Abstract
The fidelity of chromosomal duplication is monitored by cell cycle checkpoints operational during mitosis. One such cell cycle delay is invoked by microtubule-targeting agents such as nocodazole or paclitaxel (Taxol) and is mediated by mitotic checkpoint proteins that include BubR1. Relatively little is known about the regulation of expression and stability of BubR1 (or other checkpoint proteins) and how these factors dictate the durability of the cell cycle delay. We report here that treatment of HeLa cells with spindle-disrupting agents resulted in caspase activation and precipitated the cleavage of BubR1. This mechanism ultimately leads to reduced levels of full-length protein, which are accompanied by abrogation of the mitotic block; the checkpoint abrogation is substantially accelerated by inhibition of de novo protein synthesis. In contrast, inhibition of caspase activity blocked BubR1 degradation and prolonged mitosis. To confirm a direct link between caspase activity and BubR1 protein expression, we identified by site-directed mutagenesis the specific caspase cleavage sites cleaved after exposure to paclitaxel. Surprisingly, BubR1 has two sites of cleavage: primarily at Asp607/Asp610 and secondarily at Asp576/Asp579. BubR1 mutated at both locations (BubR1Δ579Δ610) was resistant to paclitaxel-induced degradation. Expression of BubR1Δ579Δ610 augmented the mitotic delay induced by spindle disruption in transfected cells as well as in clones engineered to inducibly express the mutant protein upon exposure to doxycycline and ultimately led to increased aneuploidy. Underscoring the importance of these caspase cleavage sites, both tetrapeptide motifs are identified in the amino acid sequences of human, mouse, chicken, and Xenopus BubR1. These results are potentially the first to link the control of the stability of a key mitotic checkpoint protein to caspase activation, a regulatory pathway that may be involved in killing defective cells and that has been evolutionarily conserved.
Mitosis is the visually spectacular event that marks the separation of one somatic cell into two. In order for the resultant daughter cells to remain viable and retain reproductive potential, each must receive an equal and full complement of chromosomes. The mitotic spindle checkpoint is a prime mechanism that ensures that this happens (for reviews, see references 3, 5, and 11). This cell cycle checkpoint blocks progression into anaphase until all chromosomes have completely aligned at the metaphase plate. By monitoring correct microtubule attachment to the chromosomes and the resultant tension, the spindle checkpoint is sensitive enough to delay completion of mitosis when a single chromosome is misaligned (24, 38).
Current models propose that specific checkpoint-related proteins—such as BubR1, Bub1, Bub3, Mad1, and Mad2—contribute to monitoring lack of tension or attachment between the kinetochore and microtubules of the mitotic spindle and transmit a “wait signal” to inhibit the anaphase promoting complex/cyclosome (APC/C) (for reviews, see references 29 and 37). Unaligned chromosomes preferentially accumulate these checkpoint proteins at the kinetochore; once the chromosomes become properly aligned, the kinetochore localization of these proteins diminishes. Whether or not BubR1 requires kinetochore localization for full activity remains a topic of investigation. BubR1 is clearly recruited to the kinetochores of cells in which microtubule dynamics have been disrupted by nocodazole or paclitaxel but may also participate as a component of a diffusible anaphase-wait signal. It has also been proposed that BubR1 binds to Bub1, Bub3, and the Cdc20 subunit of the APC/C, forming a complex that inhibits mitotic progression (and which has therefore been called the mitotic checkpoint complex) (4, 9, 13, 26, 28, 40, 42, 45). Meraldi and colleagues found that depletion of BubR1 or Mad2 caused acceleration of mitosis, while depletion of Mad1, Bub1, and Bub3 did not affect the timing of mitotic progression (27). Interestingly, the effect of BubR1 and Mad2 on mitotic timing appeared independent of their kinetochore localization, suggesting that cytosolic Mad2 or BubR1 can inhibit the APC before the spindle checkpoint becomes fully operational, i.e., when kinetochores are still assembling. In a separate study, a number of mitotic checkpoint complex components (Mad2, BubR1, Bub3, and Cdc20) were found to undergo rapid turnover at unattached prometaphase kinetochores, while in contrast Mad1 and Bub1 remained stable at unattached kinetochores (10). More recently, evidence has surfaced that suggests BubR1 may be required for stable microtubule attachment to chromosomes (21).
In addition to critical roles in regulating cell cycle progression, BubR1 expression also appears to influence cellular viability and organ development. Stable expression of interfering RNA targeting BubR1 resulted in protein ablation and led to massive chromosomal loss and cell death within six cell divisions (18). Studies in mice have also indicated that BubR1 is an essential protein, as complete knockout of BubR1 is embryonically lethal (46). The development of mice haplodeficient for, or with hypomorphic alleles of, the mouse Bub1b gene (which encodes BubR1 protein) has enabled assessment of the phenotypic effects of graded levels of BubR1 protein (1). Mice hypomorphic in one allele but with a knockout in the other expressed BubR1 protein at 4% of the levels in wild-type mice; however, these animals died within hours after birth. In contrast, mice hypomorphic in both alleles expressed BubR1 at 11% of the levels of wild-type animals but were viable and grew to adulthood. These mice, however, were infertile and showed marked signs of early aging, with reduced life spans, cachexia, lordokyphosis, cataracts, loss of subcutaneous fat, and impaired wound healing. This striking phenotype was felt to be likely attributable to early senescence and increased apoptosis in response to environmental stress. Mice haplodeficient for BubR1 show higher levels of BubR1 protein (25 to 50% of wild type). The haplodeficient mice are viable and grow to adulthood with no apparent shortening of their life span. However, it was noted that the embryo fibroblasts haplodeficient for BubR1 remain defective for the mitotic spindle checkpoint, and the haplodeficient mice show increased tumor formation when challenged with carcinogens (6). These reports together indicate that levels of BubR1 protein can have profound effects on the viability of cells, including response to environmental stress and microtubule disruption; impaired levels of BubR1 expression in mice are also associated with early aging, infertility, and predisposition to tumor formation.
Despite the importance of adequate expression of BubR1 on checkpoint control and cellular viability, the mechanisms that determine BubR1 protein levels remain incompletely characterized. In cells exposed to nocodazole for prolonged periods, BubR1 protein levels were found to gradually decline, which was ultimately associated with abrogation of the mitotic checkpoint (39). BubR1 protein expression levels were found to be inversely proportional to advanced age in mice, with early aging noted in BubR1-deficient mice (1). As increased caspase activity is also a hallmark of aging (14, 20, 48), we investigated whether caspase activity could be directly linked to BubR1 protein expression. We found that the time-dependent degradation of BubR1 and abrogation of the mitotic cell cycle delay after microtubule disruption were associated with caspase activation. Consistent with a role for caspases in determining duration of mitosis, inhibition of caspase activity markedly enhanced the mitotic delay. We identified, via site-directed mutagenesis, the specific caspase tetrapeptide recognition/cleavage sites in BubR1 and discovered that the two motifs are evolutionarily conserved. Finally, targeted mutation of the caspase cleavage sites of BubR1 led to delayed exit from mitosis. These observations together strongly identify caspase activation as a novel determinant of BubR1 protein levels and, consequently, mitotic progression.
MATERIALS AND METHODS
Cell culture, reagents, and drugs.
HeLa cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. Paclitaxel, vincristine, nocodazole, cycloheximide, MG-132, and aphidicolin were all from Sigma and prepared as 1,000× stock in dimethyl sulfoxide. The cell-permeable caspase inhibitors DEVD-CHO (inhibitor of caspase 3), YVAD-CHO (inhibitor of caspase 1), IETD-CHO (inhibitor of caspase 8), LEVD-CHO (inhibitor of caspase 4) and broad-spectrum (pan-) caspase inhibitor Boc-D-CMK (caspase inhibitor IV) were from either Calbiochem or Bio-Source. All inhibitors were prepared as concentrated stock solutions in dimethyl sulfoxide and were used at final concentrations of 20 μM. Transfections were performed via Lipofectamine 2000 (Invitrogen). Synchronization of cells was performed via aphidicolin as previously described (15, 16).
Analytical methods.
For immunoblotting, cells were harvested, pelleted, and resuspended, and then sonicated and resuspended again in Laemmli buffer, followed by boiling for 5 min, prior to separation via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 μg of protein per lane) and transfer to nitrocellulose membranes. After transfer, the membranes were blocked with 5% nonfat milk in phosphate-buffered saline (PBS)-Tween and then probed with the indicated primary antibodies, followed by the appropriate secondary antibodies conjugated with horseradish peroxidase. Washes were performed with PBS-Tween. Finally, membranes were processed with enhanced chemiluminescence (Amersham Biosciences) and then exposed to film. Anti-Bub1 and anti-BubR1 were generous gifts of Tim J. Yen (generated against the full-length BubR1 protein as described [13]) or Raimundo Freire, and each was used at a 1:1,000 dilution. Anti-alpha tubulin and anti-beta-actin (Sigma) were all used at a dilution of 1:1,000 for immunoblotting, while anti-caspase 3 antibody (Biomol) was used at dilution of 1:500.
