay (Promega). Ligand-dependent Notch activation was expressed as induction (n-fold) of normalized luciferase activity stimulated by ligand/L cells compared to L cells.
Northern analysis.
Northern blots with total RNA from mouse embryos were obtained from Seegene (Korea). A Pofut1 cDNA probe generated by RT-PCR using primers 525 (see above) and 599 (5′-CCACCTCTGGCAGAAAAGAAAAGGGATGTGTAAT-3′) (Fig. 1A) was labeled using Prime-It (Stratagene) with [32P]dCTP. After hybridization, the blot was finally washed with 50 ml of 0.1 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate at 65°C for 20 min.
RESULTS
Oocyte-specific deletion of the Pofut1 gene.
loxP sites were previously engineered to flank exon 2 of the Pofut1 gene by homologous recombination in WW6 mouse ES cells (46) (Fig. 1A). Females with a Pofut1 floxed allele (Pofut1F) were identified by genotyping tail DNA using Southern analysis (Fig. 1B) and RT-PCR (Fig. 1C). To eliminate maternal Pofut1 gene transcripts in oocytes, Pofut1F/F females were crossed with wild type Pofut1F/+ males bearing a ZP3Cre transgene (47). When Pofut1F/F:ZP3Cre females (n = 13) were mated to wild-type males, all pups (n = 78) were heterozygous and had a Pofut1 allele deleted, showing that the ZP3Cre transgene functioned with 100% efficiency. The ovaries of Pofut1F/F: ZP3Cre females were of normal weight and appearance and had oocytes at all stages of oogenesis in similar numbers. Most Pofut1F/F:ZP3Cre females (n = 21) had litters of the expected size (mean ± standard deviation, 7.2 ± 1.1), although a small proportion produced several litters of small size.
Pofut1Δ/Δ eggs lack Pofut1 transcripts.
A sensitive RT-PCR assay that readily detected Pofut1F/F gene transcripts in a single-egg equivalent using primers that spanned three introns was developed (Fig. 1D, lane F/F). Digestion of these Pofut1 PCR products with the restriction enzyme Sca1 produced two fragments of the predicted size from Pofut1 cDNA (Fig. 1E). By contrast, eggs from Pofut1F/F:ZP3Cre mutant females did not possess any transcripts from the floxed Pofut1 alleles. Thus, there was no maternal contribution of Pofut1 RNA. In three of the mutant samples, Pofut1Δ transcripts of the expected size for transcripts lacking exon 2 were faintly visible. These truncated Pofut1 transcripts are usually difficult to detect (46), presumably because of removal by nonsense-mediated decay. All mutant eggs gave products from β-tubulin cDNA performed in a parallel RT-PCR (Fig. 1D).
Pofut1Δ/Δ ES cells are defective in canonical Notch signaling.
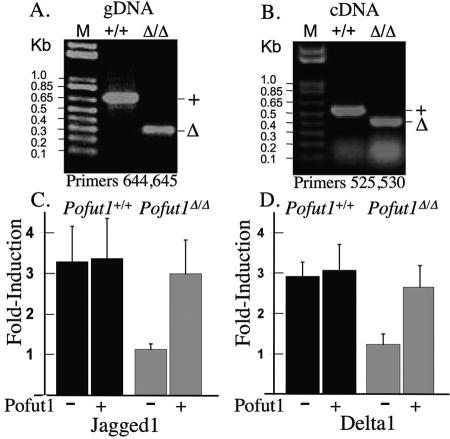
In order to determine if Notch signaling was inhibited in Pofut1Δ/Δ blastocysts, ES cells were derived from outgrowths of E3.5 blastocysts. Genotyping by PCR is shown in Fig. 2A. Pofut1Δ/Δ ES cells were found by RT-PCR to lack Pofut1 transcripts from the floxed Pofut1 alleles (Fig. 2B). However, truncated, mutant Pofut1 transcripts were evident in Pofut1Δ/Δ ES cells (Fig. 2B). The 34-amino-acid peptide encoded by these transcripts would not be expected to enter the secretory pathway, as it is largely a signal peptide (26, 33). Wild-type and mutant ES cells were tested for ligand-induced Notch signaling in a coculture reporter assay. Pofut1+/+ ES cells exhibited Notch signaling when cocultured with either of the Notch ligand-expressing cell types, Jagged1/L or Delta1/L. By contrast, mutant Pofut1Δ/Δ ES cells were not stimulated to signal when cocultured with either Jagged1/L (Fig. 2C) or Delta1/L cells (Fig. 2D). Cotransfection of a Pofut1 cDNA rescued Notch signaling in Pofut1Δ/Δ ES cells (Fig. 2C and D) showing that the lack of canonical Notch signaling in these cells was due to the absence of Pofut1. Overexpression of a Pofut1 cDNA did not, however, enhance Notch signaling in Pofut1+/+ ES cells.
