Abstract
The microbiota of the intestinal tract of chickens plays an important role in inhibiting the establishment of intestinal pathogens. Earlier culturing and microscopic examinations indicated that only a fraction of the bacteria in the cecum of chickens could be grown in the laboratory. Therefore, a survey of cecal bacteria was done by retrieval of 16S rRNA gene sequences from DNA isolated from the cecal content and the cecal mucosa. The ribosomal gene sequences were amplified with universal primers and cloned or subjected to temporal temperature gradient gel electrophoresis (TTGE). Partial 16S rRNA gene sequences were determined from the clones and from the major bands in TTGE gels. A total of 1,656 partial 16S rRNA gene sequences were obtained and compared to sequences in the GenBank. The comparison indicated that 243 different sequences were present in the samples. Overall, sequences representing 50 phylogenetic groups or subgroups of bacteria were found, but approximately 89% of the sequences represented just four phylogenetic groups (Clostridium leptum, Sporomusa sp., Clostridium coccoides, and enterics). Sequences of members of the Bacteroides group, the Bifidobacterium infantis subgroup, and of Pseudomonas sp. each accounted for less than 2% of the total. Sequences related to those from the Escherichia sp. subgroup and from Lactobacillus, Pseudomonas, and Bifidobacterium spp. were generally between 98 and 100% identical to sequences already deposited in the GenBank. Sequences most closely related to those of the other bacteria were generally 97% or less identical to those in the databases and therefore might be from currently unknown species. TTGE and random cloning indicated that certain phylogenetic subgroups were common to all birds analyzed, but sequence data from random cloning also provided evidence for qualitative and quantitative differences among the cecal microbiota of individual birds reared under very similar conditions.
The mature microbiota of the gastrointestinal tract of chickens have long been known to confer resistance to infection by Salmonella enterica (27). Beginning with the studies by Nurmi and Rantala (31), practical means were developed to promote the establishment of mature intestinal microbiota in newly hatched chicks to prevent Salmonella enterica infection. Over the years, numerous preparations of bacteria were administered to young chicks and tested for their efficacy in preventing Salmonella infection (2, 14, 37, 43, 44). The consensus derived from these studies was that complex mixtures of bacteria derived from adult chickens rather than single bacterial isolates provided the best protection against Salmonella infection. Currently both undefined, but pathogen-free mixtures and relatively well-defined competitive exclusion products are available commercially, but it is not known whether all or only some of the bacteria within these mixtures are required for effective competitive exclusion.
Our limited understanding of the contribution of the different intestinal bacteria to the competitive exclusion phenomenon is due, in part, to a paucity of data on the actual composition of the intestinal microbiota of adult chickens. Studies on the composition of the intestinal microbiota of chickens date back to 1901 (34) and were continued in the 1940s (38), but comprehensive surveys that attempted to culture as many of the intestinal bacteria as possible were not carried out until the 1970s (5, 6, 24, 36). Such studies are technically difficult since strict anaerobic conditions have to be maintained during isolation and biochemical differentiation of the bacteria. Although other parts of the digestive tract of chickens might also be important sites for pathogen-host microbiota interactions, the ceca have received most of the attention because the microbiota of the ceca is very diverse and 1 g (wet weight) of cecal content may contain 1011 bacteria (25). Potential human pathogens such as S. enterica and Campylobacter jejuni are frequently most numerous in the ceca (10, 16, 17).
Data from the earlier culture-based studies indicated that only between 10 and 60% of the bacteria in the cecum grew in culture (5, 6, 36). Therefore, the present work used molecular techniques to identify the bacteria in the cecum of commercially grown broiler chickens. Although these molecular approaches have several limitations (18, 33, 41, 42, 48, 52), including the possibility that DNA isolation, amplification, and cloning might be biased in favor of certain bacteria and sequences, they nevertheless provide an overview of the microbial diversity present in a particular sample. Data obtained allow for easy future comparison of cecal isolates without the need to resort to sometimes ambiguous comparisons of biochemical characteristics. Results of surveys of 16S rRNA gene sequences retrieved from cecal content and cecal mucus by PCR amplification and cloning or direct sequencing after temporal temperature gradient gel electrophoresis (TTGE) are presented.
MATERIALS AND METHODS
Origin and collection of cecal samples.
Broiler chickens were obtained from a commercial broiler farm or from the University of Delaware Farm where the chickens were raised under conditions identical to those found in commercial broiler operations. The broilers were not exposed to competitive exclusion preparations as newly hatched chicks and were fed a diet of commercial feed. The birds were sacrificed by cervical dislocation; the ceca were removed aseptically, clamped with forceps, and placed in sterile plastic bags on ice. In the laboratory, the narrow open ends of the ceca were cut with sterile scissors and the ceca were inverted onto sterile glass rods. Approximately one g of content was collected into a centrifuge tube containing 9 ml of sterile phosphate-buffered saline (PBS), pH 7.4, and homogenized by vortexing with glass beads (4-mm diameter) for 3 min. Debris was removed by centrifugation at 700 × g for 1 min, and the supernatant was centrifuged at 13,000 × g for 5 min. The pellet was washed twice with PBS and stored at −20°C until DNA extraction.
Mucous samples were collected from the ceca after all visible cecal lumen material had been removed by washes with PBS. The mucous layer attached to the cecal wall was gently scraped off with a small sterile spatula and mixed with 1 ml of PBS. The mixture was centrifuged at 13,000 × g for 5 min. The pellet was used directly for DNA isolation for construction of libraries 1 to 4. The pellets from all later samples were resuspended in 1 ml of PBS containing 1% Triton X-100. The suspension was incubated at 40°C for 10 min to lyse chicken cells and centrifuged at 13,000 × g for 5 min. The pellet was washed once with PBS and stored at −20°C until DNA extraction.
DNA isolation.
Bacterial genomic DNA was isolated by the method of Wilson (55) with some modifications. Cecal content or mucous samples were treated with lysozyme (final concentration of 2 mg/ml) for 30 min at 37°C, followed by treatment with sodium dodecyl sulfate (final concentration of 0.5% [wt/vol]) and proteinase K (final concentration of 0.1 mg/ml) for 2 h at 37°C. The samples were subjected to six 30-s intervals of bead beating at 5,000 rpm with zirconia-silica beads (0.1-mm diameter) on a minibead beater (Biospec Products, Inc., Bartlesville, Okla.). Completion of bacterial lysis was verified by microscopic examination. DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with isopropanol or ice-cold ethanol. The extracted DNA was treated with DNase-free RNase (Sigma Chemical Co., St. Louis, Mo.) at a final concentration of 0.2 mg/ml at 37°C for 1 h, followed by a second phenol-chloroform-isoamyl alcohol extraction and isopropanol precipitation. Finally, the DNA pellet was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), stored at −20°C, and used as template DNA in PCR to amplify the 16S ribosomal DNA rDNA for TTGE analysis and construction of 16S rDNA clone libraries. In some cases, the extracted DNA was further purified with a Geneclean Spin Kit (Bio 101, Vista, Calif.) before PCR.
PCR for TTGE analysis and construction of 16S rDNA clone libraries.
Primers used for PCR are listed in Table 1. For TTGE analysis, the variable regions V6-V8 of eubacterial 16S rDNA corresponding to positions 968 to 1401 in Escherichia coli (11) were amplified with primer pair 968F-GC/1401R (30). PCR amplifications were performed with a Robocycler Gradient 96 Temperature Cycler (Stratagene, La Jolla, Calif.). Serial dilutions of original DNA templates were tested to determine the optimal DNA concentrations for PCR by visual inspection of PCR bands on ethidium bromide-stained agarose gels. The dilution producing the cleanest PCR band was used in the subsequent amplification. After 5 min of initial denaturation at 94°C, a “touchdown” PCR was performed to increase the specificity of amplification and to reduce the formation of spurious by-products (15, 28). The initial annealing temperature (68°C) was set 10°C above the expected annealing temperature (58°C) and decreased by 1°C per cycle until a touchdown of 58°C, at which temperature 20 additional cycles were carried out. Amplification was performed at 1 min of denaturation at 94°C, 1 min of primer annealing, and 3 min of primer extension at 72°C, followed by 10 min of final primer extension. Amplification products were first analyzed by electrophoresis in 1.2% (wt/vol) agarose gels and ethidium bromide staining and then were stored at −20°C until they were used for TTGE analysis.
TABLE 1.