Immunofluorescence and fluorescence-activated cell sorting (FACS) analysis of DNA content were performed as previously described (15-17). During the FACS, no gating was performed on the propidium iodide (PI)-stained nuclei to ensure that all cells were included in the analysis, including those with sub-G1 content representative of nuclear fragmentation. Prior to staining of the nuclei, cells with sub-G1 content were confirmed to be nonviable through trypan blue and PI exclusion assays. For immunofluorescence, cells grown on coverslips were fixed in ice-cold acetone:methanol (50:50) and washed in PBS and KB buffer (50 mM Tris-HCl,pH 7.4, 150 mM NaCl, 0.1% bovine serum albumin) prior to labeling with specific antibodies. The coverslips were then mounted in 0.1% paraphenylenediamine in glycerol. Stained cells were examined with a 100× PlanNeofluor lens objective mounted on a Nikon TE-2000 microscope equipped with epifluorescence optics. Assessments of cellular viability were performed via PI exclusion in cells grown in culture dishes, via a protocol modified from Overbeeke et al. (31). Cells showing PI uptake were confirmed to be nonviable via lack of trypan blue exclusion, as well as via extended culturing; live cells are able to exclude both PI and trypan blue. The PI exclusion assays were also performed in the presence of Hoechst 33342 vital dye (Sigma), which is taken up and stains the nuclei of all cells, alive or dead. In all instances, the epifluorescent images were captured with a Hammamatsu charge-coupled-device camera that was controlled with IP LabSpectrum version 2.0.1 (Scanalytics Inc.).
Expression vectors and site-directed mutagenesis.
cDNA encoding wild-type human BubR1 fused to green fluorescent protein (GFP:BubR1) was a kind gift from Gordan Chan and Tim Yen and was targeted for mutagenesis. All mutations were made with a QuikChange site-directed mutagenesis kit (Stratagene). Oligonucleotide primer pairs used for the respective mutations were as follows (the nucleotide changed for each mutation is italicized): for the BubR1 (D73E) mutation, TAC ACT GGA AAT GAA CCT CTG GAT GTT TGG GAT AGG and CCT ATC CCA AAC ATC CAG AGG TTC ATT TCC AGT GTA; for the BubR1 (D76E) mutation, was AAT GAC CCT CTG GAA GTT TGG GAT AGG TAT ATC AGC TGG and CCA GCT GAT ATA CCT ATC CCA AAC TTC CAG AGG GTC ATT; for the BubR1 (D79E) mutation, AAT GAC CCT CTG GAT GTT TGG GAA AGG TAT ATC AGC TGG and CCA GCT GAT ATA CCT TTC CCA AAC ATC CAG AGG GTC ATT; for the BubR1 (D576E) mutation, GTG TCT CCA GAA GTT TGT GAT GAA TTT ACA GGA ATT GAA CCC and GGG TTC AAT TCC TGT AAA TTC ATC ACA AAC TTC TGG AGA CAC; for the BubR1 (D579E) mutation, GTG TCT CCA GAT GTT TGT GAA GAA TTT ACA GGA ATT GAA CCC and GGG TTC AAT TCC TGT AAA TTC TTC ACA AAC ATC TGG AGA CAC; for the BubR1 (D607E) mutation, CCT AAC CCA GAA GAA GAA ACT TGT GAC TTT GCC ACA GC and GCT CTG GCA AAG TCA CAA GTT TCT TCT GGG TTA GG; for the BubR1 (D610E) mutation, CCT AAC CCA GAA GAC ACT TGT GAA TTT GCC AGA GC and GCT CTG GCA AAT TCA CAA GTG TCT TCT GGG TTA GG; for the BubR1 (D896E) mutation, CTC AGA AAC AGA ATC CAC GAA CCC TAT GAT TGT AAG AAC AAC and GTT CTT GTT ACA ATC ATA GGG TTC GTG GAT TCT GTT TCT GAG; and for the BubR1 (D899E) mutation, CTC AGA AAC AGA ATC CAC GAT CCC TAT GAA TGT AAC AAG AAC and GTT CTT GTT ACA TTC ATA GGG ATC GTG GAT TCT GTT TCT GAG. Direct DNA sequencing was performed to confirm all engineered mutations.
Construction of doxycycline-inducible BUBR1 expression vectors and establishment of clones inducible for protein expression by doxycycline.
cDNA of human BubR1 wild type or BubR1 mutated at aspartic acid 579 (D579E) and aspartic acid 610 (D610E) was subcloned into the mammalian expression vector pTRE-Tight (Clontech) by BamHI and EcoRV sites to generate pTRE BUBR1 (wild type) or pTRE BUBR1 (D579E D610E). Nucleotide sequences of the resulting constructs were confirmed by DNA sequencing. U2OS cells transfected with these constructs were selected and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% Tet-On approved fetal bovine serum (Clontech) and in the presence of 100 μg/ml G418 and 50 μg/ml hygromycin B (Roche Applied Science). Clones were selected and individually tested for induction of protein by doxycycline (1 μg/ml).
RESULTS
The mitotic delay resulting from microtubule disruption is limited in duration but extended by caspase inhibition.
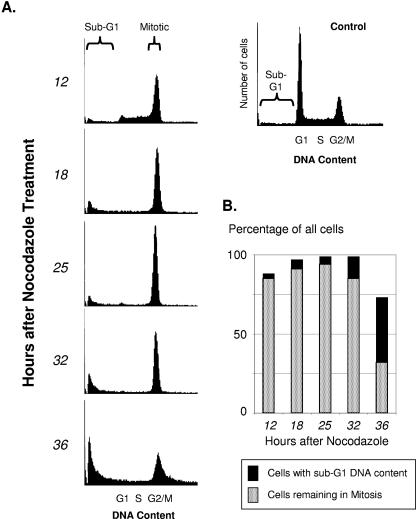
In cells with an intact spindle checkpoint, the disruption of microtubule assembly by nocodazole results in accumulation in prometaphase. The durability of the mitotic checkpoint is not infinite; with time the proportion of cells remaining blocked in mitosis is progressively reduced. To begin to investigate factors that determine the durability of the spindle checkpoint, we first established the durability of the block in exponentially growing HeLa cells. The highest proportion of cells blocked in mitosis was noted at 18 to 25 h after exposure to nocodazole, consistent with the doubling time of HeLa cells (Fig. 1). The proportion of cells blocked in mitosis decreased thereafter. By 36 h after the addition of nocodazole, less than half of the cells remained blocked in mitosis, and the reduction of mitotic cells was coincident with an increasing proportion of cells showing nuclear fragmentation and sub-G1 DNA content.
FIG. 1.
The mitotic checkpoint induced by microtubule-targeting drugs is not permanent. Parallel plates of HeLa cells were treated with nocodazole (50 ng/ml) and harvested at the times indicated for FACS analysis of cell cycle distribution. A histogram showing cell cycle distribution of control untreated cells, with the respective cell cycle phases indicated, is shown in the upper right. (A) Histograms showing cell cycle distribution of cells at each time point after nocodazole was added. The proportion of cells blocked in mitosis decreased with the passage of time, accompanied by an increasing proportion of nonviable cells with sub-G1 DNA content. (B) Bar graphs showing percentage of total cells in the histograms shown in panel A that remained either mitotic or had become nonviable with sub-G1 DNA content.
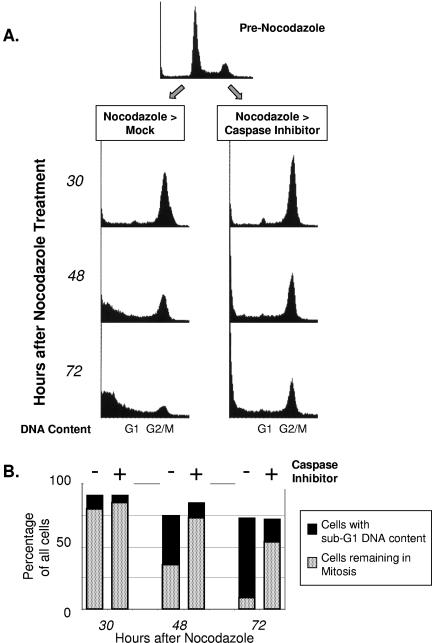
Next, we investigated the effects of inhibiting caspase activation in cells that were pretreated with nocodazole. The cells were therefore mock treated or treated with a pan-caspase inhibitor 12 h after the addition of nocodazole, and cell cycle distribution was assessed via FACS at sequential time points (Fig. 2A and B). As before, control cells that were treated with nocodazole only (Fig. 2A, nocodazole > mock) showed accumulation in mitosis that was evident at 30 h afterwards. However, the proportion of these cells that remained in mitosis thereafter progressively declined and coincided with an increasing proportion of cells with sub-G1 DNA content. In contrast, cells treated with nocodazole followed by caspase inhibitor showed a higher proportion of cells remaining in mitosis at all time points. For example, at 72 h after the addition of nocodazole, when few control cells remained in mitosis, more than half of the cells treated with caspase inhibitor after nocodazole remained in mitosis.