FIG. 2.
Pofut1Δ/Δ ES cells do not exhibit canonical Notch signaling. (A) Genomic DNA from Pofut1+/+ and Pofut1Δ/Δ ES cells was subjected to PCR with primers 644 and 645 (Fig. 1A). (B) cDNA prepared from total RNA of Pofut1+/+ and Pofut1Δ/Δ ES cells was subjected to PCR with primers 525 and 530 (Fig. 1A). (C) Induction (n-fold) (eight cultures) of Notch signaling induced by coculture of ES cells with Jagged1/L cells compared to L cells before and after transfection with mouse Pofut1 cDNA. (D) Induction (n-fold) (four cultures) of Notch signaling induced by coculture of ES cells with Delta1/L cells compared to L cells before and after transfection with mouse Pofut1 cDNA. Error bars represent standard deviation.
Pofut1Δ/Δ embryos from eggs lacking maternal Pofut1 transcripts develop indistinguishably from wild-type embryos.
To determine if fertilized eggs devoid of Pofut1 transcripts could be fertilized and develop, Pofut1F/F:ZP3Cre females were mated with Pofut1+/Δ males and embryos were examined at E9.5. Of 34 embryos from five crosses, 16 were mutants (Pofut1Δ/Δ) and 18 were heterozygous (Pofut1+/Δ). No embryos had a Pofut1F allele. Therefore, eggs lacking Pofut1 were fertilized by sperm that also lacked Pofut1 and gave the same number of E9.5 embryos as eggs fertilized with a Pofut1+ sperm.
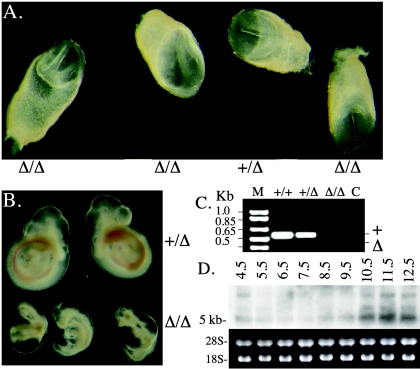
The embryos lacking both maternal and zygotic Pofut1 gene transcripts were examined at E8.0 and E9.5. Figure 3A shows that Pofut1Δ/Δ and Pofut+/Δ embryos at E8.0 from the same litter were indistinguishable from each other. However, by E9.5, the Pofut1Δ/Δ embryos were significantly smaller than the wild type, and the severe Notch signaling phenotype described in detail previously (46) was readily apparent (Fig. 3B). As observed previously (46), all Pofut1Δ/Δ E9.5 embryos were surrounded by a yolk sac with defective vascularization and had Notch signaling defects in somitogenesis, cardiogenesis, vasculogenesis, and neurogenesis (data not shown). The earliest visible Notch signaling defects were observed at approximately E8.5 in somitogenesis (somites fused and irregular) and neurogenesis (kinked neural tube), as in embryos with RBP-Jκ, Mib1, and Psen1/2 null mutations (10, 17, 22, 30, 46). Therefore, despite the absence of canonical Notch signaling, eggs were fertilized, Pofut1Δ/Δ blastocysts progressed through each stage of blastogenesis, implanted and developed in the same time and with the same morphology as heterozygous embryos derived from Pofut1Δ/Δ eggs.