Primers used for amplification and sequencing of 16S rRNA genes
| Primer designation | Sequence | Use(s) | Reference(s) |
|---|---|---|---|
| 968F-GC | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCC GCCCGAACGCGAAGAACCTTAC | TTGE analysis | 30 |
| 1401R | CGGTGTGTACAAGACCC | TTGE analysis | 30 |
| 8FPL | AGTTTGATCCTGGCTCAG | Cloning | 35a |
| 1492RPL | GGYTACCTTGTTACGACTT | Cloning | 35a |
| 515FPL | AGTGCCAGGMGCCGCGG | Cloning | 35a |
| 63F | CAGGCCTAACACATGCAAGTC | Cloning, sequencing | 22 |
| 1387R | GGGCGGWGTGTACAAGGC | Cloning, sequencing | 22 |
| 1387R-AC | ACGGGCGGWGTGTACAAGGC | Cloning | 22, this study |
| 1391R | GACGGCGGTGTGTRCA | Detection of archaea | 9, 35 |
| 23FPL | GCGGATCCGCGGCCGC | Detection of archaea | 9 |
| A109F | ACKGCTCAGTAACACGT | Detection of archaea | 19, 53 |
| A934R | GTGCTCCCCCGCCAATTCCT | Detection of archaea | 19, 53 |
| 361F | GGAATATTGGACAATGGGC | Sequencing | 54b |
| 379R | GCCCATTGTCCAATATTCC | Sequencing | 54b |
| 522F | CAGC(A/C)GCCGCGGTAAT(A/T)C | Sequencing | 54b |
| 536R | G(A/T)ATTACCGCGGC(G/T)GCTG | Sequencing | 54b |
| 750F | CTGACGCTGAGGAGCGAAAG | Sequencing | 54b |
| 769R | CTTTCGCTCCTCAGCGTCAG | Sequencing | 54b |
| 927F | GGGCCCGCACAAGCGGT | Sequencing | 54b |
| 943R | ACCGCTTGTGCGGGCCC | Sequencing | 54b |
| 1075F | TCGTGAGATGTTGGGTTAAG | Sequencing | 54b |
| 1094R | CTTAACCCAACATCTCACGA | Sequencing | 54b |
Primers based on this reference, but sequences introducing restriction sites were omitted. Primer 515FPL was synthesized such that base number 10 was either A or C.
Sequencing primers based on this reference, but with modifications to improve primer efficiency.
For construction of the 16S rRNA gene clone libraries, four sets of primers were used. Clone libraries 1A and 1C were obtained with DNA amplified with primer pair 8FPL-1492RPL, and primer pair 515FPL-1492RPL was used for amplification for libraries 1B and 1D (Table 2).
TABLE 2.
Source of DNA, PCR primers, and cloning vectors used for generation of clone libraries
| Clone library(ies) | Bird no. | Source of DNA | PCR primer pair | Cloning vector |
|---|---|---|---|---|
| 1A | 1 | Cecal content | 8FPL-1492RPL | pPCR-Script Amp SK(+) |
| 1B | 1 | Cecal content | 515FPL-1492RPL | pPCR-Script Amp SK(+) |
| 1C | 1 | Cecal mucosa | 8FPL-1492RPL | pPCR-Script Amp SK(+) |
| 1D | 1 | Cecal mucosa | 515FPL-1492RPL | pPCR-Script Amp SK(+) |
| 2 | 2 | Cecal mucosa | 63F-1387R | pCR-BluntII TOPO |
| 3 to 11 | 3 to 11 | Cecal mucosa | 63F-1387R-AC | pPCR-Script Amp SK(+) |
Primers 63F-1387R (22) were used for amplification of 16S rRNA gene sequences for library 2. Primer pair 63F-1387R-AC was used to obtain 16S rRNA gene sequence inserts for libraries 3 to 11. Primer 1387R-AC is identical to primer 1387R, except for two additional bases, AC, added at the 5′ end. The alteration at the 5′ end of primer 1387R prevented the formation of a SrfI site which would have prevented the use of SfrI to select for successful insertion of the PCR products into the cloning vector.
After 3 min of denaturation at 94°C, amplification was performed for 30 cycles to obtain sufficient DNA for cloning into pPCR-Script Amp SK(+) (Stratagene). For cloning into vector pCR-BluntII TOPO (Invitrogen), 20 cycles of amplification were performed. The cycle conditions were 30 s of denaturation at 94°C, 30 s at 55°C for annealing, and 3 min at 72°C for extension, and final extension for 10 min. Cloned Pfu DNA polymerase (Stratagene) was used for amplification. The optimal dilution of original DNA templates was determined as described above, and was used in the subsequent amplifications.
In addition to primer pair 515FPL-1492RPL, which potentially could amplify archaeal small-subunit rRNA genes, primer pairs 8FPL-1391R (9, 35), 23FPL-1391R (9), and A109F-A934 (19, 53) were used in attempts to amplify archaeal sequences from six cecal mucosa and three cecal content samples.
Amplification products were analyzed by electrophoresis in 0.7% (wt/vol) agarose gels and ethidium bromide staining, then were stored at −20°C until they were used for construction of the clone libraries.
TTGE analysis of PCR products and sequencing of TTGE bands.
PCR products generated with primer pair 968F-GC-1401R were purified and concentrated using the StrataPrep PCR Purification kit (Stratagene) as described by the manufacturer. The DNA concentration was determined by comparison with a DNA quantitation standard (GenSura Laboratories, Inc., San Diego, Calif.). Two hundred nanograms of DNA was loaded onto an 8% polyacrylamide gel (prepared from 40% [wt/vol] acrylamide-N,N′-methylenebisacrylamide stock, 29:1, and containing 8 M urea in 1.25× TAE [50 mM Tris-acetate, 1.25 mM EDTA, pH 8.0]). TTGE was performed on a Dcode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, Calif.) for 10 h at 130 V. The temperature of the gel system was programmed to increase by 1.2°C per h from a starting temperature of 55°C until the final temperature of 67°C was reached. DNA bands were visualized by silver staining, and DNA was obtained from bands in the gel by removing gel plugs with a sterile pipette tip and elution into 30 μl of 0.1× TE buffer at 4°C overnight. Ten-microliter aliquots of eluate from individual bands were used for reamplification by PCR. The amplification products were run on TTGE to ascertain that their electrophoretic mobility was the same as that of the DNA from which they were derived. The PCR products were purified using the StrataPrep PCR purification kit prior to sequence determination at the University of Delaware DNA sequencing core facility. Sequencing reactions were performed with a PE-ABI Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) as described by the manufacturer, and electrophoresis and readout were done with an ABI PRISM 3700 DNA analyzer (Applied Biosystems). Primers 986F-GC and 1401R were used in the sequencing reactions to sequence both strands of each PCR product.
Construction of 16S rDNA clone libraries and sequencing.
PCR products were purified and concentrated prior to ligation into cloning vectors. Clones to be sequenced by automated sequencing at Delaware Technology Park were obtained using the PCR-Script Amp cloning kit (Stratagene) according to the manufacturer’s instructions. For clone libraries 1A, 1B, 1C, and 1D, all white colonies originating from the four respective transformation experiments were picked by robot for template preparation and sequencing reactions. For libraries 3 to 6 and 7 to 11, 192 and 96 white colonies, respectively, were picked and subjected to automated template preparation and sequencing. One single sequence was generated per clone using the M13 reverse primer.
Clone library 2 was constructed in vector pCR-BluntII TOPO using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). Plasmid DNA was isolated manually from the transformants and used as template for reamplification of the 16S rRNA inserts for restriction enzyme analysis. The amplified insert DNA was digested simultaneously with restriction enzymes AluI and HaeIII and the resulting restriction enzyme fragment patterns were compared and sorted visually. Sequencing was done at the University of Delaware DNA sequencing core facility. A collection of conserved sequencing primers (Table 1) was used to obtain sequence data from both strands of the 16S rRNA gene inserts.
Analysis of sequence and TTGE data.
The sequences obtained by manual sequencing were assembled using the SeqMan program of the DNAStar software package (DNASTAR Inc., Madison, Wis.). The sequences were subjected to the check chimera program from the Ribosomal Database Project (21). The sequence data obtained by single-pass automated sequencing were inspected for the presence of ambiguous base assignments, and unreliable sequences at the 3′ end and occasionally at the 5′ end were removed before the sequences were submitted for similarity searches. Searches were done with the Blast program (1). For classification into phylogenetic groups or subgroups, sequences were entered into the Sequence Match program (version 2.7) from the Ribosomal Database Project (21). TTGE patterns were analyzed using the Diversity Database 2.2.0 (Bio-Rad Laboratories). Comparison of TTGE pattern profiles were performed using Dice similarity coefficient analysis and Ward’s clustering method according to the Diversity Database manual.
Nucleotide sequence accession numbers.
Representative sequences were deposited with GenBank and are available under accession numbers AF376138 to AF376466.
