Abstract
Histone deacetylases (HDACs) catalyze the removal of acetyl groups from the ɛ-amino group of distinct lysine residues in the amino-terminal tail of core histones. Since the acetylation status of core histones plays a crucial role in fundamental processes in eukaryotic organisms, such as replication and regulation of transcription, recent research has focused on the enzymes responsible for the acetylation/deacetylation of core histones. Very recently, we showed that HdaA, a member of the Saccharomyces cerevisiae HDA1-type histone deacetylases, is a substantial contributor to total HDAC activity in the filamentous fungus Aspergillus nidulans. Now we demonstrate that deletion of the hdaA gene indeed results in the loss of the main activity peak and in a dramatic reduction of total HDAC activity. In contrast to its orthologs in yeast and higher eukaryotes, HdaA has strong intrinsic activity as a protein monomer when expressed as a recombinant protein in a prokaryotic expression system. In vivo, HdaA is involved in the regulation of enzymes which are of vital importance for the cellular antioxidant response in A. nidulans. Consequently, ΔhdaA strains exhibit significantly reduced growth on substrates whose catabolism generates molecules responsible for oxidative stress conditions in the fungus. Our analysis revealed that reduced expression of the fungal catalase CatB is jointly responsible for the significant growth reduction of the hdaA mutant strains.
In eukaryotic organisms, DNA and highly basic nuclear proteins, the histones, constitute the nucleosome, which is the essential structural subunit of the chromatin. The fact that the N-terminal extensions of the core histones contain distinct sites for various posttranslational modifications alters our view of chromatin as being a static entity for the efficient packing of the genomic DNA into the nucleus of the cell. Today it is accepted that, in addition to its structural role, chromatin has an important regulatory function during DNA replication and repair, cell cycle control, cell aging, and transcription (for a review, see reference 59).
Antagonistic enzymes such as kinases/phosphatases (10, 39), histone methyltransferases/demethylases (for a review, see reference 9), and histone acetyltransferases/deacetylases (for a review, see reference 38) are responsible for a dynamic equilibrium of chromatin regulating modifications of the histone tails.
The acetylation of distinct lysine residues of H2A, H2B, H3, and H4 is the most prominent dynamic histone modification allowing or denying the access of numerous regulatory proteins, such as transcription factors, to distinct regions of genomic DNA. Although histone acetylation is by far the best- studied type of histone modification, our understanding of how this modification is linked to processes such as the regulation of transcription is still very limited. However, acetylated histones are a characteristic feature of transcriptionally active chromatin (2), and hypoacetylated histones accumulate within transcriptionally silenced domains (30, 63). Within the last decade a great number of histone acetyltransferases were analyzed and identified in many organisms as coactivators of transcription. The dynamic process of acetylation can be reversed by histone deacetylases (HDACs), and in many cases removal of acetyl groups leads to repression of transcription. Consequently, HDACs were often found as corepressors in large multiprotein complexes. However, a number of experiments during the past years with Saccharomyces cerevisiae and other model organisms provided evidence that deacetylation of histones can contribute to transcriptional activation as well (for a review, see reference 29).
HDACs are classified into three groups, which share no sequence similarities with each other: (i) the “classical” HDACs, which are divided into several subgroups (for a review, see reference 12); (ii) the silent information regulator 2 (SIR2) family (sirtuins), which are NAD+-dependent enzymes that hydrolyze one molecule of NAD+ for each lysine residue that is deacetylated, are phylogenetically conserved, and are required for transcriptional silencing in yeast (for a review, see reference 4); and (iii) the plant-specific HD2-type HDACs, which might have a function in plant-specific pathways or might have taken over functions corresponding to those carried out by other HDACs in nonplant organisms (for a review, see reference 35).
Unfortunately, nomenclature for the classical HDAC family is confusing and not yet uniform. Recent phylogenetic analyses revealed that there are at least three subgroups of classical enzymes: the RPD3 proteins (class 1), termed according to the homology to a transcriptional regulator in Saccharomyces cerevisiae (54); the HDA1 enzymes (class 2), displaying high similarity to yeast HDA1 (47); and a novel subgroup of enzymes (referred to as class 4-type enzymes) that includes, among others, human HDAC11 and HDA2 of Arabidopsis thaliana (21).
Yeasts and filamentous fungi contain members of all classical subgroups, with the exception of class 4 enzymes (for a review, see reference 20); instead, HOS3, an unusual type of enzyme which shows conserved domains primarily with class 2 HDACs but also exhibits some characteristic individual sequence motifs, was identified in yeast (47). Some of these motifs were thought to be responsible for the remarkable biochemical features of HOS3 (7). One of these features is a distinct resistance against HDAC inhibitors, such as trichostatin A.
Recently we have identified members of this HDAC type in the fungal plant pathogen Cochliobolus carbonum and in the filamentous fungi Neurospora crassa, Aspergillus fumigatus, and Aspergillus nidulans (56). We could show that expressed recombinant HosB, the HOS3 homolog in A. nidulans, not only is resistant to trichostatin A but also possesses intrinsic enzyme activity.
However, not HosB but the second class 2 type enzyme in the fungus, HdaA, was assumed to be a main contributor to HDAC activity in Aspergillus. Here we demonstrate that recombinant HdaA shows strong intrinsic HDAC activity and that disruption of the corresponding gene indeed results in the loss of the main HDAC activity of the fungus. Moreover, ΔhdaA strains display a remarkable reduction of growth under different conditions of oxidative stress. In clear contrast to the hdaA mutants, no phenotype could be observed in ΔhosB strains.
MATERIALS AND METHODS
Strains and growth conditions.
Vectors and plasmids generally were propagated in Escherichia coli DH5α cells (Life Technologies). For the production of active recombinant HdaA, E. coli BL21 (Novagen) was used as the host for expression. All Aspergillus strains used in this study were derived from A89 (biA1; argB2) for the deletion of hosB and from A768 (pyrG89 yA2; riboB2; chaA1) for the deletion of hdaA; both strains were provided by the Fungal Genetics Stock Center (Kansas City, KS). Unless otherwise noted, strains were grown at 37°C in 1-liter Erlenmeyer flasks with 250 ml minimal medium (MM) as described previously (44), supplemented as required for 18 to 24 h in shake culture or for 3 to 4 days in stationary culture. The final concentration of spores for the inoculation was 1 × 107/ml.
Disruption of hdaA and hosB and generation of hdaA/hosB deletion mutants.