FIG. 3.
Mutant embryos from Pofut1Δ/Δ eggs develop normally. E8.0 embryos (A) and E9.5 embryos (B) from a cross between a Pofut1F/F:ZP3Cre female and a Pofut1+/Δ male are shown. (C) cDNA prepared from total RNA of E6.5 embryos from a cross between Pofut1+/Δ and Pofut1+/Δ mice was subjected to PCR with primers 525 and 530 (Fig. 1A). M, molecular weight markers; C, no RT control. (D) Northern analysis with total RNA from mouse embryos at different stages postimplantation probed with a Pofut1 cDNA generated by primers 525 and 599 (Fig. 1A).
It is apparent that the ready detection of Pofut1 transcripts in Pofut1F/F eggs (Fig. 1D) and E6.5 embryos (Fig. 3C) cannot be used to predict a requirement for Pofut1 during blastogenesis or the time at which Pofut1 activity is required during postimplantation development. Northern analysis showed that Pofut1 transcripts are low just after implantation and remain barely detectable until mid-gestation (Fig. 3D). The inability to correlate transcript level with function may also apply to transcripts of other Notch pathway genes detected during blastogenesis (8, 54). In fact, not all microarray studies indicate upregulation of Notch pathway gene transcripts prior to gastrulation (15, 16, 51).
DISCUSSION
Blastocysts lacking maternal and zygotic Notch signaling develop normally.
By generating Pofut1F/F:ZP3Cre female mice, we obtained eggs that lacked maternal Pofut1 transcripts based on a sensitive RT-PCR assay. With the ZP3Cre transgene, the Pofut1 gene is inactivated at the beginning of oogenesis when an oocyte has a volume ∼200-fold less than a preovulatory oocyte. Any protein O-fucosyltransferase 1 present in oocytes before the Pofut1 gene was inactivated should be lost over the 2 to 3 weeks of oogenesis prior to ovulation. We previously showed this to be the case for another glycosyltransferase responsible for the synthesis of complex N-glycans (47). In that case, it was possible to show that Mgat1Δ/Δ eggs did not produce the glycan products synthesized by the GlcNAc-TI enzyme encoded by the Mgat1 gene. Moreover, the same strategy was used successfully by others to eliminate maternal transcripts of another glycosyltransferase gene (42). Blastocysts derived from eggs and sperm lacking Pofut1 developed normally in the absence of this essential component of the canonical Notch signaling pathway. The fact that canonical Notch signaling was inactive was shown in a coculture assay using ES cells obtained from Pofut1Δ/Δ blastocysts (Fig. 2). While Pofut1+/+ ES cells exhibited Delta1- and Jagged1-induced Notch signaling, Pofut1Δ/Δ ES cells did not. Therefore, mouse embryos lacking maternal and zygotic Pofut1 are unable to undergo canonical ligand-induced signaling through Notch receptors, and yet they develop like wild type embryos to approximately E8.0. Thus, it can be concluded that canonical Notch signaling is not required for cell lineage specifications during blastogenesis or for the formation of the ectoderm, endoderm, or mesoderm layers prior to gastrulation in the mouse embryo.
Jagged1 does not require O-fucose to function during oogenesis.