RESULTS
Over the course of this study, a total of 1,656 nucleotide sequences originating from 16S rRNA genes were retrieved from bacteria found in the cecal content and attached to and embedded in the mucosal layer of the cecum of broiler chickens. The total includes 1,358 sequences from libraries 1A to 1D and 3 to 11 obtained by automated single-pass sequencing of rRNA gene DNA cloned in plasmid vectors (average length of sequences read, 478 bases) and 33 nearly full-length 16S rRNA gene sequences determined by sequencing both strands of a rRNA gene insert (library 2). Two hundred sixty-five sequences were derived from DNA bands in TTGE gels (average length, 390 bp). All sequences were compared to 16S rRNA gene sequences in GenBank between October 2000 and January 2001 using the Blast program (1). This program identified 243 different 16S rRNA gene sequences in the databases that were the closest relatives to the cecal bacteria sequences entered. Approximately 64% of the sequences obtained with this study were between 91 and 95% identical to their closest relative in the databases; for about 28% of the sequences the identity values were between 96 and 100%. Only approximately 2% of the cecal sequences were between 85 and 87% identical to their closest relative in the database. About 6% had identity values of between 88 and 90%. In order to simplify comparisons between sequence libraries, the cecal bacteria sequences and their closest relatives from the database were classified by the online Sequence Match program (version 2.7) in the Ribosomal Database Project Web site (21). The program predicted that the cecal sequences belonged to 50 different phylogenetic groups or subgroups. Since to our knowledge the limits of the program to correctly assign phylogenetic affiliations has not been established, the classification of especially the sequences with low degrees of identity to already known sequences has to be presumed tentative. The vast majority of the sequences, however, are near or above 95% identical to their closest relatives in the databases and the classification should therefore be reliable.
Only sequences originating from eubacteria were found. Archaea have been reported to be present in fecal samples from poultry (26), but no 16S rRNA gene sequences originating from these types of prokaryotes were detected among the sequences obtained with primer pair 515FPL-1492RPL. DNA isolated from six cecal mucosal fractions and from three cecal content samples did not produce amplification products when subjected to PCR with primer pairs 8FPL-1391R or 23FPL-1391R. Primer pairs A109F-A934R, specific for the amplification of small ribosomal subunit genes from archaea and methanogens (19, 55), also did not produce amplification products from two other cecal samples. Additional studies would be required to determine if and how frequently archaea are present in the cecal environment.
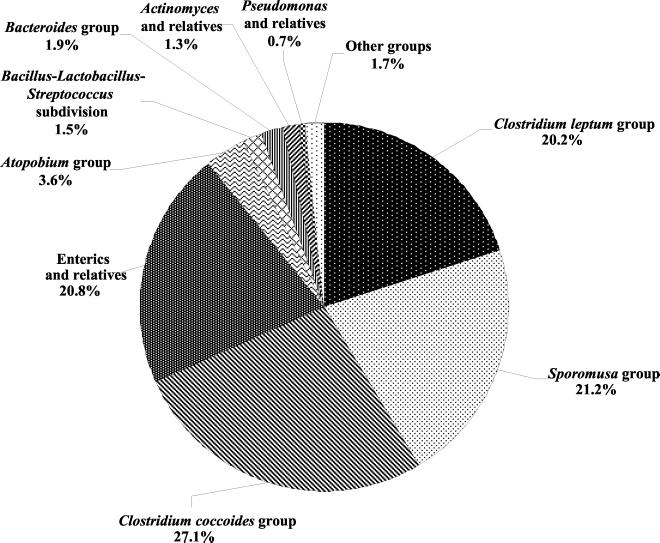
The spectrum of sequences is dominated by sequences related to those found in low-G+C gram-positive bacteria, specifically from the Clostridium leptum, Clostridium coccoides, and Sporomusa sp. groups (Fig. 1). The proteobacteria were represented mainly by members of the gamma subdivision of the division proteobacteria, especially the enterics (20.8% of sequences). Pseudomonads and their relatives accounted for less than 1% of the sequences retrieved. Also present in relatively low abundance were sequences representing the Bacteroides group (1.9%). Sequences related to those of high-G+C bacteria were categorized as members of the Atopobium group (3.6% of total) and of the actinomyces and relatives (bifidobacteria) (1.3% of total).
FIG. 1.
Percentage of the total number of sequences obtained in this study that were classified by the Sequence Match program of the Ribosomal Database Project as belonging to different phylogenetic groups or subdivisions. Sequences most closely related to those from the Eubacterium, Desulfovibrio, Clostridium propionicum, Xanthomonas, Clostridium botulinum, Acholeplasma-Anaeroplasma, Aeromonas, Rhizobium-Agrobacterium, and C. lituseburense groups were combined under “other groups.”
Percent identity values between the sequences retrieved from the cecal samples and sequences already present in GenBank ranged from 85 to 100%. In general, sequences related those of enterics, Pseudomonas, Bifidobacterium, or Lactobacillus spp. had the highest identity values, and frequently the sequences from the cecal samples were identical to those found in the databases. Cecal sequences for which the closest relatives in the databases originated from Clostridium and Eubacterium and relatives had generally lower identity values (<97%). This observation suggests that the databases currently contain few if any sequences of close relatives of these cecal bacteria.
Sequences amplified from DNA extracted from cecal content and mucosal scrapings.
On a macroscopic scale, the interior of the cecum appears to consist of the lumen filled with digesta and a mucosal layer attached to the cecal wall. The mucosal layer remains attached to the wall after washes with PBS while the visible, colored traces of the digesta are removed. For one experiment, DNA was extracted from the lumen content and from the scrapings of the cecum of one 6-week-old female broiler chicken. PCR was done on each of the two DNA extracts using two primer pairs, 8FPL-1492RPL and 515FPL-1492RPL (Table 1). The amplification products obtained were of the expected size, except for those originating from DNA isolated from the mucosal scrapings and amplified with primer pair 515FPL-1492RPL. PCR using these primers produced DNA approximately 750 bp in size in addition to the expected 900-bp DNA. The smaller DNAs originated from amplification of the mitochondrial rRNA gene of the chicken. Significantly more mitochondrial than bacterial 16S rRNA gene DNA was amplified, and it became necessary to gel purify the 900-bp DNA fragments prior to cloning. Despite the purification effort, 18 out of 109 sequences obtained from library 1D were mitochondrial 16S rRNA gene sequences. Analysis of sequences obtained from library 1C demonstrated that primer pair 8FPL/1492RPL was able to amplify nuclear DNA in the DNA extracted from mucosal scrapings, since 24 out of 79 clones from library 1C contained an 18S rRNA insert.
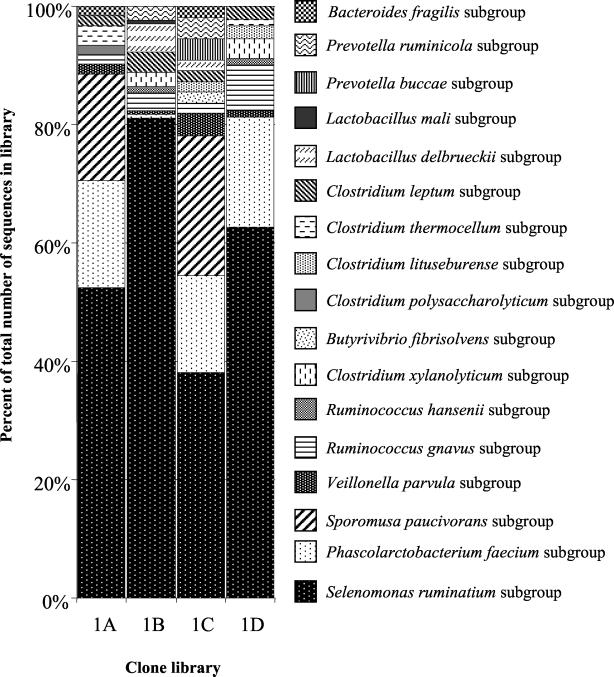
The automated sequencing system prepared DNA templates and performed a single sequencing reaction per clone from libraries 1A to -D. A total of 63 different sequences out of a total pool of 377 sequences were obtained (GenBank accession numbers AF376138 to AF376200). The sequences from each cloning experiment were compared to 16S rRNA gene sequences in GenBank and then sorted by their membership in particular bacterial subgroups (Table 3). As a percentage of the total number of sequences obtained from each library, sequences related to those of the Selenomonas ruminantium subgroup predominate (Fig. 2). These sequences were found in all four libraries and were therefore retrieved from bacteria in cecal content and the mucosa. Sequences representing the Phascolarctobacterium faecium and Veillonella parvula subgroups were also present in each of the libraries. Similarly, sequences representing the Ruminococcus gnavus subgroup of the Clostridium coccoides group of bacteria and the Clostridium leptum subgroup of the C. leptum group of bacteria were found in all four libraries. Sequences assigned to most other subgroups were generally also represented in cecal content and the cecal mucosa libraries. A small number of sequences belonging to the Butyrivibrio fibrisolvens, Clostridium lituseburense, and Prevotella buccae subgroups were found only in the mucosal libraries. One sequence belonging to the Clostridium polysaccharolyticum and the Lactobacillus mali subgroups, respectively, was only found in the cecal content libraries. Whether these findings indicate true differences in the bacterial populations or are merely the result of chance events is not known. Evidence for the occurrence of primer-related bias can be seen in Table 3. For example, sequences belonging to the S. paucivorans subgroup were found in the cecal content and the mucosal clone libraries, but only in the libraries generated with primer pair 8FPL/1492RPL. Similarly, sequences belonging to the Clostridium xylanolyticum and Ruminococcus hansenii subgroups were found only in the content and mucosal samples amplified with primer pair 515FPL/1492RPL.