FIG. 2.
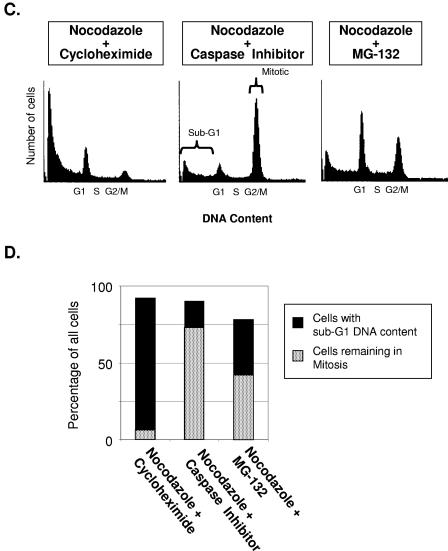
Inhibition of caspase activation prolongs mitotic cell cycle delay. (A) HeLa cells were treated with nocodazole for 12 h, followed by mock treatment or a pan-caspase inhibitor. At the times indicated after nocodazole, parallel plates of cells were harvested for cell cycle analysis via FACS. The resultant data are shown in the histograms. (B) Bar graphs showing percentages of total cells in the histograms shown in panel A that either remained mitotic or showed sub-G1 DNA content (i.e., became nonviable). Each pair of bar graphs represents either cells treated with nocodazole only or cells treated with nocodazole and then with caspase inhibitor. Inhibition of de novo protein synthesis abrogates the mitotic delay but is augmented by caspase inhibition. (C) Cells were treated with nocodazole for 12 h, followed by cycloheximide (100 μg/ml final concentration), caspase inhibitor (20 μM), or MG-132 (50 μM) for an additional 20 h (a total of 32 h in nocodazole) and then harvested for cell cycle analysis via FACS, with the resultant histograms as shown. (D) Bar graphs showing percentages of total cells in the histograms shown in panel C that either remained mitotic or showed sub-G1 DNA.
As caspases are functional proteases (cysteinyl aspartic-acid proteases), the prolongation of the mitotic delay resulting from the inhibition of caspase activity suggested a mechanism related to alterations in protein levels. In further support of this hypothesis, inhibition of de novo protein synthesis by cycloheximide substantially abrogated the mitotic delay. The abrogation of the mitotic checkpoint was evident within 6 h of adding cycloheximide (data not shown) and was virtually complete by 12 h (Fig. 2C and D).
Treatment with microtubule-targeting drugs leads to cleavage of the mitotic checkpoint protein BubR1.
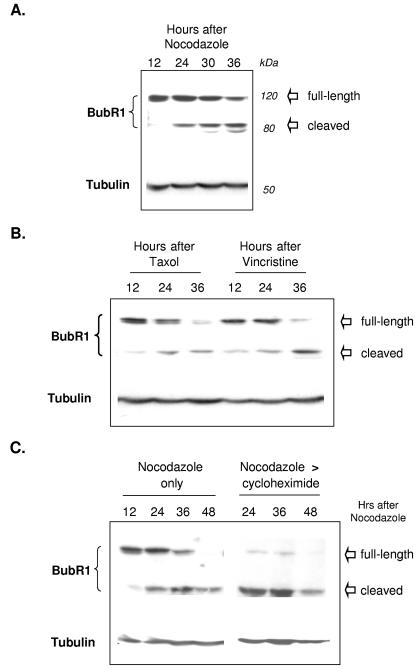
The expression levels of the mitotic checkpoint proteins seemed likely to influence duration of mitosis. We therefore harvested cells identically treated with nocodazole at sequential time points for immunoblotting for BubR1 and Bub1. In cells exposed continuously to nocodazole, BubR1 levels progressively decreased with time (Fig. 3A), as did Bub1 protein (results not shown). Interestingly, coincident with the progressive reduction in levels of full-length (120 kDa) BubR1, we noted a progressive increase of a band of 80 kDa that was also detected by the BubR1-specific antibodies (Fig. 3A). Nearly identical results were obtained with a polyclonal antibody targeting full-length BubR1 from a second source. These observations spurred us to investigate next whether levels of full-length BubR1 would be similarly affected by other microtubule-targeting drugs. Similar to nocodazole, vincristine inhibits microtubule polymerization, while paclitaxel (Taxol), in contrast, stabilizes microtubules by inhibiting depolymerization. All three drugs disrupt microtubule dynamics and prevent the assembly of the mitotic spindle. We found that exposure to paclitaxel and vincristine also led to progressive reductions in full-length BubR1 protein levels, coincident with the emergence of the lower-molecular-mass form (Fig. 3B). These results together indicate that the cleavage of BubR1 is a general consequence of microtubule disruption.
FIG. 3.
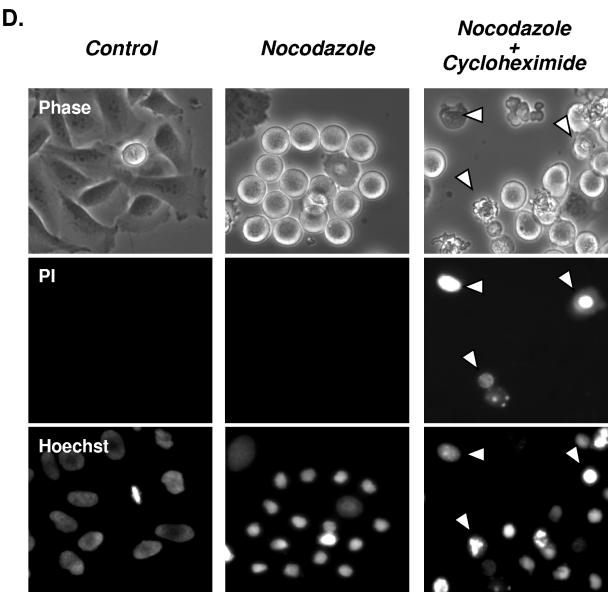
BubR1 protein undergoes progressive cleavage during the mitotic checkpoint. (A) Parallel plates of cells were treated with nocodazole and harvested at the times indicated for immunoblotting against the indicated proteins. The cell lysates were separated via SDS-PAGE, transferred to nitrocellulose membranes, and then probed for BubR1 or α-tubulin (serving as a loading control). Levels of full-length BubR1 protein progressively decreased with time, while the cleaved lower-molecular-weight form increased. (B) Paclitaxel (Taxol) and vincristine treatment also lead to decreased levels of full-length BubR1 protein and increased levels of cleaved BubR1. Parallel plates of cells were treated with concentrations of each drug that resulted in an equal degree of mitotic block and then harvested at the indicated times. (C) Mitotic levels of full-length BubR1 protein require active protein synthesis. Cells were treated with nocodazole as in panel A, with the addition of parallel plates that were treated with cycloheximide 12 h after the nocodazole had been added and an additional time point of 48 h after the addition of nocodazole. Inhibition of de novo protein synthesis accelerated the disappearance of full-length BubR1 protein (upper thick arrow), but not the lower molecular weight cleaved form of BubR1. (D) Cleavage of BubR1 precedes large-scale evidence of cell death. Cells were prepared and treated as in panel C but assessed for exclusion of PI at 24 h after the addition of nocodazole. At this time point, untreated control and cells treated only with nocodazole showed little uptake of PI. In contrast, PI uptake (examples marked by the arrowheads) was noted in a portion of cells treated with nocodazole followed by cycloheximide. Cells in each column are identical and are imaged as indicated for phase-contrast, PI uptake (nonviable cells), and Hoechst 33342 (which stains the nuclei of both live and dead cells).
The progressive decrease of full-length BubR1 protein with the passage of time suggested that its stability may be limited. To clarify the potential instability of BubR1, we inhibited de novo protein synthesis with cycloheximide (commonly utilized to assess for protein stability [25]) in cells pretreated with nocodazole. In cells treated with nocodazole only, levels of full-length BubR1 progressively decreased, again coincident with increasing levels of the lower-molecular-weight form; by 48 h after exposure to nocodazole, full-length BubR1 protein was no longer detectable (presumably all cleaved to the lower-molecular-weight form which remained detectable). In contrast to gradual loss of BubR1 with nocodazole alone, the addition of cycloheximide after nocodazole markedly and rapidly reduced levels of full-length BubR1, such that only trace levels were detectable at 24 h after nocodazole (Fig. 3C). As expected, the disappearance of full-length BubR1 after nocodazole and cycloheximide coincided with an increase in the levels of the cleaved form. Together, these findings suggest that full-length BubR1 is unstable in the presence of nocodazole and that maintaining substantial levels of this protein in the presence of nocodazole requires continuous protein synthesis.