Another conclusion from the oocyte-specific deletion of the Pofut1 gene is that O-fucose is not required on any protein with EGF-repeats containing the O-fucose consensus site (14) for functions during oogenesis, ovulation, fertilization, or early embryonic development. In situ hybridization studies have suggested that Jagged1 in the oocyte stimulates Notch receptors in cumulus cells (13, 19), and Lfng mutant studies have shown that Notch signaling modulated by Lunatic fringe in cumulus cells is required for meiosis (13). Both Serrate/Jagged and Delta Notch ligands have EGF repeats that are substrates of Pofut1 and Fringe (34). The fact that mouse oocytes in which the Pofut1 gene is inactivated at the beginning of oogenesis are not impaired in their development or ovulation suggests that Jagged1 in the oocyte does not require O-fucose to induce Notch signaling or for any other reason. This is consistent with experiments with Drosophila showing that inactivation of OFUT1 does not cause functional defects in either of the two Notch ligands Delta and Serrate (31, 40).
Roles of canonical Notch signaling are not evolutionarily conserved in early cell fate specifications.
Canonical Notch signaling is utilized in early embryonic development in several species but at different stages of embryogenesis. In C. elegans, canonical Notch signaling is involved in primitive mesoderm induction by interacting with TBX37 and TBX38, T-box genes that lack clear orthologs in other species (12). The most related T-box gene in mice is the Tbx6 gene, which is expressed in the presomitic mesoderm and is thought to work upstream of Notch signaling in influencing the formation of posterior somites (56). In C. elegans, inhibition of canonical Notch signaling in the AB cell results in retention of an ectodermal primary cell fate (38, 41). In sea urchins, LvNotch signaling determines the ectoderm-endoderm boundary (45) and altered expression or inhibition of LvNotch signaling changes that boundary. Notch action is also required for the subdivision of mesendoderm into mesenchyme and endoderm at the blastula stage in sea urchins (1, 36, 43, 44). In zebrafish, Notch/Delta signaling is involved in the regionalization of her5 gene expression by inhibiting its expression (3). her5 is the zebrafish hairy/enhancer of split-related gene, and it plays a critical role in endoderm patterning in zebrafish. Overexpression of activated Notch at an early stage in zebrafish embryos inhibits the formation of endoderm (21). However, inhibition of Notch signaling did not lead to an accumulation of endodermal precursors (21). In Drosophila, Notch signaling is utilized early in development to maintain a proneuroblast cell fate (6). We show here that, in mice, canonical Notch signaling is dispensable for early embryonic development.
During evolution, mammals may have lost the ability to use Notch signaling for early embryogenesis and the formation of the three germ layers. However, the differences in functions and stages at which canonical Notch signaling is utilized in more primitive organisms and the fact that only C. elegans and Drosophila require a maternal contribution of Notch signaling components, suggest that early embryonic roles of canonical Notch signaling may not have originated with a common ancestor. Rather, the common ancestor may have been like mammals and not used canonical Notch signaling for early cell specifications. Canonical Notch signaling may have evolved originally to function in more advanced developmental processes, such as neurogenesis and segmentation, with the use of Notch signaling in cell fate decisions being restricted to these novelties (49, 57). During subsequent evolution leading to C. elegans and sea urchins, canonical Notch signaling may have been co-opted to function also in embryonic development by interacting with different genetic networks to regulate early cell fate specifications (37, 39). This proposal is consistent with the fact that the published genomes of the unicellular protists Plasmodium falciparum and P. yeolii (http://www.tigr.org/) lack Notch signaling pathway genes. While several homologues of Notch signaling pathway genes have been found in the hydra (48), a cnidarian consisting of two layers, some two-layer species may not possess genes for Notch signaling.
Acknowledgments
We thank Suzannah Williams for reviewing ovarian sections, Gerry Weinmaster for Delta/L and Jagged/L cells, Lothar and Ursula Strobl and Georg Bornkamm for the TP-1-luciferase plasmid, Nick Baker, Robert Haltiwanger, and Ken Irvine for helpful comments on the manuscript, and Jihua Chen for technical advice.
This work was supported by grant RO1 30645 from the NIH to P.S. and partial support was provided by the Albert Einstein Cancer Center grant PO1 13330.