TABLE 3.
Classification of 16S rRNA gene sequences obtained from bacteria in cecal content and in scrapings of cecal mucosa (libraries 1A to 1D)a
| Sequence affiliationb | No. of sequences in library belonging to phylogenetic subgroup
|
|||
|---|---|---|---|---|
| 1A | 1B | 1C | 1D | |
| Sporomusa group | ||||
| S S. ruminatium | 32 | 138 | 21 | 57 |
| P. faecium | 11 | 1 | 9 | 17 |
| S. paucivorans | 11 | 13 | ||
| V. parvula | 1 | 1 | 2 | 1 |
| C. coccoides group | ||||
| R. gnavus | 1 | 5 | 1 | 7 |
| R. hansenii | 2 | 1 | ||
| C. xylanolyticum | 4 | 3 | ||
| B. fibrisolvens | 1 | |||
| C. polysaccharolyticum | 1 | |||
| C. lituseburense group (C. lituseburense) | 1 | 2 | ||
| C. leptum group | ||||
| Clostridium thermocellum | 2 | 1 | ||
| C. leptum | 1 | 6 | 1 | 2 |
| Bacillus-Lactobacillus-Streptococcus subdivision | ||||
| Lactobacillus delbrueckii | 8 | 1 | ||
| L. mali | 1 | |||
| Bacteroides group | ||||
| P. buccae | 2 | |||
| Prevotella ruminicola | 4 | 2 | ||
| Bacteroides fragilis | 1 | 1 | ||
| Total | 61 | 170 | 55 | 91 |
Source of template DNA and primers used as listed in Table 2.
Phylogenetic subgroups are listed subordinate to their group unless there is only one subgroup, in which case it is indicated parenthetically.
FIG. 2.
Percentage of sequences from clone libraries 1A to 1D most closely related to 16S rRNA gene sequences from particular phylogenetic subgroups of bacteria. The libraries were obtained from DNA extracted from cecal content (libraries 1A and 1B) and from the cecal mucosa (libraries 1C and 1D) from one 6-week-old broiler. Primers used for amplification of the 16S rRNA gene sequences are listed in Table 2.
TTGE and sequencing of DNA amplified from 16S rRNA genes in DNA from cecal content and mucosal scrapings of birds of different ages.
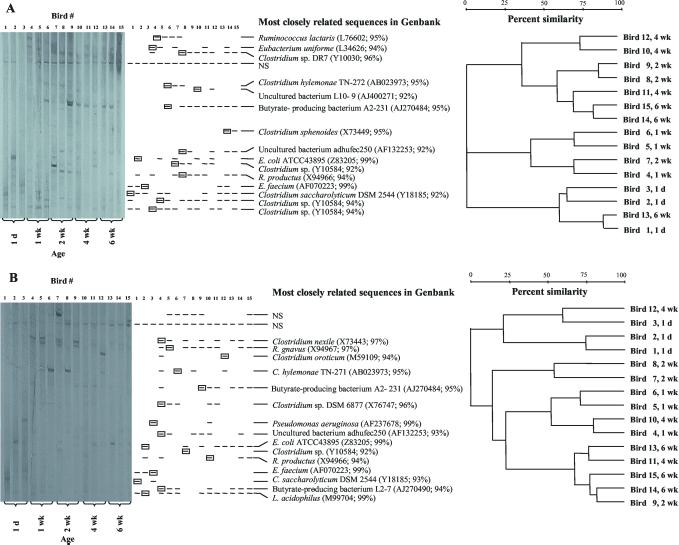
A comparison of 16S rRNA sequences retrieved from cecal content and cecal mucosa was also attempted by using TTGE and sequencing of DNA derived from cecal bacteria of chickens of different ages reared in a single flock. A gel image and a schematic drawing of the major bands seen on a TTGE gel containing samples from groups of three birds sacrificed at five different ages (1 day and 1, 2, 4, and 6 weeks) are shown in Fig. 3. The samples from the 1-day-old birds revealed the fewest bands, but there was little difference in the number of major bands visible in the samples from the older birds. Similar numbers of major bands were seen in the cecal content samples (Fig. 3A) and the cecal mucosa samples (Fig. 3B). For the sequence analysis of the DNA from the bands, DNA was eluted from the gel from only one of the bands that migrated the same distance into the gel. In total, DNA from 16 bands of the TTGE gel containing the cecal content samples and from 17 bands from the gel containing the mucosal scraping samples was eluted, and all but three of the DNA samples produced sequence data (GenBank accession numbers AF376234 to AF376253). DNA comigrating with Enterococcus faecium rRNA gene sequences was present in the cecal content and mucosa of each of the three 1-day-old birds. Corresponding bands were seen sporadically also in the content samples from older birds. Bands comigrating with a band of DNA from a Clostridium saccharolyticum strain were also present early on in the content and mucosal samples but were found only in the content samples from older birds. 16S rRNA gene DNA comigrating with DNA related to that of E. coli, Ruminococcus productus, and another Clostridium species was also found in cecal content at different ages. A number of additional bands appear in the content and mucosal samples from the older birds. Some bands are found occasionally whereas others are found in most samples of the older birds. For example, bands comigrating with the band representing a sequence related to that of butyrate-producing bacterium A2-231 (3) are seen in all but one of the cecal content samples from the 2-, 4-, and 6-week-old birds.
FIG. 3.
(A) Gel image, schematic representation, and cluster diagram of bands in TTGE gel of DNA amplified from bacteria in cecal content. Samples were prepared from 15 different birds, removed from their flock in groups of three at the age of 1 day (1 d), and 1, 2, 4, and 6 weeks (1, 2, 4, and 6 wk). One representative of each of the comigrating bands (boxed) was eluted from the gel and used for sequence determination. The name and GenBank accession number for the sequence most related to the sequence obtained from a particular band on the TTGE gels are listed to the right of the schematic representation of the banding patterns. NS denotes that no sequence data could be obtained from the eluted DNA. The dendrogram illustrates the relatedness of the TTGE banding pattern of the 15 cecal samples. The bird number in the dendrogram corresponds to the bird number indicated on the TTGE gel image. (B) Same as panel A, but samples originated from scrapings of the mucosa of the cecum.
Analysis of the patterns of bands on the TTGE gels indicates that there is only limited clustering of samples from the three different birds at a particular age. The samples from the 1-day-old birds are localized on one major branch of the dendrogram, but in both the content and mucosal samples, patterns generated from a sample from an older bird match closely with one of the patterns from the 1-day-old birds. The TTGE band patterns are therefore variable and this suggests that the microbiota of individual birds raised under identical conditions might differ.
Since no TTGE standards were available, it was not possible to directly compare the TTGE gel images from the cecal content and mucosa samples. Analysis of the sequence data obtained from bands on the gels, however, demonstrated that the major bands on the two gels contain similar 16S rRNA gene sequence (Fig. 3A). Six sequences from content and mucosal samples had the same closest relative in the database. The sequences include those of E. coli and of bacteria belonging to the C. coccoides and Enterococcus groups. Perhaps the most significant difference between the sequences obtained from the content and mucosal samples is the presence of bands in the mucosal samples presumed to be originating from bacteria closely related to Lactobacillus acidophilus.
TTGE and random cloning of 16S rRNA gene DNA from bacteria in mucosal scrapings from 5- to 7-week-old broiler chickens.
DNA extracted from scrapings of the cecal mucosa of 18 5- to 6-week-old broiler chickens was used as template for amplification with primer pair 968F-GC-1401R, and the resulting amplification products were separated by TTGE. DNA from nine of the scraping samples also served as template for amplification using primer pair 63F-1387R-AC. The amplification products were then ligated into vector pPCR-Script Amp SK(+), and a number of the 16S rRNA gene inserts were partially sequenced (libraries 3 to 11).