For cloning, standard molecular techniques were performed as described previously (49). Disruption of hdaA was achieved by targeted gene replacement. Transformation of the A. nidulans strain A768 was performed with a 3.9-kb hdaA fragment in which the HdaA-encoding sequence was replaced by the orotidine-5′-monophosphate decarboxylase-encoding gene (pyrG) of Aspergillus fumigatus as a heterologous marker in an A. nidulans pyrG uridine/uracil auxotroph. In a first step a 5.3-kb DNA fragment including the coding region of hdaA and about 900 bp of the 5′ and 1,200 bp of the 3′ noncoding regions was amplified by PCR from cosmid W27G12 of a chromosome-specific library of A. nidulans constructed in pWE15 (13) and pLORIST2 (18), respectively. Primers used were hdaAKofor (5′-GGCGCATCGATCCTTGGTCTATTAAGTTGCC) and hdaAKorev2 (5′-TATAGCGGCCGCGACAGATCGTACCACACC) with add-on restriction enzyme cleavage sites for ClaI and NotI (underlined). After digestion, the amplification product was inserted into the identically cut plasmid pBluescript-KS (Stratagene). Subsequently, the coding sequence of hdaA was excised with MluI and SpeI and was replaced by the pyrG gene of A. fumigatus from the plasmid pAfpyrG2 (61). For transformation of the fungus, the hdaA 5′ and 3′ noncoding regions encompassing the pyrG coding sequence were excised from the vector with ClaI and NotI, respectively, and gel purified. Transformation of A. nidulans A768 was carried out with 5 to 10 μg of the purified fragment as described previously (55). Protoplasts growing under selective conditions were further screened for hdaA deletion by a PCR approach. In order to obtain transformants with a homologous integration of the deletion fragment, colonies from homokaryotic spores were picked, and (single) genomic integration was confirmed by Southern blot analysis as described earlier (19).
hosB null mutants were generated using a 5.4-kb hosB fragment in which the HosB-encoding sequence was replaced by the ornithine carbamoyltransferase-encoding gene (argB) from A. nidulans (3) as an auxotrophic marker in the argB auxotrophic strain A89. In a first step the HosB-coding sequence and about 1,000 bp of each of the 5′ and 3′ noncoding regions were amplified from cosmid W25F06 as described above, using primers hosBKofor (5′-TTATTAGCGGCCGCGAGTCGACTTGAGAATC) and hosBKorev (5′-TTATTACCATGGGCCACGGGTATATCAGC) with add-on restriction enzyme cleavage sites for NotI and NcoI (underlined). The PCR product was cloned into a pGEM-5Zf(+) vector (Promega), and the hosB coding sequence was excised with EcoRI and SpeI and was replaced with the argB sequence. The transformation of strain A89 was performed after elimination of the GEM-vector with ApaI and NotI. Five to 10 micrograms of the gel-purified fragment was used for the transformation procedure. Screening of transformants was done as described above.
To generate hosB/hdaA deletion mutants, the obtained ΔhosB strains were crossed with ΔhdaA strains on agar plates containing MM without biotin and riboflavin as described previously (25). From these crosses, hdaA/hosB deletion strains were identified by a PCR approach and were confirmed by Southern blot analysis as described above.
Expression, purification, and analysis of recombinant HdaA in Escherichia coli.
For recombinant expression of full-length HdaA and the C-terminally truncated HdaA fragments in E. coli, the corresponding coding sequences were cloned in frame into a pGEX-6P-1 expression vector (Amersham Biosciences). The peptides were expressed in BL21 cells in LB medium. Two-hundred-milliliter cultures with an OD600 of 0.4 were induced with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown for 4 h at 37°C. After centrifugation of cells at 4,000 × g, the pellet was resuspended in 10 ml of glutathione S-transferase (GST) binding buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). Cells were passed through a French press (pressure setting, 1,000 lb/in2), and the resulting lysate was centrifuged at 25,000 × g for 15 min at 4°C. Aliquots of the soluble extracts were analyzed for expression products by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In a final step, the GST fusion proteins were purified from the supernatant in a batch approach using glutathione-Sepharose 4 Fast Flow beads (Amersham Biosciences), and the eluate was measured for HDAC activity.
To determine the apparent molecular weight of active recombinant HdaA, gel filtration chromatography was performed using a TSK G4000PWXL (7.8- by 300- mm) column (Tosoh Bioscience) calibrated with protein standards of the following molecular masses: 2,000 kDa (blue dextran 2000), 440 kDa (ferritin), 232 kDa (catalase), 67 kDa (bovine serum albumin), and 25 kDa (chymotrypsinogen A). Before the loading of the TSK column, the GST tag of the recombinant HdaA was cleaved for 3 h at 4°C on a GSTrap, using PreScission protease (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Recombinant cleaved HdaA was eluted with PreScission protease buffer, while the GST moiety of the fusion protein and the PreScission protease remained bound to GSTrap. Eluted cleaved HdaA was analyzed by SDS-PAGE, concentrated 15-fold with a Centriprep YM-10 centrifugal filter device (Millipore), and loaded onto the TSK column. The flow rate of the column was 0.3 ml/min at 4°C, and the absorbance was monitored at 280 nm. Finally, the HDAC activity present in individual fractions of the column was determined.
SDS-polyacrylamide gel electrophoresis and immunoblotting.
HDACs or recombinant HDACs were analyzed by SDS-PAGE (10% precast Tris-glycine polyacrylamide gels and NuPage bis-Tris gels [Invitrogen], respectively). When required, gels were blotted onto Hybond ECL nitrocellulose membranes (Amersham Pharmacia Biotech) at 25 V for 2 h and membranes were blocked with 5% (wt/vol) skim milk in Tris-buffered saline (20 mM at pH 7.4) for 2 h. Membranes were incubated with 1:1,000-diluted affinity-purified antibodies against fungal HDACs (56) in 2% skim milk in Tris-buffered saline at 4°C overnight. After washing, membranes were incubated for 2 h with alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (1:10,000; Sigma), and immunodetection was performed using the enhanced chemiluminescence detection system (ECL; Amersham Pharmacia Biotech) or the naphthol As-Mx Phosphate/Fast Red TR detection system (Sigma) according to the manufacturer's instructions.
Immunoprecipitation.
For immunoprecipitation experiments, 50 μl of partially purified HDAC fractions of A. nidulans (SourceQ chromatography) was mixed with 1.6 μl of affinity-purified HDAC antibodies (12.5 μg/μl) and 30 μl of protein A-Sepharose, equilibrated with buffer B, and incubated for 2 h with shaking at 4°C. To avoid unspecific binding, the mixture was adjusted to 300 mM NaCl. After centrifugation for 2 min at 500 × g, the supernatant was saved for HDAC assay and protein blotting. Pellets were washed three times with 500 μl of buffer B (15 mM Tris-HCl, pH 8.0, 0.25 mM EDTA, 1 mM 2-mercaptoethanol, 10% [vol/vol] glycerol)-300 mM NaCl and finally resuspended in 20 μl of buffer B. Entire pellets and 50 μl of supernatants were used for the standard HDAC assay. For immunoblotting, precipitates were mixed with SDS sample buffer, boiled, and centrifuged for 5 min at 10,000 × g, and the resulting supernatant was used for SDS-PAGE.
Analysis of oxidatively modified proteins and fluorescence detection of ROS within hyphae.
To analyze reactive oxygen species (ROS)-induced protein carbonylation, 50 mg of lyophilized mycelium was extracted in 600 μl of 50 mM Na-phosphate buffer, pH 7.0, in an MM 300 Retsch mixer mill (QIAGEN). After removal of insoluble cell debris by centrifugation (20,000 × g, 10 min, 4°C), protein carbonyl groups in the soluble cell fractions were derivatized with 2,4-dinitrophenylhydrazine (Sigma) according to a protocol described previously (32). Samples (15 μg) were separated by SDS-PAGE, and proteins were immunodetected by an antidinitrophenylhydrazine antibody (1:1,000; Dako) as described previously (5).