REFERENCES
- 1.Angerer, L. M., and R. C. Angerer. 2003. Patterning the sea urchin embryo: gene regulatory networks, signaling pathways, and cellular interactions. Curr. Top. Dev. Biol. 53:159-198. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Bally-Cuif, L., C. Goutel, M. Wassef, W. Wurst, and F. Rosa. 2000. Coregulation of anterior and posterior mesendodermal development by a hairy-related transcriptional repressor. Genes Dev. 14:1664-1677. [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, M., H. Aslam, M. Flasza, M. Fostier, J. E. Higgs, S. L. Mazaleyrat, and M. B. Wilkin. 2002. Multiple levels of Notch signal regulation. Mol. Membr. Biol. 19:27-38. [DOI] [PubMed] [Google Scholar]
- 5.Bellotto, M., D. Bopp, K. A. Senti, R. Burke, P. Deak, P. Maroy, B. Dickson, K. Basler, and E. Hafen. 2002. Maternal-effect loci involved in Drosophila oogenesis and embryogenesis: P element-induced mutations on the third chromosome. Int. J. Dev. Biol 46:149-157. [PubMed] [Google Scholar]
- 6.Campos-Ortega, J. A. 1995. Genetic mechanisms of early neurogenesis in Drosophila melanogaster. Mol. Neurobiol. 10:75-89. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., D. J. Moloney, and P. Stanley. 2001. Fringe modulation of Jagged1-induced Notch signaling requires the action of β4galactosyltransferase-1. Proc. Natl. Acad. Sci. USA 98:13716-13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier, S., S. Vandormael-Pournin, C. Babinet, and M. Cohen-Tannoudji. 2004. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr. Patterns 4:713-717. [DOI] [PubMed] [Google Scholar]
- 9.de Vries, W. N., L. T. Binns, K. S. Fancher, J. Dean, R. Moore, R. Kemler, and B. B. Knowles. 2000. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26:110-112. [PubMed] [Google Scholar]
- 10.Donoviel, D. B., A. K. Hadjantonakis, M. Ikeda, H. Zheng, P. S. Hyslop, and A. Bernstein. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dosch, R., D. S. Wagner, K. A. Mintzer, G. Runke, A. P. Wiemelt, and M. C. Mullins. 2004. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev. Cell 6:771-780. [DOI] [PubMed] [Google Scholar]
- 12.Good, K., R. Ciosk, J. Nance, A. Neves, R. J. Hill, and J. R. Priess. 2004. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development 131:1967-1978. [DOI] [PubMed] [Google Scholar]
- 13.Hahn, K. L., J. Johnson, B. J. Beres, S. Howard, and J. Wilson-Rawls. 2005. Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development 132:817-828. [DOI] [PubMed] [Google Scholar]
- 14.Haines, N., and K. D. Irvine. 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4:786-797. [DOI] [PubMed] [Google Scholar]
- 15.Hamatani, T., M. G. Carter, A. A. Sharov, and M. S. Ko. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6:117-131. [DOI] [PubMed] [Google Scholar]
- 16.Hamatani, T., T. Daikoku, H. Wang, H. Matsumoto, M. G. Carter, M. S. Ko, and S. K. Dey. 2004. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. USA 101:10326-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herreman, A., D. Hartmann, W. Annaert, P. Saftig, K. Craessaerts, L. Serneels, L. Umans, V. Schrijvers, F. Checler, H. Vanderstichele, V. Baekelandt, R. Dressel, P. Cupers, D. Huylebroeck, A. Zwijsen, F. Van Leuven, and B. De Strooper. 1999. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. USA 96:11872-11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ioffe, E., Y. Liu, M. Bhaumik, F. Poirier, S. M. Factor, and P. Stanley. 1995. WW6: an embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc. Natl. Acad. Sci. USA 92:7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J., T. Espinoza, R. W. McGaughey, A. Rawls, and J. Wilson-Rawls. 2001. Notch pathway genes are expressed in mammalian ovarian follicles. Mech. Dev. 109:355-361. [DOI] [PubMed] [Google Scholar]