Cleavage of BubR1 occurs prior to evidence of cell death.
As shown by the experiments depicted in Fig. 1 and 2, the mitotic block induced by nocodazole eventually dissipates after prolonged exposure, coincident with an increasing proportion of likely nonviable cells with sub-G1 DNA content, which is accelerated by the addition of cycloheximide. To determine whether cleavage of full-length BubR1 protein preceded or was a late consequence of cell death, we assessed the ability to exclude PI of control cells or cells treated with nocodazole alone or nocodazole followed by cycloheximide. As expected, no evidence of PI uptake was detected in control HeLa cells (Fig. 3D). PI uptake was also not detected in cells treated with nocodazole for 24 h (Fig. 3D), a time when cleavage of BubR1 was detectable (Fig. 3A and C). PI uptake in cells treated with nocodazole was particularly evident only after further prolonged exposure, e.g., after 36 h (data not shown). In contrast, PI uptake was already evident in a proportion of cells treated for 12 h with nocodazole followed by the addition of cycloheximide for another 12 h (Fig. 3D; nonviable cells showing PI uptake are marked by arrowheads). Interestingly, at this time point, some of the cells treated with nocodazole and cycloheximide remained similar in appearance to cells treated only with nocodazole and retained the ability to exclude PI, even though immunoblotting indicated complete or near complete cleavage of BubR1 after combined nocodazole and cycloheximide (Fig. 3C). These results together suggest that cleavage of BubR1 precedes large-scale evidence of cell death.
Inhibition of caspase activity blocks cleavage of BubR1 protein.
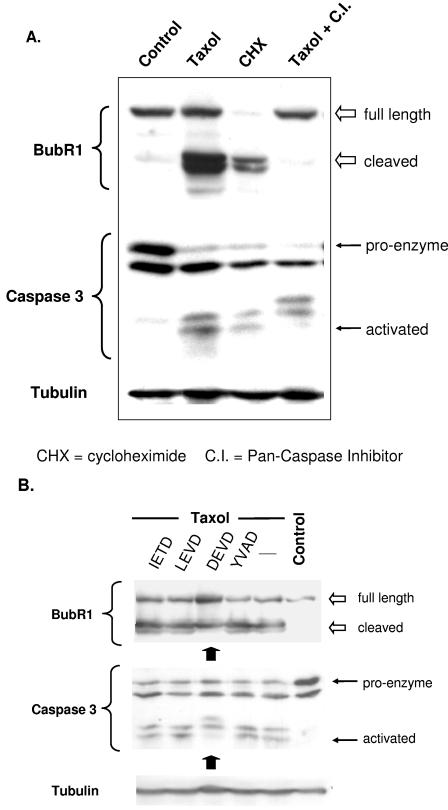
The preceding observations suggested that the increased durability of mitosis seen after the inhibition of caspase activity may be related to effects on the cleavage of key proteins involved in the checkpoint, such as BubR1. To begin to test this hypothesis, we assessed whether cleavage of BubR1 might be influenced by modulation of caspase activity. As expected, exposure to paclitaxel and cycloheximide led to the appearance of the cleaved form of BubR1, with cycloheximide leading to the complete disappearance of the full-length BubR1 due to inhibition of de novo synthesis. Both agents led to conversion of caspase 3 proenzyme to the activated, lowest-molecular-weight form (Fig. 4A). In contrast to these effects, concomitant treatment with a pancaspase inhibitor during exposure to paclitaxel completely inhibited BubR1 cleavage and the final caspase activation step (Fig. 4A, far right lane; note the absence of the cleaved form of BubR1 as well as the lowest-molecular-weight form of caspase 3). Taken together, these results suggest that exposure of cells to paclitaxel leads to BubR1 cleavage but that overall levels of the full-length protein initially remain unchanged, most likely due to continued protein synthesis. Blocking de novo protein synthesis, however, reveals the instability of BubR1, and all full-length protein is converted to the cleaved form. The cleavage of BubR1 likely involves caspase-mediated mechanisms, as inhibiting caspase activation completely prevents the appearance of the lower-molecular-weight cleaved form of BubR1 protein.
FIG. 4.
Caspase-dependent degradation of BubR1 protein. (A) Cells were mock treated (Control), treated with the paclitaxel (Taxol), cycloheximide, or paclitaxel preceded 1 h by pancaspase inhibitor (Taxol+C.I.) and then harvested 24 h later. The cell lysates were separated via SDS-PAGE, transferred to nitrocellulose, and immunoblotted against the proteins indicated. Probing for α-tubulin served as a loading control. Paclitaxel and cycloheximide led to degradation of BubR1 (full-length and cleaved forms are indicated), likely due to caspase activation (the caspase activation results from cleavage of the larger proenzyme of caspase 3). In contrast, the degradation (and the final caspase activation step) is blocked in the presence of pancaspase inhibitor. (B) Cells were treated with the indicated caspase inhibitor for 1 h, followed by paclitaxel (Taxol) for 24 h. Control cells were mock treated. All cells were then harvested, and the cell lysates were immunoblotted for BubR1, total caspase 3, and α-tubulin. Only the caspase 3 inhibitor DEVD reduced the level of cleavage of full-length BubR1. As expected, DEVD inhibited the final cleavage step that fully activates caspase 3.
Having identified caspase activity as a mechanism that regulates BubR1 degradation, we performed a preliminary screen in the hope of identifying specific caspases that are involved in mediating BubR1 cleavage. We mock treated or treated cells with paclitaxel alone or concomitantly with cell-permeable inhibitors of caspase 8 (IETD), caspase 4 (LEVD), caspase 3 (DEVD), or caspase 1 (YVAD). Of these, only the inhibitor of caspase 3 was found to have an appreciable effect in partially reducing the amount of cleaved BubR1 (Fig. 4B). These results support a potential role for caspase 3 in the cleavage of BubR1 but do not exclude the possibility that other caspases not tested might be involved as well.
Identification of potential caspase recognition/cleavage sites in BubR1.
The pharmacologic studies described above suggested a role for caspase activity in degrading BubR1 but did not indicate whether the mechanism was direct or indirect. In other words, it may be possible that caspase activation precipitates other non-caspase proteases to cleave BubR1. To test whether BubR1 was a substrate cleaved directly by caspases, we searched the amino acid sequence of BubR1 for potential caspase recognition/cleavage sites.
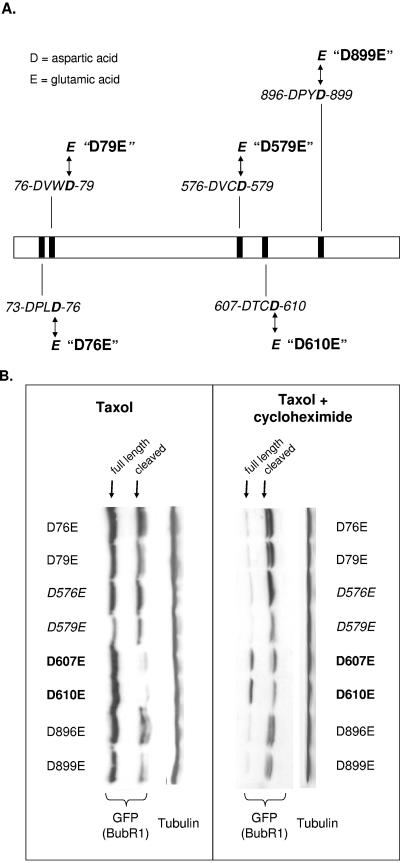
All known caspases share a singular specificity for cleaving substrates after an aspartic acid residue. Effector caspases, such as caspase 3, preferentially target a tetrapeptide sequence consisting of a D-X-X-D motif, in which the first and fourth amino acid positions are occupied by aspartic acid (2, 7). We identified five such tetrapeptide motifs in BubR1, as shown in Fig. 5A. For each of these candidate tetrapeptide caspase cleavage sites, we mutated the first (i.e., proximal) or the fourth (i.e., distal) aspartic acid to glutamic acid (Fig. 5A). This resulted in a total of nine mutations (aspartic acid 76 is the distal aspartic acid of 73-DPLD-76 but is also the proximal aspartic acid of 76-DVWD-79). The mutations were thereafter referred to as D73E, D76E, D79E, D576E, D579E, D607E, D610E, D896E, and D899E. These mutants were then fused to GFP to enable easy detection from endogenous BubR1.
FIG. 5.