To analyze ROS within hyphae, 1.5 × 106 spores were inoculated in petri dishes containing 15 ml complete medium with coverslips on the bottom of the dishes, and mycelia were grown on these slips overnight without shaking at 30°C. Subsequently, 5 μM dihydroethidium (DHE) (Invitrogen) or 10 μM 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) (Invitrogen) was added to the medium and left for 45 min. For positive controls, 5 μM of the fungal antibiotic nystatin (Sigma) was additionally added to the medium 20 min prior to washing the hyphae. Subsequently, coverslips with attached mycelia were washed two times with phosphate-buffered saline (pH 7.4), and 10 μl Vectashield mounting medium for fluorescence (Vector Laboratories, Inc.) was pipetted onto the hyphae. Mycelia were viewed with a fluorescence microscope (Zeiss).
Partial purification of histone deacetylase activity and histone deacetylase assay.
Mycelia of Aspergillus were collected, washed in a sintered glass filter, thoroughly dried with filter paper, and immediately frozen in liquid nitrogen for subsequent lyophilization. Five grams of lyophilized mycelium was ground to powder in an IKA grinding machine, and the powder was suspended in 40 ml of buffer B containing one protease inhibitor tablet (Complete; Roche). The mixture was stirred on ice for 10 min and then centrifuged for 15 min at 32,000 × g at 4°C. The supernatant was used for SourceQ anion-exchange chromatography.
The protein extracts were transferred to a fast protein liquid chromatography column (1.6 by 15 cm) with 13 ml of Source 15Q anion-exchange medium (Amersham Pharmacia Biotech) equilibrated with buffer B. Elution of proteins was performed with 50 ml of a linear gradient of 10 mM to 500 mM NaCl in buffer B at a flow rate of 1 ml/min. Fractions of 1.5 ml were collected and assayed for HDAC activity.
HDAC activity was determined as described previously (52), using [3H]acetate-prelabeled chicken reticulocyte histones as the substrate. Sample aliquots of 50 μl of the chromatography fractions were mixed with 10 μl of total [3H]acetate-prelabeled chicken reticulocyte histones (1 mg/ml), resulting in a concentration of 11 μM. After incubation for 1 h at 25°C, the reaction was stopped by addition of 50 μl of 1 M HCl-0.4 M acetylacetate and 0.8 ml ethyl acetate. After centrifugation at 10,000 × g for 5 min, an aliquot of 600 μl of the upper phase was counted for radioactivity in 3 ml liquid scintillation cocktail.
Determination of CatB activity.
For the determination of catalase activity, native polyacrylamide gels were loaded with protein extracts (25 μg) of strains grown in MM for 18 h at 37°C in shake culture and for an additional 3 h with or without hydrogen peroxide (HP). CatB activity was determined by staining as described previously (8). As a loading control, aliquots of the protein extract were loaded onto a SDS-polyacrylamide gel and stained with Coomassie blue.
Northern blot analysis.
Transcription levels of HDACs in the deletion strains were analyzed by Northern hybridization. RNA preparation, blotting, and hybridization were done as described earlier (19). For studies of expression of detoxifying enzymes of the cellular antioxidant response under oxidative stress conditions, the strains were grown in MM for 18 h at 37°C in shake culture and for an additional 3 h with various concentrations of menadione or H2O2. Transcripts on the autoradiograms were quantified with a Personal Densitometer SI (Amersham Biosciences) equipped with Image-Quant software. Results were corrected for loading and blotting of mRNA by using the γ-actin gene of A. nidulans (14) as an internal control (data not shown). Reliability of the obtained densitometric results was confirmed by measurements of different amounts of RNA.
RESULTS
Disruption of class 2 HDAC genes in A. nidulans.
Disruption of hosB, hdaA, and hosB/hdaA was done as described in Materials and Methods. In order to confirm putative deletion mutants and to determine the number of additional randomly integrated deletion constructs, Southern blot analyses were performed (data not shown). In the case of the hdaA transformants, 3 of 26 independent transformants (H4, H6, and H14) exhibited the expected pattern of DNA hybridization for a single gene replacement. In two of the ΔhdaA strains, additional fragments were integrated. Three strains (H1, H7, and H8) carried a randomly integrated selection marker without a deletion of hdaA.
Southern analysis of hosB transformants revealed two ΔhosB strains (B6 and B14) with a single integrated marker (data not shown). Seven strains carried one or more randomly integrated fragments without an hosB deletion.
Crosses of ΔhdaA with ΔhosB strains resulted in eight independent hdaA/hosB double mutants. Two of these strains (HB20 and HB44) were further analyzed.
In all analyses, prototrophic strains with a single randomly integrated selection marker without deletion of HDAC genes served as controls and are characterized as wild type (wt). Finally, all deletion strains were checked for a loss of expression of the corresponding transcripts by Northern blot analysis. In contrast to the controls, no transcripts could be detected in the deletion strains (data not shown).
In order to find evidence for a possible compensation of lost HDAC activities in the deletion strains by other HDACs, Northern blot analyses were performed with mRNAs of the deletion mutants, using probes against all classical HDACs. However, no change in the transcription rates could be observed in the deletion mutants compared with the wt strains (data not shown).
HdaA is responsible for the main HDAC activity in A. nidulans.
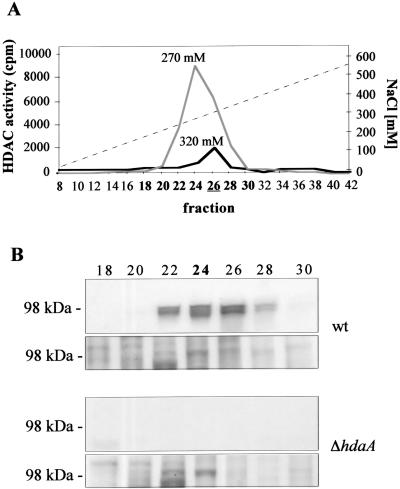
To determine to what extent the HDAC activity of the fungus was affected by the deletion of hosB or hdaA, protein extracts of the corresponding null mutants and controls were subjected to SourceQ anion-exchange chromatography. Identical elution profiles of HDAC activity were observed in the controls and ΔhosB strains. In both cases, the main HDAC activity eluted at a salt concentration of 270 mM as described previously (56). In contrast, the activity peak of the hdaA deletion strains (H4, H6, and H14) was significantly reduced and eluted at 320 mM (Fig. 1A). Western blot analysis with an anti-HdaA antibody revealed that the loss of the core activity was accompanied by a loss of the HdaA-specific signal at 98 kDa in fractions 22 to 28 (Fig. 1B). The intensity of the HdaA signal in the controls exactly corresponded to the main HDAC activity (peak fraction 24), confirming that HdaA accounts for the predominant HDAC activity of the fungus. All data shown in Fig. 1 are derived from H4 as the ΔhdaA strain and H1 as the wt strain. However, the same results were achieved with two other independent deletion strains and controls (data not shown).
FIG. 1.