- 20.Kemler, R., A. Hierholzer, B. Kanzler, S. Kuppig, K. Hansen, M. M. Taketo, W. N. de Vries, B. B. Knowles, and D. Solter. 2004. Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development 131:5817-5824. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, Y., H. Verkade, J. F. Reiter, C. H. Kim, A. B. Chitnis, A. Kuroiwa, and D. Y. Stainier. 2004. Notch signaling can regulate endoderm formation in zebrafish. Dev. Dyn. 229:756-762. [DOI] [PubMed] [Google Scholar]
- 22.Koo, B. K., H. S. Lim, R. Song, M. J. Yoon, K. J. Yoon, J. S. Moon, Y. W. Kim, M. C. Kwon, K. W. Yoo, M. P. Kong, J. Lee, A. B. Chitnis, C. H. Kim, and Y. Y. Kong. 2005. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development, 132:3459-3470. [DOI] [PubMed]
- 23.Lan, Z. J., X. Xu, and A. J. Cooney. 2004. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 71:1469-1474. [DOI] [PubMed] [Google Scholar]
- 24.Lei, L., A. Xu, V. M. Panin, and K. D. Irvine. 2003. An O-fucose site in the ligand binding domain inhibits Notch activation. Development 130:6411-6421. [DOI] [PubMed] [Google Scholar]
- 25.Levine, M., and E. H. Davidson. 2005. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102:4936-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, Y., and R. S. Haltiwanger. 2005. O-fucosylation of notch occurs in the endoplasmic reticulum. J. Biol. Chem. 280:289-294. [DOI] [PubMed] [Google Scholar]
- 27.Luo, Y., W. Vornman, V. Panin, S. Shi, P. Stanley, and R. S. Haltiwanger. 2003. Identification of a novel enzyme responsible for O-fucosylation of thrombospondin type 1 repeats. Glycobiology 13:873. [Google Scholar]
- 28.Moskowitz, I. P., and J. H. Rothman. 1996. lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are regulated by maternal GLP-1 signaling in Caenorhabditis elegans. Development 122:4105-4117. [DOI] [PubMed] [Google Scholar]
- 29.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 30.Oka, C., T. Nakano, A. Wakeham, J. L. de la Pompa, C. Mori, T. Sakai, S. Okazaki, M. Kawaichi, K. Shiota, T. W. Mak, and T. Honjo. 1995. Disruption of the mouse RBP-J. kappa gene results in early embryonic death. Development 121:3291-3301. [DOI] [PubMed] [Google Scholar]
- 31.Okajima, T., and K. D. Irvine. 2002. Regulation of Notch signaling by O-linked fucose. Cell 111:893-904. [DOI] [PubMed] [Google Scholar]
- 32.Okajima, T., A. Xu, and K. D. Irvine. 2003. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J. Biol. Chem. 278:42340-42345. [DOI] [PubMed] [Google Scholar]
- 33.Okajima, T., A. Xu, L. Lei, and K. D. Irvine. 2005. Chaperone activity of protein O-fucosyltransferase 1 promotes Notch receptor folding. Science 307:1599-1603. [DOI] [PubMed] [Google Scholar]
- 34.Panin, V. M., L. Shao, L. Lei, D. J. Moloney, K. D. Irvine, and R. S. Haltiwanger. 2002. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 277:29945-29952. [DOI] [PubMed] [Google Scholar]
- 35.Pelegri, F., M. P. Dekens, S. Schulte-Merker, H. M. Maischein, C. Weiler, and C. Nusslein-Volhard. 2004. Identification of recessive maternal-effect mutations in the zebrafish using a gynogenesis-based method. Dev. Dyn. 231:324-335. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, R. E., and D. R. McClay. 2005. A Fringe-modified Notch signal affects specification of mesoderm and endoderm in the sea urchin embryo. Dev. Biol. 282:126-137. [DOI] [PubMed] [Google Scholar]
- 37.Pires-daSilva, A., and R. J. Sommer. 2003. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4:39-49. [DOI] [PubMed] [Google Scholar]