Mutants to evaluate potential caspase cleavage sites in BubR1. (A) Schematic of BubR1 protein, showing the location of DxxD tetrapeptide motifs representing potential caspase recognition/cleavage sites. Mutants were generated in which each aspartic acid (D) was changed to glutamic acid (E), e.g., D610E represents aspartic acid 610 mutated to glutamic acid. Targeted mutations of aspartic acids at the fourth position of the candidate tetrapeptide motifs are shown in this figure, but the aspartic acids at the first position were also mutated (see text and panel B). (B) Stability of BubR1 mutated at potential caspase-cleavable aspartic acids. For each of the candidate caspase cleavage sites that we identified in BubR1, the indicated aspartic acids were individually changed to glutamic acid, and fused to GFP, and the GFP-tagged mutant was expressed in HeLa cells. Twelve hours after transfection, the cells were mock treated or treated with cycloheximide for an additional 24 h. The lysates of all cells were immunoblotted for GFP to detect the mutant protein and for α-tubulin as a loading control (left blot). D607E and D610E are the most stable, but degraded protein from each is detectable when cycloheximide was also added (right blot). To enable easier reading, the Western blots are rotated 90 degrees from the direction of separation on SDS-PAGE.
Identification of Asp607/Asp610 and Asp576/Asp579 as the prime caspase cleavage sites in BubR1.
To evaluate the stability of each mutant, the mutants were individually expressed in HeLa cells, followed by exposure to paclitaxel. All mutants except for D607E and D610E conspicuously showed the lower-molecular-weight degraded form of the protein (Fig. 5B, left). Expression levels of the full-length form for many of the mutants were, however, similar to those of D607E and D610E, most likely due to continued protein synthesis in the presence of paclitaxel. To better determine the stability of each mutant, the experiment was repeated, this time in the presence of cycloheximide. This resulted in the complete abrogation of the full-length forms of all mutants except for D607E and D610E (Fig. 5B, right). These results together suggested that the 607-DTCD-610 tetrapeptide sequence was the prime caspase recognition sequence and that its aspartic acids were the prime caspase cleavage sites in BubR1. These results further suggested that mutation of either the proximal or distal aspartic acid was sufficient to inhibit cleavage at the other aspartic acid of the tetrapeptide sequence.
We were surprised, however, to note that with the addition of cycloheximide, the degraded forms of D607E and D610E were also increased. A closer inspection of D607E and D610E treated with paclitaxel alone also revealed the faint presence of the degraded form. These observations seemed to suggest that a second cleavage site might be present. By further increasing caspase activity, cycloheximide might accelerate cleavage of BubR1 at the second cleavage site. As a further indication that two cleavage sites might be present, we also noted that the degraded form in many immunoblots, including that of the BubR1 mutants, actually appeared as double bands (Fig. 5B; the double bands of the degraded forms can also be seen in Fig. 4A). An exception seemed to be the D576E and D579E mutants, in which only the upper band of the pair of degraded forms was evident (Fig. 5B, after paclitaxel alone as well as paclitaxel plus cycloheximide). This suggested that D576E/D579E might encompass a second tetrapeptide sequence that was cleavable by caspases.
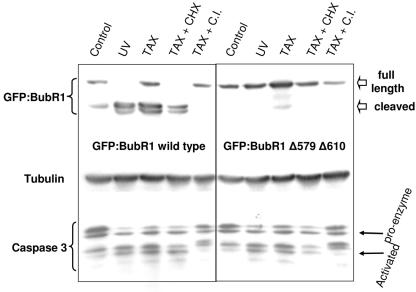
To investigate the possibility that BubR1 might contain two caspase recognition/cleavage sites, a mutant was created in which both Asp579 and Asp610 were changed to glutamic acids. This double mutant (hereafter referred to as GFP:BubR1Δ579Δ610) was expressed in HeLa cells, which were then mock-treated, or treated with paclitaxel alone, paclitaxel simultaneously with cycloheximide, or simultaneously with pan-caspase inhibitor. For an additional control with activated caspases, cells expressing GFP:BubR1 wild type or the double mutant were also UV irradiated. The result of this experiment showed that the double mutant was completely stable after all treatments, including after UV irradiation, paclitaxel, or the simultaneous administration of paclitaxel and cycloheximide (Fig. 6). Under the same treatment conditions, the wild-type GFP:BubR1 fusion protein clearly showed the pair of lower-molecular-weight forms representing the cleaved forms of the protein, except when caspase activity was inhibited. GFP:BubR1Δ579Δ610 stability was also stable after nocodazole treatment and remained impervious to cleavage for at least 120 h of drug treatment (results not shown). These results together strongly support the existence of two distinct caspase cleavage sites in BubR1.
FIG. 6.
Mutating both caspase cleavage sites of BubR1 renders it ultrastable. GFP fused to wild-type BubR1 (GFP:BubR1 wild type) or BubR1 with Asp579 and Asp610 changed to glutamic acids (GFP:BubR1Δ579Δ610) were expressed in HeLa cells. Twelve hours after transfection, the cells were mock treated or treated as indicated and then harvested an additional 24 h later. Cell lysates were then immunoblotted for BubR1, caspase 3, or α-tubulin (as a loading control). UV irradiation (60 J) and paclitaxel alone (TAX) and with cycloheximide (TAX+CHX) activated caspase 3 in all cells and led to degradation of wild-type BubR1 but not the BubR1 doubly mutated at the two caspase cleavage sites. The addition of caspase inhibitor (which prevented the final step of the conversion of caspase proenzyme to the fully activated cleaved form) concurrently with paclitaxel (TAX+C.I.) likewise prevented the degradation of full-length BubR1.
Expression of BubR1Δ579Δ610 results in a more durable mitotic delay in the presence of microtubule-targeting drugs.
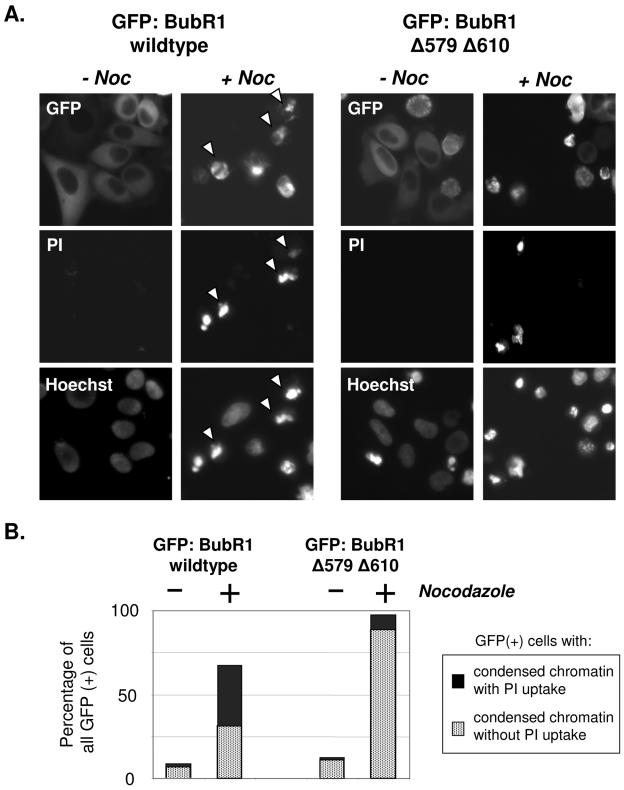
When expressed in HeLa cells that were then treated with nocodazole or paclitaxel, GFP:BubR1Δ579Δ610 (as well as GFP:BubR1 wild type) localized to paired kinetochores in mitotic cells, suggesting that it may be functional (see Supplemental Fig. 1 at http://www.xrt.upenn.edu/radiation_biology/Kao_Research.html). We therefore sought to test the effects of expressing either wild-type or mutated GFP:BubR1. Cells expressing both GFP:BubR1 wild type and GFP:BubR1Δ579Δ610 were treated with nocodazole, which resulted in the accumulation of cells expressing either form of protein in mitosis. However, with prolonged exposure to nocodazole, nuclear morphology alone was insufficient to distinguish mitotic from apoptotic cells, as either would show condensed chromatin. We therefore assessed for cellular viability via the nucleic acid stain PI, which is excluded by viable cells. These assays were also performed in the presence of Hoechst 33342 vital dye, which stains all nuclei, both viable and nonviable, and which facilitated the detection of condensed chromatin. In the absence of nocodazole, few GFP:BubR1 wild-type- or BubR1 mutant-expressing cells showed either PI uptake or condensed chromatin (Fig. 7A and B, −Noc). With prolonged exposure to nocodazole, an increasing proportion of cells expressing GFP:BubR1 wild type was unable to exclude PI, and the majority of these showed condensed chromatin as well. Under identical treatment conditions, a consistently greater proportion of HeLa cells expressing GFP:BubR1Δ579Δ610 showed condensed chromatin consistent with mitosis and retained the ability to exclude PI (Fig. 7A and B and data not shown).
FIG. 7.