The patterns of bands seen with TTGE for each of the 18 samples contained numerous bands that were barely visible even with silver staining. In addition, 10 to 20 dark bands were present in each of the samples derived from birds 3 to 20, and sequence data could be obtained from the DNA of most of the dark bands. Blast searches of the sequence data showed that the 16S rRNA gene sequences obtained represented bacteria from only three phylogenetic groups (C. coccoides, C. lituseburense, and enterics) (Table 4). Sequences representing the C. coccoides group were present in all 18 samples analyzed, and in all but four of the samples, every band examined contained DNA representing the C. coccoides group. In two samples, a band representing the C. lituseburense group was found, and in three birds, a band representing the enterics group was present. The TTGE-based analysis of the samples from 18 birds thus revealed the presence of only a narrow spectrum of bacteria in the mucosal scrapings.
TABLE 4.
Sequences obtained after TTGE of PCR-amplified 16S rRNA gene DNA obtained from the cecal mucosa of 18 5- to 7-week-old broiler chickens
| Sequence affiliationa | No. of sequences obtained from cecal mucosa of bird no.:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| C. coccoides group | ||||||||||||||||||
| R. gnavus | 6 | 9 | 7 | 3 | 8 | 2 | 5 | 5 | 8 | 6 | 3 | 5 | 4 | 7 | 4 | 4 | 8 | 9 |
| R. hansenii | 6 | 5 | 5 | 4 | 1 | 4 | 5 | 3 | 4 | 4 | 2 | 7 | 6 | 4 | 5 | 5 | 3 | |
| C. xylanolyticum | 3 | 3 | 4 | 1 | 3 | 3 | 1 | 1 | 3 | 5 | 3 | 2 | 1 | 2 | 1 | 1 | ||
| C. polysaccharolyticum | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | ||||||
| B. fibrisolvens | 1 | |||||||||||||||||
| C. aminovalericum | 1 | |||||||||||||||||
| E. ventriosum | 1 | |||||||||||||||||
| Enterics and relatives (Escherichia) | 1 | 1 | 1 | |||||||||||||||
| C. lituseburense group (C. glycolicum) | 1 | 1 | ||||||||||||||||
See footnote b to Table 3.
A certain degree of uniformity among the 18 bird samples is still reflected at the subgroup level. Sequences representing the R. gnavus subgroup were found in all 18 samples, and sequences representing the R. hansenii subgroup were found in 17 of the 18 samples. The C. xylanolyticum subgroup was represented in 16 of the 18 samples. Twelve samples contained sequences representing the C. polysaccharolyticum subgroup. A sequence representing the Eubacterium ventriosum, Clostridium aminovalericum, and the B. fibrisolvens subgroups were found in the sample from one bird each. In two of the 18 samples, sequences from the Clostridium glycolicum subgroup of the C. lituseburense group of bacteria were present. Representative sequences obtained from TTGE bands are available from GenBank under the accession numbers AF376254 to AF376302.
Random cloning of 16S rRNA gene sequences was done using DNA extracted from cecal samples from birds 3 to 11. The primer pair used for the amplification of the 16S rRNA gene sequences was 63F-1387R-AC. Preliminary experiments had shown amplification to be more robust with primer pair 63F-1387R described by Marchesi et al. (22) than with primer pair 8FPL-1492RPL or 515FPL-1492RPL. Primer pair 63F/1387R was then tested on a DNA sample derived from the cecal mucosa of bird 2 (Table 2). The amplified DNA was ligated into vector pPCR-BluntII TOPO since the 1387R primer sequence is not compatible with the cloning mechanism for vector pPCR-Script AMP SK(+). Automated sequencing was attempted for the pPCR-BluntII TOPO clones but was not successful. Therefore, a total of 152 clones were manually picked from plates, plasmid DNA was isolated, and the 16S rRNA gene inserts were amplified by PCR to obtain DNA that could be digested with restriction enzymes AluI and HaeIII. Thirty-three restriction enzyme fragment patterns could be distinguished. The nearly full-length sequence of one clone representing each of the 33 restriction enzyme digest patterns was determined. (GenBank accession numbers AF376201 to AF376233). Blast search results are listed in Table 5. Most of the closest relatives in the databases belong to as of yet uncultured bacteria from human feces (47). Except for the sequence related to Pseudomonas fluorescens, the level of identity between the sequences in the databases and the sequences determined with this experiment ranged from 90 to 97%.
TABLE 5.
Blast search results for 16S rRNA gene sequences obtained from the cecal mucosa of a 6-week-old bird (library 2)
| Phylogenetic affiliation (subgroup) and no. of clones | Closest relative in GenBank (accession no.) | % Identity |
|---|---|---|
| Clostridium leptum | ||
| 13 | Uncultured bacterium adhufec218 (AF132246) | 91–95 |
| 3 | Elbe river snow isolate Iso155 (AF150697) | 91 |
| 2 | Uncultured bacterium adhufec168 (AF132242) | 94–96 |
| 2 | Fusobacterium prausnitzii (X85022) | 94 |
| 2 | Uncultured bacterium adhufec13 (AF132237) | 96 |
| 1 | Butyrate-producing bacterium L2-6 (AJ270470) | 97 |
| 1 | Butyrate-producing bacterium A2-20 (AJ270471) | 90 |
| 1 | Uncultured bacterium A03 (AF052408) | 97 |
| 1 | Unidentified eubacterium clone vadinBA08 (U81642) | 92 |
| 1 | Uncultured bacterium adhufec101 (AF132235) | 92 |
| 1 | Uncultured rumen bacterium 4C28d-4 (AB034125) | 93 |
| 1 | Ruminococcus flavefaciens (AF104834) | 91 |
| 1 | Uncultured bacterium adhufec365 (AF132265) | 95 |
| Clostridium polysaccharolyticum (n = 1) | Eubacterium halii (L34621) | 94 |
| Pseudomonas azotoformans (n = 1) | Pseudomonas fluorescens (AF228367) | 100 |
| Bacteroides fragilis (n = 1) | Uncultured bacterium adhufec77.25 (AF153865) | 92 |
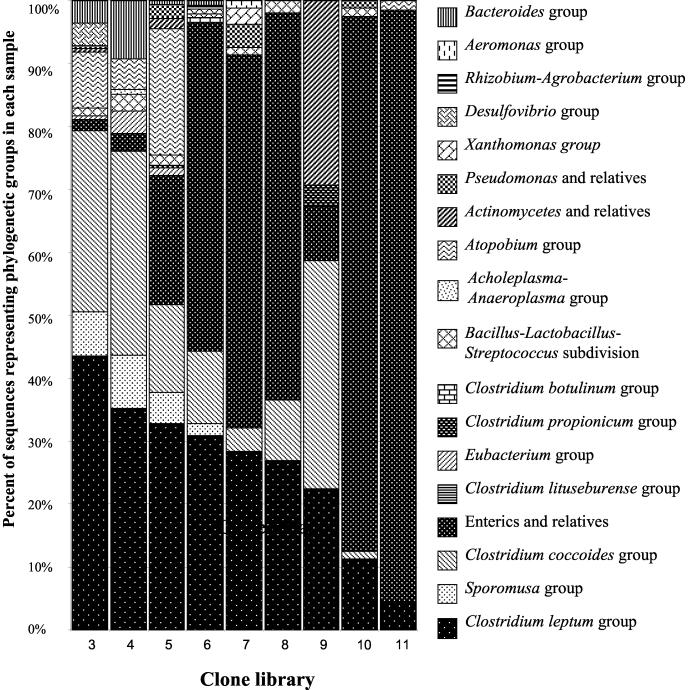
In contrast to primer pair 8FPL-1492RPL, primer pair 63F-1387R apparently did not amplify 18S rRNA gene sequences to a detectable degree. Since the preliminary experiment had also demonstrated that 63F-1387R amplified diverse sequences from mucosal bacteria, 16S rRNA gene amplification from samples from birds 3 to 11 was carried out with this primer pair. Also because automated sequencing had only been successful for 16S rRNA gene inserts in pPCR-Script AMP SK(+), the PCR-Script cloning strategy was applied to the construction of libraries 3 to 11. Primer pair 63F-1387R was adapted for use with this cloning system by the addition of two bases, A and C, to the 5′ end of primer 1387R. This 5′ addition was unlikely to change the universality of the primer, especially since these bases are present at the corresponding positions in the 16S rRNA genes of virtually all bacteria examined (22). Automated sequencing was successful for the clone libraries derived from cecal mucosal samples from birds 3 to 11. The sequences were compared to sequences in GenBank and then assigned to phylogenetic groups and subgroups (Table 6). Representative sequences were deposited with GenBank (accession numbers AF376303 to AF376466). The sequences belonged to bacteria from 17 phylogenetic subdivisions or groups. Sequences representing the C. leptum group of bacteria were found in all nine samples. The percentage of enteric sequences of the total number of sequences in each library ranges from a few percent to over 90% (Fig. 4). With respect to enteric 16S rRNA gene sequences, random cloning thus produced results that are different from what was observed with the TTGE approach since DNA sequence data from only one of the major bands (sample 11) were closely related to those of enteric sequences (Table 4). Random cloning and TTGE approaches also produced a different result for sample 11 with regards to sequences belonging to the C. coccoides phylogenetic group. Several major TTGE bands originating from sample 11 contained DNA sequences closely related to those from the C. coccoides group (Table 4), but none of the sequences in the random cloning library 11 were closely related to sequences from this group of bacteria (Table 6). The overall differences in retrieval of sequences with the random cloning and the TTGE methods are listed in Table 7. There is very limited agreement between the results of both methods, and the random cloning approach clearly presents a picture of greater diversity than does the TTGE/sequencing method.