HDAC activities of A. nidulans wt and hdaA deletion strains. (A) Elution profile of HDAC activity after Source 15Q anion-exchange chromatography of protein extracts of A. nidulans wt (gray line) and ΔhdaA (black line) strains. Elution was performed with 50 ml of a linear gradient (dashed line) of 10 to 500 mM NaCl in buffer B at a flow rate of 1 ml/min. Fractions of 1.5 ml were collected and assayed for HDAC activity. Fraction 26 of the ΔhdaA strain, used for subsequent immunoprecipitation with an RpdA antibody, is underlined. Fractions used for immunoblotting are shown in boldface. (B) Immunological detection of HdaA in Source Q fractions 18 to 30. Aliquots of each fraction were subjected to SDS-PAGE with subsequent blotting onto nitrocellulose membranes. A specific antibody against HdaA was used for immunodetection. The fraction with the main HDAC activity is shown in boldface. All data shown in this figure are derived from H4 as the ΔhdaA strain and H1 as a control representing the wt. As a loading control, aliquots used for immunodetection were stained with Coomassie blue (lower panel).
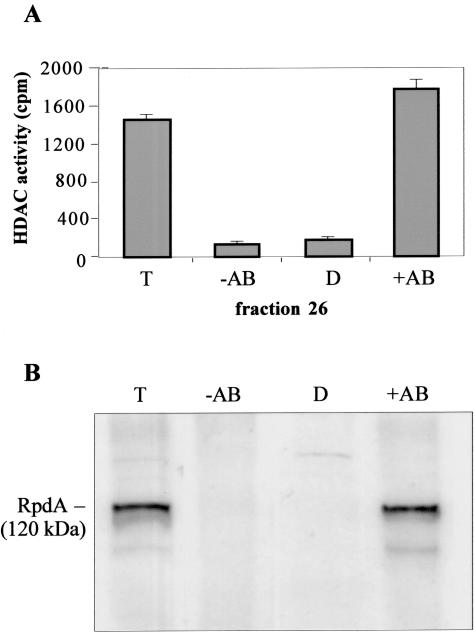
In order to substantiate an earlier assumption that the remaining HDAC activity in the ΔhdaA strain comes exclusively from the class 1 enzyme RpdA (56) and to rule out the possibility that activities of other enzymes (HosA or HosB) contribute to the activity peak in the ΔhdaA strains, immunoprecipitation experiments were performed. In the control strains, the HDAC activity of fraction 24 was quantitatively precipitated with the anti-HdaA antibody, whereas the activity of fraction 26 could be only partially depleted. However, subsequent immunoprecipitation by an RpdA-specific antibody raised against the nonconserved C-terminal tail of this enzyme resulted in a complete loss of activity in the supernatant of the wt strain (data not shown). In contrast, in ΔhdaA strains the activity of fraction 26 was quantitatively precipitated by anti-RpdA (Fig. 2A); only background activity could be detected in the supernatant after precipitation. Subsequent immunoblot analysis revealed a clear RpdA signal at the expected size of 120 kDa in the pellet (Fig. 2B).
FIG. 2.
Immunoprecipitation of RpdA in hdaA deletion strains. Aliquots (50 μl) of fraction 26 of the Source 15Q anion-exchange chromatography were incubated with or without 1.6 μl (20 μg) of immunopurified anti-RpdA antibody and with 30 μl of protein A-Sepharose; incubation was performed for 2 h with continuous shaking at 4°C in buffer B. After centrifugation, the supernatant was directly used for the HDAC assay and immunoblotting. Pellets were washed three times and finally were resuspended in 20 μl of buffer B. (A) Fifty microliters of the depleted supernatant (D), the resuspended pellets obtained with (+AB) or without (−AB) antibodies, and 50 μl of fraction 26 (total input, T) were assayed for HDAC activity. HDAC activity data represent results of three independent experiments performed with three independent hdaA mutant strains. Error bars indicate standard deviations of three independent experiments. (B) Ten microliters of the supernatant, 10 μl of the resuspended pellets, and 10 μl of fraction 26 were used for Western blot analysis. Immunoblotting was carried out using the anti-RpdA antibody. The molecular mass of RpdA is indicated.
These results clearly demonstrate that under the growth conditions used, HosA and HosB obviously do not account for detectable HDAC activity in the fungus.
HdaA has intrinsic activity as a monomer when expressed in E. coli.
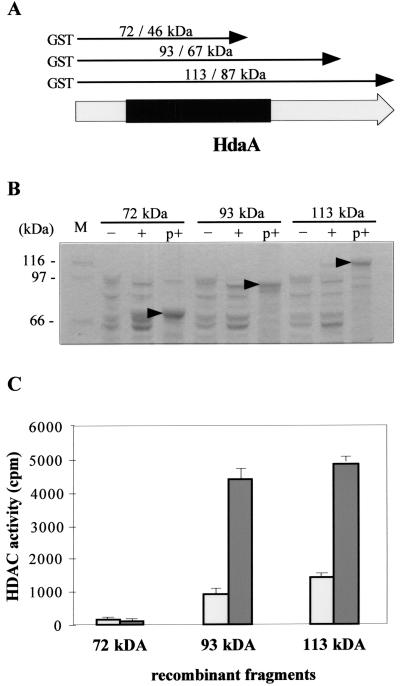
With only a few exceptions (7, 53), purified recombinant HDACs did not exhibit intrinsic enzymatic activity. In order to address the question for HdaA, the full-length enzyme and two C-terminally truncated fragments were expressed as GST fusion proteins in E. coli BL21 (Fig. 3A and B) and affinity purified with glutathione-Sepharose. Interestingly, significant enzyme activity was detected in the full-length enzyme and the 93-kDa fragment but not in the truncated 72-kDa fragment lacking a part of the conserved N terminus typical for class 2 HDACs (Fig. 3C). Weak activities above the background in noninduced control strains indicate a leakage of the tac promoter used in the pGEX expression vector (Fig. 3C).
FIG. 3.
Recombinant expression of HdaA and C-terminally truncated fragments and determination of their HDAC activity. (A) Schematic representation of HdaA. The conserved part of the enzyme comprising motifs essential for HDAC activity is indicated in black. Arrows illustrate the recombinant fragments produced in E. coli. The predicted molecular masses with and without the fused GST tag, respectively, are given. (B) SDS-PAGE of (purified) recombinant HdaA. Fifteen microliters of induced (+) or noninduced (−) E. coli extracts or aliquots of the purified GST fusion proteins (p+) were subjected to SDS-PAGE and stained with Coomassie blue. Molecular masses of marker proteins are indicated; arrows mark the purified recombinant products. Aliquots of the purified fractions were used for the HDAC assay. (C) HDAC activity of purified recombinant HdaA fragments. Fifteen microliters of purified HDAC fragments was used for the HDAC assay (dark bars). As controls, extracts from noninduced cultures were subjected to the same procedures as described for panel B and tested for HDAC activity (light bars). HDAC activity data represent results of three independent experiments. Error bars indicate standard deviations.
To determine whether HdaA is active as a monomer, gel filtration chromatography was performed with the recombinant full-length HdaA (data not shown). Prior to loading, the GST tag was cleaved with PreScission protease as described in Materials and Methods, and cleavage was analyzed by SDS-PAGE with subsequent Coomassie blue staining. Cleaved recombinant HdaA was then concentrated and loaded onto a TSK G4000 column. The activity peak eluted at an apparent molecular mass of 80 to 110 kDa (data not shown), which is in accordance with the predicted molecular weight of 98,000. Therefore, we conclude that recombinant HdaA is active as a monomer.