Expression of GFP:BubR1Δ579Δ610 mutant protein leads to a more durable mitotic cell cycle delay in the presence of nocodazole. (A) GFP:BubR1 wild-type protein or mutated for the caspase cleavage sites was expressed in HeLa cells, and the cells expressing either protein were then mock treated or treated with nocodazole for 24 h. The cells of all four treatment groups were then assessed for expression of the GFP-fusion promoter, PI exclusion to test for viability, and degree of nuclear condensation with Hoechst 33342 (which stains the nuclei of all cells, living or dead). Representative cells of each treatment group are shown (at least 300 cells of each group were assessed), with each respective column showing images of the same cells. Expression of either wild-type or mutant GFP:BubR1 did not noticeably impair cellular viability in the absence of nocodazole, as indicated by the absence of PI-positive cells (first and third columns of cells). After nocodazole, many cells expressing GFP:BubR1 wild type showed condensed chromatin visible via Hoechst but were unable to exclude PI, suggestive of cell death (arrowheads, second column). In contrast, in the presence of nocodazole, the majority of cells expressing GFP:BubR1Δ579Δ610 mutant protein were able to exclude PI, even those with condensed chromatin (thereby suggestive of mitotic cells). (B) Quantitation of the percentage of GFP-expressing cells in each treatment group described in panel A that showed, respectively, condensed chromatin with PI uptake (suggestive of cell death) or condensed chromatin while still excluding PI (suggestive of mitotic cells). The experiment was repeated three times with similar results.
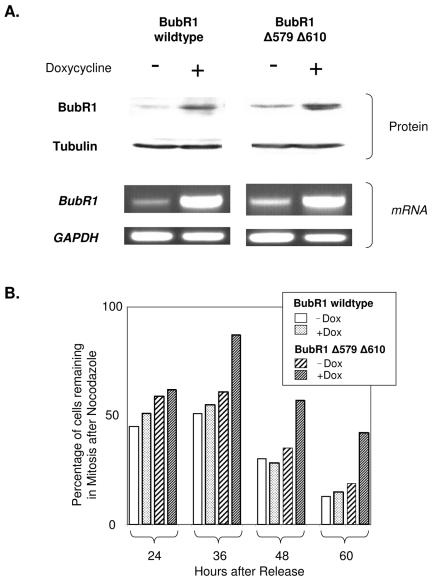
We further tested the effects of the BubR1Δ579Δ610 protein by placing its expression under the control of a doxycycline-inducible promoter, by introducing the engineered vectors into U2OS cells containing the appropriate transactivator (Tet-On; Invitrogen), and by subsequently isolating clones that stably expressed the BubR1 mRNA and protein upon exposure to doxycycline. This was done for both BubR1 wild-type and BubR1Δ579Δ610 double-mutant proteins. Exposure to doxycycline led to the expression of BubR1 mRNA and protein levels that were greater than five times that of uninduced cells (Fig. 8A shows the characterization of representative clones). Induced wild-type and mutant BubR1 protein was readily detectable and was appropriately phosphorylated (23, 44) in cells treated with nocodazole (albeit not all induced protein was phosphorylated, presumably due to exceeding the kinase activity available for phosphorylation and/or cells not yet in mitosis) (see Supplemental Fig. 2 at the URL mentioned above).
FIG. 8.
Induced expression of BubR1Δ579Δ610 mutant protein leads to a more durable mitotic cell cycle delay in the presence of nocodazole. (A) Tet-On U2OS cells that were stably inducible with doxycycline (i.e., expressing pTRE-Tight BubR1 wild type or BubR1Δ579Δ610) for either BubR1 wild type or BubR1Δ579Δ610 were mock induced or induced with doxycycline. Twelve hours later, parallel plates of each treatment group were harvested for cell lysates to be immunoblotted for BubR1 or α-tubulin protein (loading control) or were harvested for total mRNA, which then served as the template mRNA for reverse transcriptase PCR using primers specific for BubR1 or GAPDH (glyceraldehyde-3-phosphate dehydrogenase; assay control). Results from representative clones are shown. (B) Induction of BubR1Δ579Δ610 leads to delayed exit from the mitotic delay induced by nocodazole. The cells described in panel A were synchronized in G1/S and mock induced (−Dox) or induced with doxycycline (+Dox) 3 h after release, and nocodazole was added to all cells 6 h after release. Parallel plates of each treatment group were harvested at the indicated times after release to assess for cell cycle distribution via FACS. The bar graphs show the proportion of cells of each treatment group that remained in mitosis at the indicated times following release.
In control untreated cells (i.e., not exposed to nocodazole), induction of either wild-type or mutant protein modestly increased the proportion of cells remaining in G1 and was mainly associated with a growth delay, with little detectable cell death (data not shown; see Supplemental Fig. 3, 4, and 5 at the URL mentioned above). To assess for specific effects of wild-type or mutant BubR1 on mitotic cell cycle progression, the cells were therefore synchronized and released from G1/S block, and the protein was induced only after the cells had fully exited from G1. This enabled the induction of protein that persisted but that did not impede the entry of the cells into mitosis nor the induction of the mitotic block by nocodazole (Fig. 8B; see Supplemental Fig. 6 at the URL mentioned above). Under these conditions, the most conspicuous effect of the induction of double-mutant BubR1 was increased mitotic cell cycle delay under sustained nocodazole exposure. This was evident at 36 h after release (and 30 h after nocodazole was added) as well as at all other time points. In contrast, the induction of wild-type BubR1 did not substantially perturb the proportion of cells in mitosis. By 60 h after release (i.e., 54 h after nocodazole treatment), a greater proportion of cells induced for BubR1Δ579Δ610 remained in mitosis compared to the cells in the other treatment groups (Fig. 8B). Interestingly, while doxycycline-induced induction of double-mutant BubR1 led to the greatest proportion of cells remaining in mitosis in the presence of nocodazole, this proportion decreased with time, suggesting that mechanisms beyond BubR1 expression or stability may also be in effect to control the duration of the mitotic block.
Mutation of caspase cleavage sites in BubR1 leads to increased aneuploidy after nocodazole exposure.
What is the biological consequence of blocking caspase cleavage of BubR1? Cells expressing either wild-type or caspase-mutated BubR1 readily entered mitosis, so we surmised that biological consequences may instead be more related to the fate of the cells after mitosis. As noted previously, the appearance of nonviable cells largely coincided with exit from the mitotic block induced by nocodazole. We therefore hypothesize that augmentation of the mitotic block by mutation of the caspase cleavage sites in BubR1 may reduce the rate of cell death. This appeared to be the case, as reflected by fewer cells showing PI uptake after exposure to nocodazole (Fig. 9A). While the majority of cells in which wild-type BubR1 was induced and subsequently exposed to nocodazole for a prolonged period died with condensed chromatin (Fig. 9B, arrowheads), we noted that a subset of cells reattached and flattened; interestingly, many of these were found to have fragmented nuclei by Hoechst staining (Fig. 9B, dashed boxes in lower left column). The factors that dictate either of these cell fates remain unclear, but cells with fragmented nuclei were not observed to undergo subsequent cell division or colony formation even after prolonged cell culture (data not shown), so it seems likely that these cells are also incapable of subsequent proliferation (similar findings of “abnormal mitoses” and flattened cells with fragmented nuclei have been noted after prolonged exposure to microtubule-targeting drugs [30, 41]; prolonged culture or clonogenic survival assays would be required to confirm loss of proliferative potential).
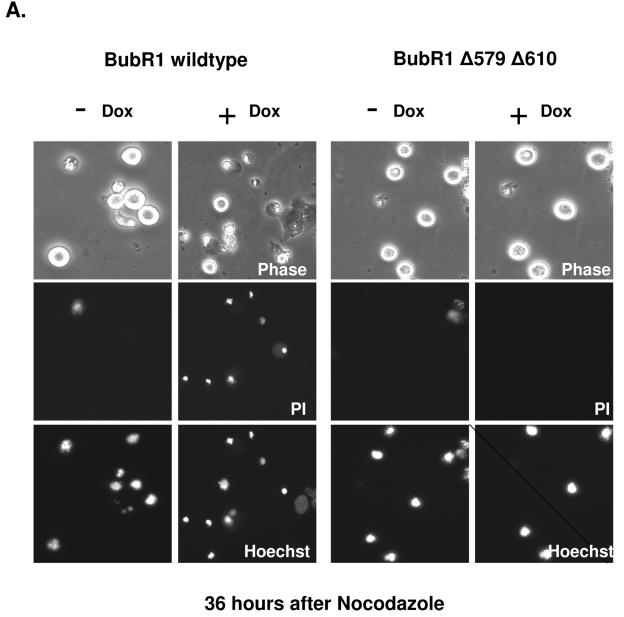
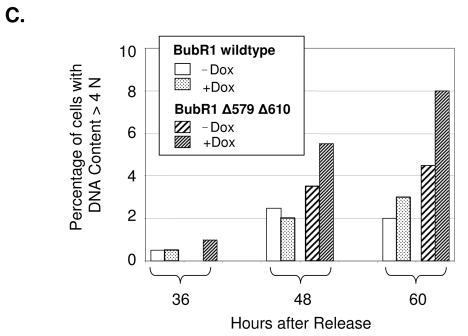
FIG. 9.