TABLE 6.
Classification of 16S rRNA gene sequences from 1 cecal mucosa of nine 5- to 7-week-old broiler chickens (libraries 3 to 11)
| Sequence affiliationa | No. of sequences in library belonging to phylogenetic subgroup:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Enterics and relatives | |||||||||
| Escherichia | 2 | 4 | 37 | 73 | 46 | 31 | 4 | 65 | 61 |
| Enterobacter asburiae | 4 | 3 | |||||||
| Salmonella enteritidis | 1 | 1 | 1 | ||||||
| Klebsiella pneumoniae | 1 | 1 | 1 | ||||||
| Proteus vulgaris | 2 | ||||||||
| Pasteurella anatis | 1 | ||||||||
| C. leptum group | |||||||||
| C. leptum | 65 | 48 | 55 | 45 | 23 | 14 | 13 | 8 | 3 |
| C. thermocellum | 9 | 2 | 4 | 1 | 1 | ||||
| C. coccoides group | |||||||||
| R. gnavus | 30 | 29 | 15 | 10 | 3 | 2 | 17 | 1 | |
| C. xylanolyticum | 4 | 10 | 5 | 2 | 1 | ||||
| C. polysaccharolyticum | 11 | 3 | 3 | 1 | 1 | 2 | |||
| R. hansenii | 4 | 4 | 2 | 4 | 2 | ||||
| B. fibrisolvens | 1 | ||||||||
| Lachnospira multipara | 1 | ||||||||
| Atopobium group (Atopobium minutum) | 15 | 7 | 36 | 1 | 1 | ||||
| Sporomusa group (P. faecium) | 12 | 12 | 9 | 3 | |||||
| Actinomyces and relatives (Bifidobacterium infantis) | 1 | 3 | 17 | ||||||
| Bacteroides group | |||||||||
| Porphyromonas macacae | 1 | 7 | 1 | ||||||
| Rikenella microfusus | 4 | 2 | |||||||
| B. fragilis | 4 | 1 | |||||||
| P. buccae | 1 | ||||||||
| Bacillus-Lactobacillus-Streptococcus subdivision | |||||||||
| L. delbrueckii | 1 | 3 | 3 | ||||||
| Aneurinibacillus aneurinilyticus group | 1 | ||||||||
| Virgibacillus pantothenticus | 1 | ||||||||
| Enterococcus group | 1 | ||||||||
| Lactobacillus reuteri | 1 | ||||||||
| Lactobacillus plantarum | 1 | ||||||||
| L. mali | 1 | ||||||||
| Pseudomonas and relatives | |||||||||
| P. azotoformans | 1 | 1 | 1 | ||||||
| P. tolaasii | 2 | 1 | |||||||
| P. stutzeri | 2 | ||||||||
| P. taetrolens | 1 | ||||||||
| Eubacterium group | |||||||||
| Clostridium aminobutyricum | 1 | 3 | 1 | ||||||
| Eubacterium brachy | 2 | 1 | |||||||
| Desulfovibrio group (Desulfovibrio desulfuricans) | 6 | ||||||||
| Clostridium propionicum group | |||||||||
| C. propionicum | 2 | ||||||||
| Clostridium colinum | 1 | ||||||||
| Xanthomonas group (Stenotrophomonas maltophilia) | 2 | ||||||||
| Clostridium botulinum group (Clostridium aurantibutyricum) | 1 | ||||||||
| Acholeplasma-Anaeroplasma group (Eubacterium cylindroides) | 1 | ||||||||
| Aeromonas group (Aeromonas hydrophila) | 1 | ||||||||
| Rhizobium-Agrobacterium group (Caulobacter vibrioides) | 1 | ||||||||
| Total | 170 | 143 | 180 | 149 | 81 | 52 | 58 | 80 | 68 |
See footnote b to Table 3.
FIG. 4.
Percentage of sequences obtained by automated single-pass sequencing and belonging to particular phylogenetic groups or subdivisions. Libraries 3 to 11 were generated after amplification of 16S rRNA gene sequences with primers 63F-1387R-AC.
TABLE 7.
Frequency of detection of sequences belonging to different groups of bacteria by cloning and sequencing versus TTGE and sequencing
| Sequence affiliation | No. of sequences detected by sequencing and:
|
|
|---|---|---|
| Cloning | TTGE | |
| Acholeplasma-Anaeroplasma group | 1 | 0 |
| Actinomyces and relatives | 21 | 0 |
| Aeromonas group | 1 | 0 |
| Atopobium group | 60 | 0 |
| Bacillus-Lactobacillus-Streptococcus subdivision | 13 | 0 |
| Bacteroides group | 21 | 0 |
| C. botulinum group | 1 | 0 |
| C. coccoides group | 168 | 230 |
| C. leptum group | 291 | 0 |
| C. lituseburense group | 0 | 2 |
| C. propionicum group | 3 | 0 |
| Desulfovibrio group | 6 | 0 |
| Enterics and relatives | 339 | 3 |
| Eubacterium group | 8 | 0 |
| Pseudomonas and relatives | 9 | 0 |
| Rhizobium-Agrobacterium group | 1 | 0 |
| Sporomusa group | 36 | 0 |
| Xanthomonas group | 2 | 0 |
| Total | 981 | 235 |
The random cloning method coupled with automated single-pass sequencing allowed relatively rapid surveys of the bacterial diversity of samples but has the potential to cause an overestimation of the diversity. Since the cloning involved blunt-end ligation, the sequencing data obtained will be either from the 5′ or the 3′ end of the 16S rRNA insert depending on the orientation of the insert with respect to the primer binding site on the plasmid. Potentially, sequences from the 3′ and 5′ end of the same clone could yield two different sequences with the highest identity scores when a Blast search is performed. Two analyses were done to better assess the impact of insert orientation on the diversity analysis. Sequences obtained from clone libraries 3 to 11 were sorted based on whether they originated from the 5′ or 3′ end of the cloned 16S rRNA gene inserts. Surprisingly, a significant deviation from the expected 1:1 ratio was observed since out of a total of 979 sequences, 345 were from the 5′ end and 634 sequences were from the 3′ end of the 16S rRNA genes (ratio, 1:1.9). The reason for this ratio is not known. For the individual libraries, the ratios ranged from 1:1.1 to 1:3.7. There was no obvious correlation between the sequence composition of the individual libraries and the ratio of sequences from the 5′ or 3′ end. The location of the sequences within the 16S rRNA genes had essentially no influence on which phylogenetic groups or subdivisions were detected. There was also little difference at the level of the phylogenetic subgroups (data not shown); however, a noticeable difference in the percentage of sequences belonging to individual groups can be observed. For example, 29.6% of the sequences derived from the 3′ end of 16S rRNA genes were most closely related to those of enterics, but 44% of the sequences from the 5′ end belonged to this group of sequences. The difference for sequences belonging to the C. leptum phylogenetic group was also noticeable. Thirty-four percent of the sequences from the 3′ end but only 21% of the sequences from the 5′ end of the 16S rRNA gene inserts belonged to this phylogenetic group. This quantitative bias, however, was not strong enough to alter the spectrum of the major phylogenetic groups or subgroups that were detected.