Lack of HdaA results in a remarkable reduction of growth under conditions of oxidative stress.
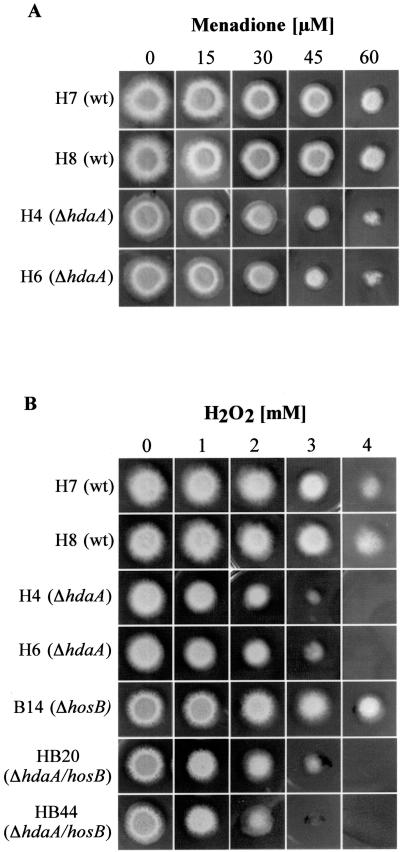
In order to analyze possible physiological consequences caused by the loss of the HDAC genes, deletion strains and corresponding controls were grown on media with different carbon sources (glucose, xylose, or xylan), different nitrogen sources (ammonium tartrate or sodium nitrate), or different amino acids (proline or glutamine) as nitrogen and carbon sources. Moreover, strains were grown in shake culture or stationary culture with or without light at a temperature of 25°C, 37°C, or 42°C. Subsequently strains were compared with respect to germination of spores, extension and branching of the filaments, conidiation, or formation of cleistothecia. Whereas no significant differences between controls and mutant strains were found for most of the substrates used, addition of paraquat (PQ) (data not shown) or menadione (MD) (Fig. 4A) led to a distinct inhibition of growth in the HdaA null mutants. Substances such as PQ and MD are known as drugs generating free radicals such as superoxide anions, hydrogen peroxide (HP), and other reactive oxygen species (ROS) and therefore induce oxidative stress.
FIG. 4.
Growth phenotypes of HDAC deletion strains under conditions of oxidative stress. Independent A. nidulans ΔhdaA strains (H4 and H6) were tested for their sensitivity against menadione (A). Moreover, these strains, one of the ΔhosB strains (B14), and two ΔhosB/hdaA double mutants (HB20 and HB44) were tested for their sensitivity against hydrogen peroxide (B). As controls, wt strains H7 and H8 with different random integrations of the pyrG marker were used. (A) Sensitivity of ΔhdaA strains to increasing concentration of MD. Conidiospores (104) of the corresponding strains were point inoculated on minimal medium plates containing various concentrations of MD and were grown at 37°C for 72 h. (B) Sensitivity of ΔhdaA, ΔhosB, and ΔhdaA/ΔhosB strains to increasing concentrations of HP. Conidiospores of strains were incubated as described above at 37°C for 18 h. Subsequently, mycelia were overlaid with top agar (0.25%) containing increasing concentrations of HP and were incubated for another additional 30 h at 37°C.
CatB activity is significantly decreased in hdaA null mutants under conditions of oxidative stress.
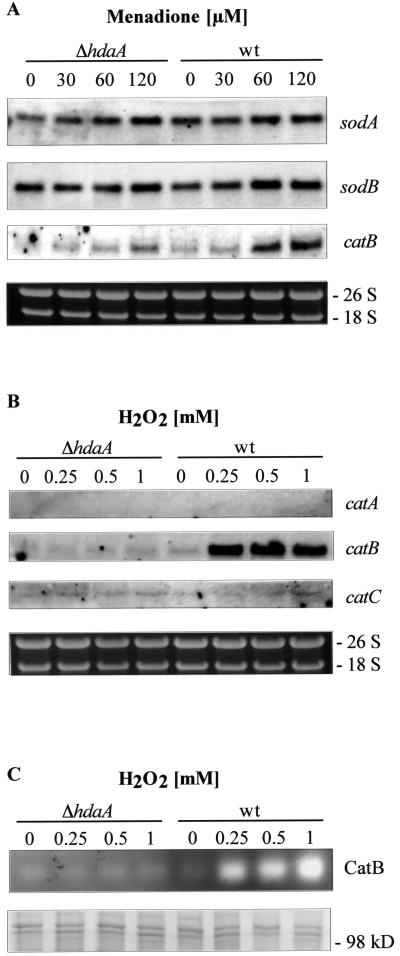
Changes in the level of intracellular free radicals in general affect defense enzymes such as superoxide dismutases and catalases. To examine whether the reduction of growth of the ΔhdaA strains under stress-inducing conditions is due to a diminished availability of one or more of these defense factors, we analyzed the transcriptional responses of known superoxide dismutases (40, 41) and catalases (26) of the fungus with different concentrations of MD. Whereas no significant effects on the transcription of most of the detoxifying enzymes were observed, a considerable accumulation of catB transcripts was found with increasing concentrations of MD in the controls but not in the hdaA null mutant (Fig. 5A). Since CatB is known to be a typical catalase, which detoxifies HP in Aspergillus (27), transcriptional analysis of catA, catB, and catC was performed with RNAs of strains growing with different concentrations of HP. As shown in Fig. 5B, 0.25 mM HP led to a strong upregulation of catB transcription in the controls. However, in our hdaA deletion mutant strain even a concentration of 1 mM HP did not significantly increase the catB transcription rate, confirming the assumption that HdaA is involved in the expression of CatB under conditions of oxidative stress. In order to confirm these data, CatB activity in protein extracts of the hdaA deletion strains and controls was determined under the same growth conditions as employed for the Northern blot analysis (Fig. 5C). The zymogram analysis clearly shows that catalase activity increases with the HP concentration in the control strains. As expected, no increase of CatB activity was detectable in the ΔhdaA strain.
FIG. 5.
Regulation of detoxifying enzymes of the cellular antioxidant response in hdaA deletion strains. H4 as a ΔhdaA strain and H1 as a control representing the wt were used. (A) Transcription rates of the coding sequences of superoxide dismutases A and B and catalase B at various concentrations of menadione. Northern analyses were performed with 10 μg of total RNA isolated from strains grown for 18 h in minimal medium and for a further 3 h in the presence of the corresponding MD concentration. Transcripts were detected with digoxigenin-labeled probes specific for the respective sequences. (B) Transcription rates of three catalase genes (catA, catB, and catC) in the presence of different concentrations of H2O2. Strains were grown for 18 h in minimal medium and for a further 3 h in the presence of the corresponding H2O2 concentration. Ethidium bromide-stained rRNA was used as a quality and loading control in panels A and B. (C) CatB activity in protein extracts from H4 (ΔhdaA) and H1 (wt) growing under conditions as described for panel B. Protein extracts were loaded onto a native polyacrylamide gel, and CatB activities were determined by staining as described in Materials and Methods. As a loading control, aliquots used for the zymogram analysis were loaded onto an SDS-polyacrylamide gel and stained with Coomassie blue (lower panel).