Induced expression of BubR1Δ579Δ610 ultimately leads to decreased death and increased aneuploidy in the presence of nocodazole. Tet-On U2OS cells stably inducible for either BubR1 wild type or BubR1Δ579Δ610 were synchronized and mock induced or induced with doxycycline 3 h after release from G1/S block, and nocodazole was added to all cells 7 h after release. All groups were assessed for PI exclusion and DNA content after release via FACS. (A) Representative cells of each treatment group are shown (with at least 300 cells of each group assessed) at 36 h after exposure to nocodazole. Each respective column shows images of the same cells, taken under phase-contrast, for PI, and for nuclei with the nuclear stain Hoechst 33342. Each column shows images of the same cells, and representative cells of each treatment group are shown (out of at least 300 cells of each group). (B) Images at 72 h after release of cells induced for either BubR1 wild type or BubR1Δ579Δ610 and exposed continuously to nocodazole. Most cells expressing BubR1 wild type have died with condensed chromatin (examples marked by arrowheads), but adherent cells are detectable that exclude PI but show fragmented micronuclei. Adherent cells induced for BubR1Δ579Δ610 are also notable and also exclude PI, but many show a single, intact nucleus. Cells demarcated by the dashed boxes are shown below at higher magnification. (C) Cells mock induced or induced for either BubR1 wild type or BubR1Δ579Δ610 and exposed continuously to nocodazole were assessed at the indicated times after release from G1/S block, and the percentages of cells with DNA content > 4N are shown.
In contrast to cells in which wild-type BubR1 was expressed, fewer of the cells in which BubR1Δ579Δ610 was induced were found to die in mitosis. At 72 h after nocodazole, 15% of cells induced for BubR1Δ579Δ610 showed PI uptake, compared to 58% of BubR1 wild-type-expressing cells. Most of the cells expressing mutant BubR1 that reattached showed an intact, single nucleus (Fig. 9B, dashed boxes in lower right column). We repeated the analysis of DNA content via FACS of cells induced for wild-type or caspase-mutated BubR1 and subsequently exposed to nocodazole, but this time we focused on the proportion of cells with greater than 4N DNA content. This analysis showed that after exposure to nocodazole, a higher proportion of cells induced for caspase-mutated BubR1, compared to those expressing wild-type BubR1, showed DNA content greater than 4N (Fig. 9C; see Supplemental Fig. 7 at the URL mentioned above). Taken together, these results suggest that expression of BubR1 resistant to caspase cleavage leads to reduced cell death during mitosis and increased aneuploidy.
The caspase cleavage sites in BubR1 are evolutionarily conserved.
Having established a functional role for the caspase cleavage sites of BubR1 in human cells, we were curious whether similar motifs could be identified in the BubR1 protein of other species. We searched the amino acid sequences of mouse, chicken, and Xenopus (African clawed frog) BubR1 (available through the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov/entrez/). In each species, the tetrapeptide representing the prime caspase cleavage motif (DTCD) was preserved intact (Fig. 10). The tetrapeptide representing the secondary caspase cleavage (DVCD in human BubR1) was also identifiable in each species. Interestingly, the aspartic acids in the first and fourth positions of both caspase cleavage sites remained intact in all species, with the sole exception of chicken BubR1, in which the fourth position of the first tetrapeptide was occupied by histidine, which, like aspartic acid, is also a polar-charged amino acid.
FIG. 10.
The caspase cleavage sites of BubR1 are evolutionarily conserved. The corresponding amino acid sequences of human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), and frog (Xenopus laevis) BubR1 are shown. The distal tetrapeptide caspase recognition motif (DTCD) is completely conserved among all species, while homology in the proximal caspase tetrapeptide motif (DVCD in human BubR1) has been retained.
DISCUSSION
We describe here the systematic investigation that led to the identification of two distinct caspase cleavage sites in BubR1. The caspase cleavage sites in BubR1 consist of two tetrapeptide motifs, each flanked by aspartic acids—at 607-DTCD-610 and 576-DVCD-579—and both motifs are evolutionarily conserved. Treatment of cells with microtubule-targeting agents led to caspase activation, which in turn resulted in cleavage of BubR1 at these sites. Inhibition of de novo protein synthesis under these conditions led to rapid cleavage and depletion of full-length BubR1 protein, which was then associated with abrogation of the mitotic spindle checkpoint. In contrast, mutation of these caspase cleavage sites at either of the flanking aspartic acids stabilized the protein, which then led to a more durable mitotic checkpoint in the presence of microtubule-targeting drugs and which was associated with reduced cell death and increased aneuploidy.
While exposure of cells to microtubule-targeting drugs has previously been reported to increase caspase activity (12, 19, 32, 33, 35, 36, 47), our findings to our knowledge are the first to establish a direct link during mitosis between the caspase protease pathway and a protein component of the mitotic spindle checkpoint machinery. Our findings, including our discovery that inhibition of de novo protein synthesis accelerates abrogation of the mitotic delay, suggest that the duration of the mitotic delay is determined at least in part by the balance between caspase-mediated cleavage and ongoing synthesis of key protein(s). These results are therefore in accordance with and extend the relevance of previous reports in which the knockdown or knockout of BubR1, both leading to reduced BubR1 protein levels, led to abrogation of the mitotic checkpoint (6, 8, 18, 21, 22, 43).
The mitotic checkpoint has been postulated as a mechanism to ensure the fidelity of chromosomal duplication and segregation. A delay in the completion of mitosis may therefore allow additional time for repair of damaged or broken chromosomes that otherwise results in defective attachment to the microtubules of the mitotic spindle, lack of microtubule tension, or misalignment during metaphase. On the other hand, cells in which repair is not possible or sufficient should be targeted for death to prevent the deleterious effects of polyploidy or aneuploidy. We propose that control of the levels of a protein(s) necessary for the spindle checkpoint may help time the length of the mitotic delay. Linking processes for the time-dependent degradation of protein with those capable of killing defective cells through caspase activation may therefore provide an elegant way to help ensure that diploid cells incapable of completing mitosis are not allowed to convert to polyploidy or aneuploidy. A mechanism that provides for the cleavage of the proteins that maintain the mitotic checkpoint may therefore help ensure the complete execution of targeted cells. It is therefore tempting to speculate that the consequence of full caspase activation includes the abrogation of the mitotic checkpoint, possibly then facilitating mitotic catastrophe (18). In contrast, blocking caspase activity or the targets of caspase cleavage during mitosis might increase the risk of aneuploidy. These possibilities may underlie the importance of caspase-mediated degradation of BubR1 and the high degree of conservation of the two caspase cleavage sites throughout evolution (Fig. 10).
It has been proposed that fragments of BubR1, possibly generated through proteolytic cleavage, may have cytotoxic properties (39). Our results do not exclude and may therefore be in accordance with the possibility that the reduced cell death noted after exposure to spindle-disrupting agents in cells expressing BubR1 mutated at the caspase cleavage sites may be related to reduced levels of such cytotoxic fragments. Our results also do not exclude the possibility that caspase-independent “backup” mechanisms may be in play that regulate BubR1 levels (e.g., transcriptional repression) or that other protein components influencing the mitotic checkpoint may also be regulated through caspase-mediated cleavage. While the present article was undergoing revision, Perera and Freire reported that the spindle checkpoint protein Bub1 also undergoes caspase-mediated cleavage at specific recognition sites (34). Whether such cleavage occurs during mitosis or after exposure to microtubule-targeting drugs was not determined, but this supports the presence of multiple intersections between the caspase activation and mitotic checkpoint machineries.
Investigations such as that described here will likely also be useful in elucidating the translational significance of mechanisms that control BubR1 protein expression levels and potentially the integrity of the mitotic checkpoints mediated by BubR1. For example, aging is associated with decreased BubR1 protein expression, which in turn is associated with defective mitotic checkpoint control (1, 6). Our findings described here may suggest that the increased caspase activity associated with the aging process (14, 20, 48) might be contributory factors leading to decreased BubR1 protein levels and ultimately diminished checkpoint function in aged cells.
Acknowledgments
We are grateful to other members of the Kao Laboratory for expert assistance, especially K. Ranh Voong and Jessica Liao. We thank Tim J. Yen and Gordon Chan for the gifts of anti-BubR1 and anti-Bub1 antibody and of BubR1 cDNA. We acknowledge the support of W. Gillies McKenna, Anna Kennedy, and Larry Solin. We are grateful to Raimundo Freire for helpful comments and generous gifts of reagents.
S.E.P. was supported by National Institutes of Health Training Grant 5T32CA009677. This work was also supported by the Office of Research and Development Medical Research Service, Department of Veterans Affairs (Advanced Career Research Award), the National Institutes of Health (CA107956-01), and funds from the University of Pennsylvania Research Foundation and the Breast Cancer Research Foundation.