- 38.Priess, J. R., H. Schnabel, and R. Schnabel. 1987. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell 51:601-611. [DOI] [PubMed] [Google Scholar]
- 39.Raff, R. A., and B. J. Sly. 2000. Modularity and dissociation in the evolution of gene expression territories in development. Evol. Dev. 2:102-113. [DOI] [PubMed] [Google Scholar]
- 40.Sasamura, T., N. Sasaki, F. Miyashita, S. Nakao, H. O. Ishikawa, M. Ito, M. Kitagawa, K. Harigaya, E. Spana, D. Bilder, N. Perrimon, and K. Matsuno. 2003. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development 130:4785-4795. [DOI] [PubMed] [Google Scholar]
- 41.Schnabel, R., and J. R. Priess. 1997. Specification of cell fates in the early embryo, vol. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 42.Shafi, R., S. P. Iyer, L. G. Ellies, N. O'Donnell, K. W. Marek, D. Chui, G. W. Hart, and J. D. Marth. 2000. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97:5735-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwood, D. R., and D. R. McClay. 1997. Identification and localization of a sea urchin Notch homologue: insights into vegetal plate regionalization and Notch receptor regulation. Development 124:3363-3374. [DOI] [PubMed] [Google Scholar]
- 44.Sherwood, D. R., and D. R. McClay. 1999. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 126:1703-1713. [DOI] [PubMed] [Google Scholar]
- 45.Sherwood, D. R., and D. R. McClay. 2001. LvNotch signaling plays a dual role in regulating the position of the ectoderm-endoderm boundary in the sea urchin embryo. Development 128:2221-2232. [DOI] [PubMed] [Google Scholar]
- 46.Shi, S., and P. Stanley. 2003. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 100:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, S., S. A. Williams, A. Seppo, H. Kurniawan, W. Chen, Z. Ye, J. D. Marth, and P. Stanley. 2004. Inactivation of the Mgat1 gene in oocytes impairs oogenesis, but embryos lacking complex and hybrid N-glycans develop and implant. Mol. Cell. Biol. 24:9920-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steele, R. E. 2002. Developmental signaling in Hydra: what does it take to build a “simple” animal? Dev. Biol. 248:199-219. [DOI] [PubMed] [Google Scholar]
- 49.Stollewerk, A., M. Schoppmeier, and W. G. Damen. 2003. Involvement of Notch and Delta genes in spider segmentation. Nature 423:863-865. [DOI] [PubMed] [Google Scholar]
- 50.Strobl, L. J., H. Hofelmayr, C. Stein, G. Marschall, M. Brielmeier, G. Laux, G. W. Bornkamm, and U. Zimber-Strobl. 1997. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology 198:299-306. [DOI] [PubMed] [Google Scholar]
- 51.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sweet, H. C., M. Gehring, and C. A. Ettensohn. 2002. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development 129:1945-1955. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, D. S., R. Dosch, K. A. Mintzer, A. P. Wiemelt, and M. C. Mullins. 2004. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev. Cell 6:781-790. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Q. T., K. Piotrowska, M. A. Ciemerych, L. Milenkovic, M. P. Scott, R. W. Davis, and M. Zernicka-Goetz. 2004. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell. 6:133-144. [DOI] [PubMed] [Google Scholar]
- 55.Weinmaster, G., and C. Kintner. 2003. Modulation of notch signaling during somitogenesis. Annu. Rev. Cell Dev. Biol. 19:367-395. [DOI] [PubMed] [Google Scholar]
- 56.White, P. H., D. R. Farkas, E. E. McFadden, and D. L. Chapman. 2003. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development 130:1681-1690. [DOI] [PubMed] [Google Scholar]
- 57.Wodarz, A., and W. B. Huttner. 2003. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech. Dev. 120:1297-1309. [DOI] [PubMed] [Google Scholar]