Since inhibition of catB transcription actually was more pronounced with HP than with MD, we checked possible consequences of increased HP concentrations on growth of the hdaA mutants. In fact, 1 mM HP already led to a significant reduction of growth, while 3.5 mM was definitely lethal for the hdaA null mutant (Fig. 4B). This contrasts to the case for the wt strains (H7 and H8), both of which were able to survive with concentrations of up to 4 to 5 mM HP. To test if the sensitivity against HP is further increased in hdaA mutant strains with an additional deletion of hosB, hdaA/hosB double mutants were generated, grown under the HP conditions described above, and compared with hdaA and hosB single mutants. As shown in Fig. 4B, no further increase in HP sensitivity could be detected in the double mutants; the resistance of hosB deletion strains did not differ significantly from that of the controls. These data clearly demonstrate that HosB neither is involved in the antioxidant defense of A. nidulans nor is able to compensate the sensitivity against oxidative stress in the hdaA null mutants.
hdaA deletion mutants show symptoms of oxidative stress even under “nonstress” conditions.
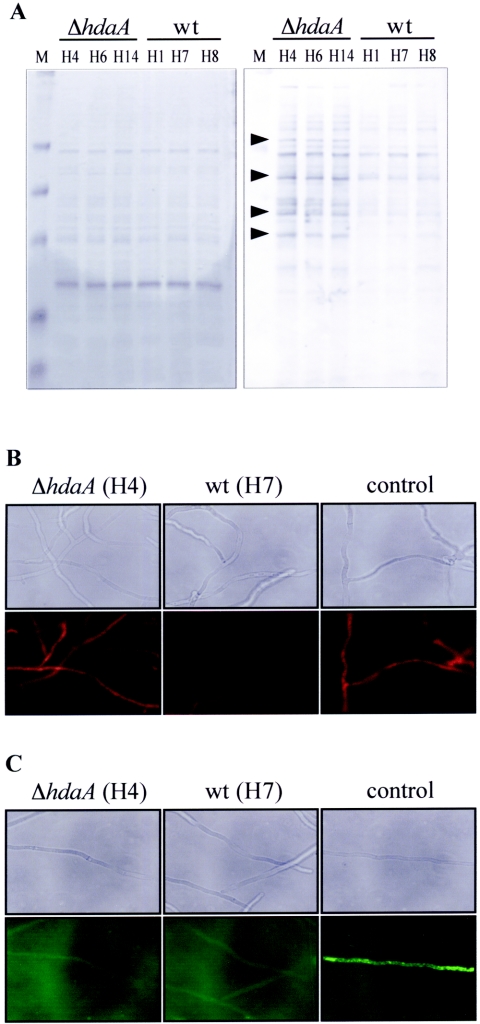
Since our mutant strains obviously exhibit a lower stress tolerance in the presence of drugs generating free radicals, we were interested in whether intracellular ROS are accumulated within the cells under growth conditions which do not yet show any phenotype. To address this question, ΔhdaA strains and controls were grown in MM without ROS-generating drugs, and subsequently proteins of the cell extracts were analyzed for their carbonylation status. Oxidative stress in general leads to damaged molecules and a high percentage of carbonylated proteins within cells (32). As shown in Fig. 6A, a significant increase in protein carbonylation was evident in the hdaA mutants compared to the wt or the ΔhosB strains (data not shown), indicating that hdaA null mutants obviously suffer from oxidative stress even under “nonstress” conditions. To further confirm this indication, mycelia of ΔhdaA strains and controls were incubated with DHE, a redox-sensitive probe which has been widely used to detect intracellular ROS (28, 36). Interestingly, the fluorescence intensity of hyphae of the ΔhdaA strains indeed was increased compared to that of the wt (Fig. 6B). This result together with the data obtained from the carbonylation assay strongly suggests an increased oxidative stress of the hdaA mutants even when no ROS-generating drugs are present in the medium. In order to check if these stress symptoms are due to an already-affected CatB, we performed another fluorescence-based technique using H2DCFDA as fluorescent dye. H2DCFDA is specific for the detection of HP and secondary and tertiary peroxides. Since HP is the major peroxide within cells, H2DCFDA assays in whole cells primarily measure this molecule (46). As shown in Fig. 6C, no significant differences could be observed between ΔhdaA strains and the wt. These data and the similar low expression rates of CatB in both the mutants and the wt (Fig. 5C) suggest that under “nonstress” conditions, ROS other than HP or HP-derived oxidants are accumulated in the ΔhdaA cells.
FIG. 6.
Determination of oxidative stress symptoms in strains grown under “nonstress” condition. (A) Pattern of oxidatively damaged proteins. Different ΔhdaA strains and controls were grown in shake culture for 18 h in minimal medium, and carbonyl groups of soluble protein extracts were derivatized with 2,4-dinitrophenylhydrazine. Subsequently, proteins were separated by SDS-PAGE and either stained with Coomassie blue (left panel) or blotted onto a Hybond nitrocellulose membrane. Immunodetection was performed with an antidinitrophenylhydrazine antibody (right panel). HDAC deletion strains and the corresponding wt controls are indicated; arrowheads mark typical examples of proteins with oxidative modifications in ΔhdaA strains. (B and C) ROS within hyphae of a ΔhdaA strain (H4) and a wt strain (H7) were detected by fluorescence-based techniques. DHE is oxidized to ethidium by ROS-like superoxide anions, which was visualized in hyphae as red fluorescence using fluorescence mi- croscopy (B). Peroxides were detected through the use of the nonfluorescent dye H2DCFDA, which is hydrolyzed within the cells to 2,7-dichlorodihydrofluorescein. In hyphae, 2,7-dichlorodihydrofluorescein interacts with peroxides to form 2,7-dichlorofluorescein, which was visualized as green fluorescence in the fluorescence microscope (C). As a positive control, strains were grown in medium containing 5 μM of the fungal antibiotic nystatin, as described in Materials and Methods.
DISCUSSION
During the last decade it has become clear that histone modifications constitute an important part of a cell's epigenetic toolbox. Therefore, much effort has been made to identify and characterize enzymes involved in this process, and consequently, the number of identified HDACs has increased during the last years. In human cells not fewer than 11 different classical HDACs were found, which can be categorized into several classes (21). The situation has become even more complex because proteins with high sequence similarities to HDACs are also present in archaea and bacteria, strongly suggesting that in eukaryotes the targets of HDACs are not confined to core histones.
As a model organism for lower eukaryotes, the filamentous fungus A. nidulans has only four classical HDAC genes: one gene for each of the class 1 enzymes RpdA and HosA (19) and one gene each for HdaA and HosB, both of which are class 2 HDACs (20). However, in A. nidulans enzymatic activity was demonstrated only for RpdA and HdaA, both of which act in large enzymatic protein complexes (56).
Here we demonstrate that deletion of hdaA leads to a significant decrease of HDAC activity in the fungus and that the residual activity indeed can be assigned exclusively to RpdA. No further global HDAC activity was detectable in the ΔhdaA strains.