REFERENCES
- 1.Baker, D. J., K. B. Jeganathan, J. D. Cameron, M. Thompson, S. Juneja, A. Kopecka, R. Kumar, R. B. Jenkins, P. C. de Groen, P. Roche, and J. M. van Deursen. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744-749. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, M., A. Degterev, and J. Yuan. 2004. Caspases: an ancient cellular sword of Damocles. Cell Death Differ. 11:29-37. [DOI] [PubMed] [Google Scholar]
- 3.Chan, G. K., and T. J. Yen. 2003. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog. Cell Cycle Res. 5:431-439. [PubMed] [Google Scholar]
- 4.Chen, R. H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 6.Dai, W., Q. Wang, T. Liu, M. Swamy, Y. Fang, S. Xie, R. Mahmood, Y. M. Yang, M. Xu, and C. V. Rao. 2004. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 64:440-445. [DOI] [PubMed] [Google Scholar]
- 7.Degterev, A., M. Boyce, and J. Yuan. 2003. A decade of caspases. Oncogene 22:8543-8567. [DOI] [PubMed] [Google Scholar]
- 8.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell, B. J., B. Moree, E. M. Farrar, S. Stewart, G. Fang, and E. D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953-964. [DOI] [PubMed] [Google Scholar]
- 11.Hoyt, M. A. 2000. Exit from mitosis: spindle pole power. Cell 102:267-270. [DOI] [PubMed] [Google Scholar]
- 12.Ibrado, A. M., C. N. Kim, and K. Bhalla. 1998. Temporal relationship of CKD1 activation and mitotic arrest to cytosolic accumulation of cytochrome C and caspase-3 activity during Taxol-induced apoptosis of human AML Hl-60 cells. Leukemia 12:1930-1936. [DOI] [PubMed] [Google Scholar]
- 13.Jablonski, S. A., G. K. Chan, V. A. Cooke, W. C. Earnshaw, and T. J. Yen. 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107:386-396. [DOI] [PubMed] [Google Scholar]
- 14.Joaquin, A. M., and S. Gollapudi. 2001. Functional decline in aging and disease: a role for apoptosis. J. Am. Geriatr. Soc. 49:1234-1240. [DOI] [PubMed] [Google Scholar]
- 15.Kao, G. D., W. G. McKenna, A. Maity, K. Blank, and R. J. Muschel. 1997. Cyclin B1 availability is a rate-limiting component of the radiation-induced G2 delay in HeLa cells. Cancer Res. 57:753-758. [PubMed] [Google Scholar]
- 16.Kao, G. D., W. G. McKenna, and T. J. Yen. 2001. Detection of repair activity during the DNA damage-induced G2 delay. Oncogene 20:3486-3496. [DOI] [PubMed] [Google Scholar]
- 17.Kao, G. D., W. G. McKenna, M. G. Guenther, R. J. Muschel, M. A. Lazar, and T. J. Yen. 2003. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 160:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kops, G. J., D. R. Foltz, and D. W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA 101:8699-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kottke, T. J., A. L. Blajeski, L. M. Martins, R. W. Mesner, Jr., N. E. Davidson, W. C. Earnshaw, D. K. Armstrong, and S. H. Kaufmann. 1999. Comparison of paclitaxel-, 5-fluoro-2′-deoxyuridine-, and epidermal growth factor (EGF)-induced apoptosis. J. Biol. Chem. 274:15927-15936. [DOI] [PubMed] [Google Scholar]
- 20.Lacelle, C., S. Xu, and E. Wang. 2002. Identification of high caspase-3 mRNA expression as a unique signature profile for extremely old individuals. Mech. Ageing Dev. 123:1133-1144. [DOI] [PubMed] [Google Scholar]
- 21.Lampson, M. A., and T. M. Kapoor. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93-98. [DOI] [PubMed] [Google Scholar]
- 22.Lee, E. A., M. K. Keutmann, M. L. Dowling, E. Harris, G. Chan, and G. D. Kao. 2004. Inactivation of the mitotic checkpoint as a determinant of the efficacy of microtubule-targeted drugs in killing human cancer cells. Mol. Cancer Ther. 3:661-662. [PubMed] [Google Scholar]
- 23.Li, W., Z. Lan, H. Wu, S. Wu, J. Meadows, J. Chen, V. Zhu, and W. Dai. 1999. BUBR1 phosphorylation is regulated during mitotic checkpoint activation. Cell Growth Differ. 10:769-775. [PubMed] [Google Scholar]
- 24.Li, X., and R. B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature 373:630-632. [DOI] [PubMed] [Google Scholar]
- 25.Liu, F., M. Dowling, X. J. Yang, and G. D. Kao. 2004. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279:34537-34546. [DOI] [PubMed] [Google Scholar]
- 26.Mao, Y., A. Abrieu, and D. W. Cleveland. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114:87-98. [DOI] [PubMed] [Google Scholar]
- 27.Meraldi, P., V. M. Draviam, and P. K. Sorger. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7:45-60. [DOI] [PubMed] [Google Scholar]
- 28.Millband, D. N., and K. G. Hardwick. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22:2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 30.Nitta, M., O. Kobayashi, S. Honda, T. Hirota, S. Kuninaka, T. Marumoto, Y. Ushio, and H. Saya. 2004. Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents. Oncogene 23:6548-6558. [DOI] [PubMed] [Google Scholar]
- 31.Overbeeke, R., M. Yildirim, C. P. Reutelingsperger, C. Haanen, and I. Vermes. 1999. Sequential occurrence of mitochondrial and plasma membrane alterations, fluctuations in cellular Ca2+ and pH during initial and later phases of cell death. Apoptosis 4:455-460. [DOI] [PubMed] [Google Scholar]
- 32.Oyaizu, H., Y. Adachi, S. Taketani, R. Tokunaga, S. Fukuhara, and S. Ikehara. 1999. A crucial role of caspase 3 and caspase 8 in paclitaxel-induced apoptosis. Mol. Cell Biol. Res. Commun. 2:36-41. [DOI] [PubMed] [Google Scholar]
- 33.Park, S. J., C. H. Wu, J. D. Gordon, X. Zhong, A. Emami, and A. R. Safa. 2004. Taxol induces caspase-10-dependent apoptosis. J. Biol. Chem. 279:51057-51067. [DOI] [PubMed] [Google Scholar]
- 34.Perera, D., and R. Freire. 2005. Human spindle checkpoint kinase Bub1 is cleaved during apoptosis. Cell Death Differ. 12:827-830. [DOI] [PubMed] [Google Scholar]
- 35.Perkins, C., C. N. Kim, G. Fang, and K. L. Bhalla. 1998. Overexpression of Apaf-1 promotes apoptosis of untreated and paclitaxel- or etoposide-treated Hl-60 cells. Cancer Res. 58:4561-4566. [PubMed] [Google Scholar]
- 36.Perkins, C. L., G. Fang, C. N. Kim, and K. N. Bhalla. 2000. The role of Apaf-1, caspase-9, and Bid proteins in etoposide- or paclitaxel-induced mitochondrial events during apoptosis. Cancer Res. 60:1645-1653. [PubMed] [Google Scholar]
- 37.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 38.Rieder, C. L., R. W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin, H. J., K. H. Baek, A. H. Jeon, M. T. Park, S. J. Lee, C. M. Kang, H. S. Lee, S. H. Yoo, D. H. Chung, Y. C. Sung, F. McKeon, and C. W. Lee. 2003. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 4:483-497. [DOI] [PubMed] [Google Scholar]
- 40.Sudakin, V., G. K. Chan, and T. J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J. Cell Biol. 154:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudo, T., M. Nitta, H. Saya, and N. T. Ueno. 2004. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 64:2502-2508. [DOI] [PubMed] [Google Scholar]
- 42.Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-independent inhibition of APC Cdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1:227-237. [DOI] [PubMed] [Google Scholar]
- 43.Tanudji, M., J. Shoemaker, L. L'Italien, L. Russell, G. Chin, and X. M. Schebye. 2004. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15:3771-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, S. S., D. Hussein, Y. Wang, S. Elderkin, and C. J. Morrow. 2001. Kinetochore localization and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385-4395. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, S. S., E. Ha, and F. McKeon. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Q., T. Liu, Y. Fang, S. Xie, X. Huang, R. Mahmood, G. Ramaswamy, K. M. Sakamoto, Z. Darzynkiewicz, M. Xu, and W. Dai. 2004. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood 103:1278-1285. [DOI] [PubMed] [Google Scholar]
- 47.Wieder, T., F. Essmann, A. Prokop, K. Schmelz, K. Schultz-Osthoff, R. Beyaert, B. Dorken, and P. T. Daniel. 2001. Activation of caspase-8 in drug-induced apoptosis of B-lymphoid cells is independent of CD95/Fas receptor-ligand interaction and occurs downstream of caspase-3. Blood 97:1378-1387. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, J. H., Y. Zhang, and B. Herman. 2003. Caspases, apoptosis and ageing. Ageing Res. Rev. 2:357-366. [DOI] [PubMed] [Google Scholar]