Sequence data from library 2 were used to further assess how the input of relatively short (∼350 bp) sequences from either the 5′ or 3′ end might affect the evaluation of sequence diversity. The results of Blast searches done with the full-length sequences (Table 5) were compared with results of searches done with only 350 bases of sequence from either the 5′ or the 3′ end of the full-length sequence. The results indicated that regardless of the sequence used, the Blast search always produced top-scoring sequences from the same phylogenetic subgroup of bacteria. This result suggests that placements into phylogenetic subgroups based on the short sequences produced by automated sequencing can be made with confidence. A determination of whether or not a sequence obtained from the 5′ or 3′ end came from the same 16S rRNA gene sequence, however, cannot be made with certainty since only 30% of the 350-bp sequences from the 5′ end produced exactly the same top-scoring sequence as the full-length sequence in a Blast search. The corresponding percentage for the 3′-end sequences was 51%. The number of different sequences obtained by random cloning and automated sequencing and assigned different relatives in the databases is therefore probably larger than the actual number of different bacteria present.
DISCUSSION
Earlier studies that involved culturing of bacteria from the cecum had demonstrated that this intestinal site harbored a complex microbiota (5, 6, 24, 36). Using an approach that did not require growing the cecal bacteria, we were able to confirm those observations. The earlier identifications methods rarely allowed definitive determinations of bacterial isolates, and often did not allow identification to even the genus level. These limitations therefore make it difficult to compare the culture-based data with the 16S rRNA gene-based data, but it appears that both approaches detected some of the same groups of bacteria. Among the culturable bacteria from the cecum, gram-positive cocci were most numerous, and bacteria identified as Eubacterium and Clostridium sp. were also prominent (5, 6). Phylogenetically, most of these bacteria would presumably be classified as low-G+C gram-positive bacteria. Sequences representing this group of bacteria were the most common sequences recovered in the present study. Budding cocci and a budding bacterium, Gemmiger formicilis, also featured prominently in the culturable population (5, 6, 13). It is not known which of the currently recognized bacterial groups were represented by the group of budding cocci. None of the sequences recovered in this study appears to belong to Bergey’s Group 13, budding and/or appendaged bacteria. Bifidobacterium spp. were also represented significantly among the culturable anaerobes (5, 6, 32). Sequences originating from bacteria related to Bifidobacterium (actinomycetes and relatives) were detected in this study, but they were relatively rare (Fig. 1). It is possible that biases inherent in the genetic approach could have caused an underrepresentation of the Bifidobacterium-related sequences. Similarly, there are relatively few sequences related to those of members of the Bacteroides group among the sequences detected in this study. The culture-based approaches, on the other hand, identified Bacteroidaceae as one of the main groups of culturable anaerobes from the cecum. Lactobacilli and enterics were cultured from the cecum, and sequences related to those from these bacteria were also seen in the present study.
Although in low numbers, sequences of bacteria related to Pseudomonas spp. were found in this study. These bacteria were not among the dominant culturable cecal bacteria in the earlier studies. It is perhaps surprising to find evidence of generally aerobic bacteria in a cecal environment, but it is known that some Pseudomonas species are capable of anaerobic respiration with nitrate or nitrite (51) and of slow growth in rich medium containing arginine under anaerobic conditions (50). It is also possible that the Pseudomonas sequences found originate from bacteria that are only transient, and perhaps their 16S rRNA genes were preferentially amplified. Interestingly, the competitive exclusion preparation originating from cecal bacteria and developed into Preempt was reported to contain one Pseudomonas species (12). A number of the bacteria reported to be part of Preempt belong to groups whose 16S rRNA sequences were also found in this study. The exceptions were Propionibacterium and Lactococcus species. Several of these bacteria are apparently in the competitive exclusion product, but sequences from these types of bacteria were not found in this study.
The earlier culture-based studies generally isolated cecal bacteria from the contents of the ceca, and several of the competitive exclusion preparations studied over the years were also based on bacteria from the content of the gut (7, 29, 31). One preparation (45, 46), however, is based on scrapings of the mucosal wall of the cecum. The bacteria inhabiting this location could be of significant value to competitive exclusion preparation since they are presumably adapted to this particular environment and could form a strong barrier against bacteria capable of invading the tissue of the cecum. Our studies indicate that it is difficult to detect differences in the bacterial populations of the cecal wall and the cecal content. Possibly, the techniques used for separation of lumen and mucosa were not effective. Microscopic examination of cecal content and mucosal scrapings consistently indicated that there are significantly fewer bacteria in the mucosal scrapings than in the cecal content. Even small amounts of content matter retained in the scraping fraction could thus contribute more bacteria to the scraping fraction than the mucosa itself. Although the cecal wall appears to be relatively smooth on a macroscopic level, the cecum contains many crypts that might harbor content bacteria that cannot be easily washed off. These bacteria might contaminate mucosal bacteria during scraping. Alternatively, many of the bacteria that inhabit the lumen of the cecum might also be capable of adhesion to or even penetration of the mucosa.
The microbiota on and in the mucosa will be influenced by the biochemical processes occurring in the digesta and by factors originating from the animal host. Knowledge of the bacterial metabolism in the cecum is still incomplete, but fermentative processes appear to predominate (reviewed in reference 25). The production and utilization of organic acids (acetic, propionic, butyric, valeic, lactic, and succinic acids) in the cecum has been described (8), and degradation of uric acid ingressing into the cecum has also been proposed based on the consistent observation of uric acid degrading bacteria among cecal isolates (23, 24). The birds used for this study were fed a standard commercial diet composed of plant- and animal-derived ingredients, nutritional additives such as certain amino acids, and an anticoccidial compound. Although the birds originated from different flocks, it was assumed that the birds did not differ in their dietary status. Some studies have indicated that the microbiota of the cecum is relatively stabile, and even changes from low- to high-protein diets have been reported to have little influence on the microbial populations of the cecum (4, 49). The largely fermentative activity of the cecum and the relatively uniform diets might explain why relatively few major phylogenetic groups or subdivisions of bacteria were found in the cecal samples.
Earlier culture-based studies found that host factors such as age influence the composition of the cecal microbiota (5, 6). Our TTGE data are in agreement with the previous observations that population changes occur within one week after hatching (Fig. 3). The data also suggest that birds reared under identical conditions might not harbor the same bacteria in the same proportions. The TTGE-based survey of 18 5- to 7-week-old broilers indicated that certain subgroups of the C. coccoides group of bacteria might have been inhabiting all the birds examined in numbers sufficient for detection by TTGE (Table 4). This method of analysis, however, might underestimate the diversity of bacteria present at the cecal sites, since sequences belonging to more and different phylogenetic groups were detected by random cloning and sequencing (Table 6). Although sequences from the C. coccoides group were also present in most of the clone libraries, sequences from other groups, such as the C. leptum group and the enterics and relatives were also present in high numbers. Differences between the outcomes of the TTGE-sequencing and the cloning-automated sequencing approach might be due to the use of different primer pairs for PCR amplification and/or some other bias inherent to the techniques.
The limitation of PCR and cloning to provide accurate, quantitative measurements of the actual composition of a microbial community are known (18, 48, 52), but it is unlikely that the quantitative and qualitative differences between samples from different birds illustrated in Fig. 4 were solely caused by technical bias. The differences probably reflect variations in the cecal microbiotas of the individual birds. The preponderance of sequences from enterics, primarily Escherichia, in several of the libraries is striking. If indeed reflective of the actual bacterial population in the ceca, such high counts of enterics might be temporary and reflect the nutritional or health status of a particular bird, but perhaps such levels are within the range of variation in the microbiota of chickens of similar age and dietary status. Little is known about the scope of such variability in commercially raised chickens. Differences in the degree of efficacy of competitive exclusion preparations derived from different birds of a flock (40) are supportive of the notion that significant differences exist between the cecal microbiota of adult birds.
Evidence for relatively stabile, but distinct intestinal microbiota in human individuals (56) and in pigs (39) was obtained by molecular analyses of fecal samples. Despite such observed individual and interspecies differences in intestinal bacterial populations, some bacterial species are probably common inhabitants of many animal species. Some cecal sequences such as those of enterics and Bifidobacterium, Pseudomonas, and Lactobacillus species were virtually identical to sequences from other animal or even human sources. On the other hand, the sequence data also suggest that at least part of the bacterial population of the intestinal tract of different animal species might be unique. Sequences related to those of Clostridium and Eubacterium and relatives are generally only about 97% or less identical to sequences in the database. Many of the sequences in the databases originate from studies of human or rumen populations, and the sequence differences might suggest that related, but distinct species or genera inhabit the intestinal tract of different animal species. Such differences might be of practical importance as illustrated by Impey et al. (20) who showed that a competitive exclusion mixture that is effective in chickens did not show the same efficacy in turkeys.