These results are in contrast to earlier data, where deletion of yeast HDA1 led to a strong reduction of the major enzyme peak but ΔHDA1/RPD3 double mutant cells still possessed residual deacetylase activity (47). It was discussed that the remaining activity may be due to one of three putative HOS HDACs in yeast. Although two of these enzymes (HosA and HosB) are also present in Aspergillus, no further activity could be detected in partially purified protein extracts of this fungus. For the lack of HOS-related HDAC activity in Aspergillus, two explanations are possible. (i) Under the growth conditions used, these enzymes are expressed at very low levels and therefore are below the limit of detection of our HDAC activity assay. In the case of HosB, the absence of an apparent phenotype of the corresponding deletion mutant and a very low hosB transcription in wt strains (56) would argue for this possibility. (ii) HOS proteins have other, still unknown targets rather than histones and therefore cannot be detected with the HDAC assay performed, where in vivo-labeled core histones are used as the substrate. These targets could be similar to those of the HDAC-related enzymes in prokaryotic organisms. Indeed, it was shown recently that members of another HDAC family, the SIR2-related enzymes, have additional roles beyond the deacetylation of histones and mediate diverse other biological functions in eukaryotic and prokaryotic cells (4, 37). This could be also true for (one of the) HOS proteins. In this case, they would be out of the question to act as a kind of safeguard enzymes when other HDACs are inhibited or deleted. Since a yeast ΔHDA1/RPD1 double mutant does not increase its acetylation state more than either single mutant does, a compensation of the deleted activity by HOS enzymes was hypothesized earlier (47). Similar conclusions were considered for the inhibitor-resistant HDACs of the HOS3 group in A. nidulans and Cochliobolus carbonum (1, 56). However, in this study we neither observed increased transcription of one of the hos genes (hosA or hosB) nor detected an enhanced or novel activity peak in the ΔhdaA strain.
Recently, HOS3 from S. cerevisiae and HosB from Aspergillus were found to exhibit enzyme activity without complex partners when produced as recombinant proteins (7, 56). Relatively little is known about specific conditions required for enzymatic activity of HdaA. Recent observations, however, provide evidence that HdaA acts within a high-molecular-weight complex (molecular weight of about 450,000) in vivo (56), and other eukaryotic class 2 HDACs were described to be active only in the presence of specific complex partners (15). Therefore, they do not display activity as recombinant proteins and are strictly dependent on their ability to interact with certain complex partners to exhibit HDAC activity. Based on this and other observations (22, 34, 57, 58, 62), it was assumed that HDAC4 by itself is not a functional enzyme but serves to recruit preexisting MRT/N-CoR complexes with the class 1 enzyme HDAC3 as the enzymatically active protein.
Our investigation demonstrates that Aspergillus HdaA is active as a recombinant monomer. Moreover, truncated N-terminal fragments of the enzyme are enzymatically active as long as the conserved part of the enzyme remains intact. This intrinsic activity together with earlier results for active recombinant HosB indicates that complex partners are not necessarily required for the enzymatic function of class 2 HDACs in A. nidulans. Recently, a kinetic study on six human HDACs yielded similar results. The data suggest that protein cofactors in HDAC corepressor complexes are not in general required for an enzymatic function of most of the enzymes but may be important for advanced stability of the activity (51). However, it is undebatable that cofactors play an essential role in the targeting of HDACs within the cell.
Identification of the target genes and biological function of HDACs in various cellular pathways was the object of numerous studies (24, 48, 53, 60) as well as an important aim of this work. Our investigations revealed that oxidative stress-inducing substances such as MD or PQ led to a remarkable reduction of growth in HdaA mutant strains, and relatively low concentrations of HP turned out to be lethal. Oxidative stress is always the result of an imbalance between pro-oxidant species and the levels of antioxidant defenses, resulting from the generation of ROS. In addition to nonenzymatic protective molecules such as glutathione, thioredoxin, vitamins, or uric acids, activities of detoxifying enzymes are involved in the antioxidant defenses of biological systems. In contrast to the case for bacteria (11, 17) and yeasts (16, 23, 31), few data are available on the antioxidative defense system of filamentous fungi. However, several detoxifying enzymes were identified in A. nidulans, including the superoxide dismutases SodA and SodB (40, 41), three catalases (26), and a putative catalase-peroxidase, CpeA (50). Expression analysis revealed that one of these enzymes, CatB, is upregulated with increasing concentrations of ROS in wild-type but not in ΔhdaA strains. Since CatB is known to be particularly responsible for the detoxification of HP in hyphae and during conidiation (27), we hypothesize that the failure to upregulate CatB expression is a major reason for the sensitivity of the ΔhadΑ strains against ROS. This assumption is substantiated by the fact that growing colonies from ΔcatB strains showed an increased sensitivity against HP and drugs facilitating intracellular HP formation (27). Since histone acetylation is generally linked to induction of the corresponding target genes, a direct action of HdaA on the catB locus seems to be unlikely. We suppose, rather, that transcription of catB is indirectly regulated by HdaA, but only limited data about specific regulators (and in particular about repressors) of fungal catalases are yet available. Moreover, it is important to note that biochemical symptoms of oxidative stress were observed in the hdaA mutants even under “nonstress” conditions, where CatB expression is very similar to that of the wt strains. These results strongly suggest that in addition to catB, other, still-unknown targets of HdaA that confer protection against oxidative stress might exist. Genome-wide microarray deacetylation maps of S. cerevisiae confirm this suggestion; it was demonstrated that HDA1, the HdaA homolog in yeast, in general has a preference for genes involved in detoxification and stress response of cells (45). This is also an interesting aspect in filamentous fungi, since oxidative stress has been implicated in playing a distinct role in the pathogenicity of Aspergillus fumigatus in immunocompromised patients. Upon infection, the pathogen is attacked by ROS produced by alveolar phagocytic cells of the host, which contribute to elimination of the fungus in the respiratory tract (6, 33, 43). The detoxification of ROS was proposed as one way to overcome this host response, and indeed, a recent study revealed that a CAT1/CAT2 double mutant of A. fumigatus shows a significantly slower development in the lungs of immunosuppressed rats (42). On the other hand, strong evidence also was found that in addition to catalases, other, still-unknown mechanisms protect A. fumigatus against ROS (42). Virulence analyses of Aspergillus hdaA deletion strains in animal models will help to determine whether a recently identified HdaA homolog of A. fumigatus (56) is involved in the regulation of these unknown protection mechanisms.
Acknowledgments
This work was supported by grants from the Austrian Science Foundation to S.G. (P15439) and from the Austrian National Bank to P.L. (P-9000).
We thank Karin Mair for excellent technical assistance and Alexandra Lusser for critical reading of the manuscript.
REFERENCES
- 1.Baidyaroy, D., G. Brosch, S. Graessle, P. Trojer, and J. D. Walton. 2002. Characterization of inhibitor-resistant histone deacetylase activity in plant-pathogenic fungi. Eukaryot. Cell 1:538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 3.Berse, B., A. Dmochowska, M. Skrzypek, P. Weglenski, M. A. Bates, and R. L. Weiss. 1983. Cloning and characterization of the ornithine carbamoyltransferase gene from Aspergillus nidulans. Gene 25:109-117. [DOI] [PubMed] [Google Scholar]
- 4.Buck, S. W., C. M. Gallo, and J. S. Smith. 2004. Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 75:939-950. [DOI] [PubMed] [Google Scholar]
- 5.Cabiscol, E., E. Piulats, P. Echave, E. Herrero, and J. Ros. 2000. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 275:27393-27398. [DOI] [PubMed] [Google Scholar]
- 6.Calera, J. A., S. Paris, M. Monod, A. J. Hamilton, J. P. Debeaupuis, M. Diaquin, R. Lopez-Medrano, F. Leal, and J. P. Latge. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect Immun. 65:4718-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmen, A. A., P. R. Griffin, J. R. Calaycay, S. E. Rundlett, Y. Suka, and M. Grunstein. 1999. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96:12356-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chary, P., and D. O. Natvig. 1989. Evidence for three differentially regulated catalase genes in Neurospora crassa: effects of oxidative stress, heat shock, and development. J. Bacteriol. 171:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, P., and P. Lau. 2005. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 19:563-573. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 11.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 12.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, G. A., and G. M. Wahl. 1987. Cosmid vectors for genomic walking and rapid restriction mapping. Methods Enzymol. 152:604-610. [DOI] [PubMed] [Google Scholar]
- 14.Fidel, S., J. H. Doonan, and N. R. Morris. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283-293. [DOI] [PubMed] [Google Scholar]
- 15.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 16.Flattery-O'Brien, J., L. P. Collinson, and I. W. Dawes. 1993. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J. Gen. Microbiol. 139:501-507. [DOI] [PubMed] [Google Scholar]
- 17.Fridovich, I. 1998. Oxygen toxicity: a radical explanation. J Exp. Biol. 201:1203-1209. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, T. J., A. R. Coulson, J. E. Sulston, and P. F. Little. 1987. Lorist2, a cosmid with transcriptional terminators insulating vector genes from interference by promoters within the insert: effect on DNA yield and cloned insert frequency. Gene 53:275-281. [DOI] [PubMed] [Google Scholar]
- 19.Graessle, S., M. Dangl, H. Haas, K. Mair, P. Trojer, E. Brandtner, J. D. Walton, P. Loidl, and G. Brosch. 2000. Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta 1492:120-126. [DOI] [PubMed] [Google Scholar]
- 20.Graessle, S., P. Loidl, and G. Brosch. 2001. Histone acetylation: plants and fungi as model systems for the investigation of histone deacetylases. Cell Mol. Life Sci. 58:704-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 22.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 23.Jamieson, D. J., D. W. Stephen, and E. C. Terriere. 1996. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 138:83-88. [DOI] [PubMed] [Google Scholar]
- 24.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawasaki, L., D. Wysong, R. Diamond, and J. Aguirre. 1997. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, G. W., T. Kondo, N. Noshita, and P. H. Chan. 2002. Manganese superoxide dismutase deficiency exacerbates cerebral infarction after focal cerebral ischemia/reperfusion in mice: implications for the production and role of superoxide radicals. Stroke 33:809-815. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 30.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J., I. W. Dawes, and J. H. Roe. 1995. Adaptive response of Schizosaccharomyces pombe to hydrogen peroxide and menadione. Microbiology 141:3127-3132. [DOI] [PubMed] [Google Scholar]
- 32.Levine, R. L., J. A. Williams, E. R. Stadtman, and E. Shacter. 1994. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 233:346-357. [DOI] [PubMed] [Google Scholar]
- 33.Levitz, S. M., and R. D. Diamond. 1985. Mechanisms of resistance of Aspergillus fumigatus conidia to killing by neutrophils in vitro. J. Infect. Dis. 152:33-42. [DOI] [PubMed] [Google Scholar]
- 34.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loidl, P. 2004. A plant dialect of the histone language. Trends Plant Sci. 9:84-90. [DOI] [PubMed] [Google Scholar]
- 36.Luo, J., N. Li, J. P. Robinson, and R. Shi. 2002. The increase of reactive oxygen species and their inhibition in an isolated guinea pig spinal cord compression model. Spinal Cord 40:656-665. [DOI] [PubMed] [Google Scholar]
- 37.Marmorstein, R. 2004. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem. Soc. Trans. 32:904-909. [DOI] [PubMed] [Google Scholar]
- 38.Marmorstein, R., and S. Y. Roth. 2001. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11:155-161. [DOI] [PubMed] [Google Scholar]
- 39.Murnion, M. E., R. R. Adams, D. M. Callister, C. D. Allis, W. C. Earnshaw, and J. R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656-26665. [DOI] [PubMed] [Google Scholar]
- 40.Oberegger, H., M. Schoeser, I. Zadra, B. Abt, and H. Haas. 2001. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41:1077-1089. [DOI] [PubMed] [Google Scholar]
- 41.Oberegger, H., I. Zadra, M. Schoeser, and H. Haas. 2000. Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in Aspergillus. FEBS Lett. 485:113-116. [DOI] [PubMed] [Google Scholar]
- 42.Paris, S., D. Wysong, J. P. Debeaupuis, K. Shibuya, B. Philippe, R. D. Diamond, and J. P. Latge. 2003. Catalases of Aspergillus fumigatus. Infect. Immun. 71:3551-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philippe, B., O. Ibrahim-Granet, M. C. Prevost, M. A. Gougerot-Pocidalo, M. Sanchez Perez, A. Van der Meeren, and J. P. Latge. 2003. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 71:3034-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 1953:141-238. [DOI] [PubMed] [Google Scholar]
- 45.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 46.Royall, J. A., and H. Ischiropoulos. 1993. Evaluation of 2′,7′-dichlorofluorescin and dihydrorhodamine 123 as fluorescent probes for intracellular H2O2 in cultured endothelial cells. Arch. Biochem. Biophys. 302:348-355. [DOI] [PubMed] [Google Scholar]
- 47.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Scherer, M., H. Wei, R. Liese, and R. Fischer. 2002. Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 1:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz, B. E., S. Misialek, J. Wu, J. Tang, M. T. Conn, R. Tahilramani, and L. Wong. 2004. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry 43:11083-11091. [DOI] [PubMed] [Google Scholar]
- 52.Sendra, R., I. Rodrigo, M. L. Salvador, and L. Franco. 1988. Characterization of pea histone deacetylases. Plant Mol. Biol. 11:857-866. [DOI] [PubMed] [Google Scholar]
- 53.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 54.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 55.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trojer, P., E. M. Brandtner, G. Brosch, P. Loidl, J. Galehr, R. Linzmaier, H. Haas, K. Mair, M. Tribus, and S. Graessle. 2003. Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res. 31:3971-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 58.Urnov, F. D., J. Yee, L. Sachs, T. N. Collingwood, A. Bauer, H. Beug, Y. B. Shi, and A. P. Wolffe. 2000. Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J. 19:4074-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaquero, A., A. Loyola, and D. Reinberg. 2003. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003:RE4. [DOI] [PubMed]
- 60.Verdone, L., J. Wu, K. van Riper, N. Kacherovsky, M. Vogelauer, E. T. Young, M. Grunstein, E. Di Mauro, and M. Caserta. 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 62.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolffe, A. P., and D. Pruss. 1996. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell 84:817-819. [DOI] [PubMed] [Google Scholar]