Overall the genetic techniques in this study presented a picture of the cecal microbiota that is characterized by the presence of 16S rRNA gene sequences indicative of many genera and species, yet at a higher phylogenetic level, relatively few of the bacterial groups are represented. The data will be useful for future studies relating to competitive exclusion and for experiments evaluating the impact of, for example, growth promoters, anticoccidial compounds or stress on the intestinal microbiota. Clearly, the choice of technique used to analyze the bacterial populations influenced the data obtained in this study, and even the choice of primer pairs made a difference. There is clearly a need to complement the earlier culture-based data and the data generated with techniques based on PCR amplification with data that are independent of amplification bias, such as in situ hybridization.
Acknowledgments
This study was supported by grants from the University of Delaware Research Foundation, the USDA-NRI (980230), Hatch Project no. DEL00383, and by the Delaware Biotechnology Institute.
Footnotes
Published as paper no. 1700 in the Journal Series of the Delaware Agricultural Experiment Station.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. S. 1987. Factors affecting microbial competitive exclusion in poultry. Food Technol. 41:88–92. [Google Scholar]
- 3.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, E. M. 1972. The avian intestinal flora with particular reference to the possible ecological significance of the cecal anaerobic bacteria. Am. J. Clin. Nutr. 25:1475–1497. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, E. M., G. C. Mead, D. A. Barnum, and E. G. Harry. 1972. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci. 13:311–326. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, E. M. 1979. The intestinal microflora of poultry and game birds during life and after storage. J. Appl. Bacteriol. 46:407–419. [DOI] [PubMed] [Google Scholar]
- 7.Barnes, E. M., C. S. Impey, and D. M. Cooper. 1980. Manipulation of the crop and intestinal flora of the newly hatched chick. Am. J. Clin. Nutr. 33:2426–2433. [DOI] [PubMed] [Google Scholar]
- 8.Barnes, E. M., C. S. Impey, and B. J. H. Stevens. 1979. Factors affecting the incidence and anti-salmonella activity of the anaerobic caecal flora of the young chick. J. Hyg. 82:263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrow, P. A., M. B. Huggins, M. A. Lovell, and J. M. Simpson. 1987. Observation on the pathogenicity of experimental Salmonella typhimurium infection in chickens. Res. Vet. Sci. 42:194–199. [PubMed] [Google Scholar]
- 11.Brosius, J., T. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801–4805. [DOI] [PubMed] [Google Scholar]
- 12.Corrier, D. E., D. J. Nisbet, C. M. Scanlan, A. G. Hollister, and J. R. Deloach. 1995. Control of Salmonella typhimurium colonization in broiler chicks with a continuous-flow characterized mixed culture of cecal bacteria. Poult. Sci. 74:916–924. [DOI] [PubMed] [Google Scholar]
- 13.Croucher, S. S., and E. M. Barnes. 1983. The occurrence and properties of Gemmiger formicilis and related anaerobic budding bacteria in the avian caecum. J. Appl. Bacteriol. 54:7–22. [Google Scholar]
- 14.DeLoach, J. R. 1998. Development of PREEMPT™: the first FDA approved CE product to reduce Salmonella in poultry, p.123–126. In Technology Transfer Society 1998 Annual Summit Proceedings. Technology Transfer Society, Chicago, Ill.
- 15.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle, M. P. 1991. Colonization of chicks by Campylobacter jejuni. In L. C. Blankenship (ed.), Colonization control of human bacterial enteropathogens in poultry. Academic Press, Inc.
- 17.Duchet-Suchaux, M., P. Lechopier, J. Marly, P. Bernadet, R. Delaunay, and P. Pardon. 1995. Quantification of experimental Salmonella enteritidis carrier state in B13 leghorn chicks. Avian Dis. 39:796–803. [PubMed] [Google Scholar]
- 18.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Impey, C. S., G. C. Mead, and S. M. George. 1984. Evaluation of treatment with defined and undefined mixtures of gut microorganisms for preventing Salmonella colonization in chicks and turkey poults. Food Microbiol. 1:143–147. [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. L. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mead, G. C. 1974. Anaerobic utilization of uric acid by some group D streptococci. J. Gen. Microbiol. 82:421–423. [DOI] [PubMed] [Google Scholar]
- 24.Mead, G. C., and B. W. Adams. 1975. Some observation on the caecal microflora of the chick during the first two weeks of life. Br. Poult. Sci. 16:169–176. [DOI] [PubMed] [Google Scholar]
- 25.Mead, G. C. 1997. Bacteria in the gastrointestinal tract of birds, p.216–240. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology, vol. 2. Chapman & Hall, New York, N.Y.
- 26.Miller, T. L., M. J. Wolin, and E. A. Kusel. 1986. Isolation and characterization of methanogens from animal feces. Syst. Appl. Microbiol. 8:234–238. [Google Scholar]
- 27.Milner, K. C., and M. F. Schaffer. 1952. Bacteriological studies of experimental salmonella infections in chicks. J. Infect. Dis. 90:81–96. [DOI] [PubMed] [Google Scholar]
- 28.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nisbet, D. J., D. E. Corrier, C. M. Scanlan, A. G. Hollister, R. C. Beier, and J. R. DeLoach. 1993. Effect of a defined continuous-flow derived bacterial culture and dietary lactose on Salmonella typhimurium colonization in broiler chickens. Avian Dis. 37:1017–1025. [PubMed] [Google Scholar]
- 30.Nubel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nurmi, E., and M. Rantala. 1973. New aspects of salmonella infection in broiler production. Nature 241:210–211. [DOI] [PubMed] [Google Scholar]
- 32.Ochi, Y., T. Mitsuoka, and T. Sega. 1964. Untersuchungen ueber die Darmflora des Huhnes. Zentbl. Bakteriol. Parasitenkd. 193:80–95. [PubMed] [Google Scholar]
- 33.Qui, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahner, R. 1901. Bakteriologische Mitteilungen ueber die Darmbakterien der Huehner. Zentbl. Bakteriol. Parasitenkd. 80:239–244. [Google Scholar]
- 35.Reysenbach, A.-L., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salanitro, J. P., I. G. Fairchilds, and Y. D. Zgornicki. 1974. Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl. Microbiol. 27:678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleifer, J. H. 1985. A review of the efficacy and mechanism of competitive exclusion for the control of salmonella in poultry. World’s Poult. Sci. J. 41:72–83. [Google Scholar]
- 38.Shapiro, S. K., and W. B. Sarles. 1949. Microorganisms in the intestinal tract of normal chickens. J. Bacteriol. 58:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snoeyenbos, G. H., O. M. Weinack, and C. F. Smyer. 1978. Protecting chicks and poults from salmonellae by oral administration of ‘normal’ gut flora. Avian Dis. 22:273–287. [PubMed] [Google Scholar]
- 41.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. D. Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stackebrandt, E., and B. M. Grobel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence analysis on the present species identification in bacteriology. Int. J. Syst. Bacteriol. 44:846–849. [Google Scholar]
- 43.Stavric, S. 1987. Microbial colonization control of chicken intestine using defined cultures. Food. Technol. 41:93–98. [Google Scholar]
- 44.Stavric, S., and J. Y. D’Aoust. 1993. Undefined and defined bacterial preparations for the competitive exclusion of Salmonella in poultry—a review. J. Food Prot. 56:173–180. [DOI] [PubMed] [Google Scholar]
- 45.Stern, N. J. 1994. Mucosal competitive exclusion to diminish colonization of chickens by Campylobacter jejuni. Poult. Sci. 73:402–407. [DOI] [PubMed] [Google Scholar]
- 46.Stern, N. J., S. Bailey, N. A. Cox, Jr., and L. C. Blankenship. September 1995. Mucosal competitive exclusion flora. U.S. patent 5,451,400.
- 47.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi, M., M. Kametaka, and T. Mitsukota. 1982. Influence of diets low in protein or lysine on the intestinal flora of chicks with reference to cecal content. J. Nutr. Sci. Vitamin 28:501–510. [DOI] [PubMed] [Google Scholar]
- 50.Vander Wauven, C., A. Pierard, M. Raymann-Kley, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Hartingsveldt, J., and A. H. Stouthamer. 1973. Mapping and characterization of mutants of Pseudomonas aeruginosa affected in nitrate respiration in aerobic or anaerobic growth. J. Gen. Microbiol. 74:97–106. [DOI] [PubMed] [Google Scholar]
- 52.Von Wintzingerode, F., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229. [DOI] [PubMed] [Google Scholar]
- 53.Whitehead, T. R., and M. A. Cotta. 1999. Phylogenetic diversity of methanogenic Archaea in swine waste storage pits. FEMS Microbiol. Lett. 179:223–226. [DOI] [PubMed] [Google Scholar]
- 54.Wilmotte, A., G. van der Auwere, and R. De Wachter. 1993. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96–100. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p.2.4.1–2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., Brooklyn, N.Y. [DOI] [PubMed]
- 56.Zoetendal, E., A. Akkermans, and W. deVos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]