Abstract
Cryptococcus gattii has recently emerged as a pathogen of humans and animals in the temperate climate of Vancouver Island, British Columbia (B.C.). The majority (∼95%) of the isolates from the island belong to the VGII molecular type, and the remainder belong to the VGI molecular type. The goals of this study were to compare patterns of molecular variation among C. gattii isolates from B.C. with those from different areas of the world and to investigate the population structure using a comparative gene genealogy approach. Our results indicate that the C. gattii population in B.C. comprises at least two divergent lineages, corresponding to previously identified VGI and VGII molecular types. The genealogical analysis of strains suggested a predominantly clonal population structure among B.C. isolates, while there was evidence for sexual recombination between different molecular types on a global scale. We found no geographic pattern of strain relationships, and nucleotide sequence comparisons revealed that genotypes among isolates from B.C. were also present among isolates from other areas of the world, indicating extensive strain dispersal. The nucleotide sequence diversity among isolates from B.C. was similar to that among isolates from other areas of the world.
The basidiomycete fungus Cryptococcus gattii is a primary pathogen of humans and animals. C. gattii, previously recognized as Cryptococcus bacillisporus and Cryptococcus neoformans var. gattii, has traditionally been associated with tropical and subtropical climates (20) and with the infection of immunocompetent hosts (30, 32). The latter feature distinguishes C. gattii from the related pathogens C. neoformans var. neoformans and C. neoformans var. grubii, which are opportunistic pathogens of immunocompromised hosts, including AIDS patients (6). An unprecedented emergence of C. gattii infection in humans and many animal species has occurred on Vancouver Island in British Columbia (B.C.), Canada, over the past 6 years (15, 33; M. Fyfe et al., unpublished), and there is no evidence of C. gattii infection in B.C. prior to 1999 (S. E. Kidd, unpublished results). This emergence is striking not only because of the temperate climate of this region but because the reported infection rate is currently the highest in the world, e.g., 37 times greater than that reported in Australia, where C. gattii is considered endemic (5, 15). Environmental sampling revealed that C. gattii has colonized trees and soil on Vancouver Island and that the fungus can readily be detected in air samples (15; K. H. Bartlett, L. MacDougall, S. Mak, C. Duncan, S. Kidd, and M. Fyfe, Abstr. 16th Biometeorol. Aerobiol. Meet. 2004, abstr. 5.5, 2004 [http://ams.confex.com/ams /pdfpapers/80027.pdf]). For this study, we were interested in the patterns of molecular variation among C. gattii isolates from B.C. as well as in their potential origins and mode of reproduction in nature.
Traditionally, cryptococcal isolates have been classified into five groups, i.e., A, B, C, D, and AD, by serotyping of the capsular polysaccharide (8, 37). More recently, PCR fingerprinting, restriction fragment length polymorphism (RFLP) analysis, and amplified fragment length polymorphism (AFLP) analysis have been used extensively in genotyping studies of the C. neoformans species complex that includes C. gattii. It has been demonstrated that eight major molecular types exist within the species complex, initially defined according to distinct PCR fingerprinting and randomly amplified polymorphic DNA profiles (26, 27, 29, 31) and supported by a number of different molecular typing techniques (17). C. neoformans var. grubii (serotype A) isolates correspond to molecular types VNI and VNII; C. neoformans var. neoformans (serotype D) corresponds to molecular type VNIV; the serotype AD hybrid corresponds to molecular type VNIII; and C. gattii (serotypes B and C) corresponds to four molecular types, namely, VGI, VGII, VGIII, and VGIV. Many genetic subtypes exist within each molecular type, representing different strains of the organism. In this context, a pilot study using isolates from clinical and environmental sources on Vancouver Island (collected between 1999 and 2002) revealed that approximately 95% of the isolates belong to the VGII molecular type (15). PCR fingerprinting and AFLP analysis also revealed two VGII subtypes among the Vancouver Island isolates. These were designated VGIIa/AFLP6A and VGIIb/AFLP6B, with approximately 90% of VGII isolates belonging to the VGIIa/AFLP6A subtype (15). In terms of the potential origins of the VGII strains in B.C., we note that a single C. gattii strain, NIH444 (also known as CBS6956 and ATCC 32609), isolated from a human in Seattle, Wash., circa 1971, has also been typed as VGII/AFLP6 (1, 15). Washington State is geographically proximal to Vancouver Island and the B.C. mainland and shares a similar climate.
Besides the VGII isolates, one or two VGI isolates have been isolated each year since 2001 from clinical sources in B.C. (approximately 5% of typed cases) (15; S. E. Kidd, unpublished results), but no analogous environmental isolates of this molecular type were found. Many of the cases were associated with a travel history, leaving unclear the validity of VGI as an endemic molecular type for B.C. However, two VGI strains were recently (2004 and 2005) isolated from environmental sources in B.C. The first isolate was included in this study and came from an arbutus tree on Saltspring Island, located in the Strait of Georgia between Vancouver Island and the B.C. lower mainland (S. E. Kidd and K. H. Bartlett, unpublished results). The second isolate was obtained from a different tree on Saltspring Island following the completion of the analyses in this study. This finding indicates that a colonized source of the VGI molecular type may exist in B.C. and suggests that a deeper analysis of the population structure of C. gattii in B.C. is needed.
C. gattii is capable of both sexual and asexual (clonal) reproduction, and sexual recombination could potentially be occurring within the population on Vancouver Island, even though all isolates examined to date from B.C. have been of the alpha mating type (MATα) (12, 15). Such a biased distribution of mating type alleles has also been observed in populations of C. gattii in other parts of the world (19, 25, 40). The skewed mating type distribution suggests that the B.C. population of C. gattii might be predominantly clonal. However, conclusive evidence is lacking at this time.
The genealogy of a given gene may be used to approximate the evolutionary history of an organism. However, comparative analyses of multiple gene genealogies can provide insight into the mode of reproduction that facilitates evolutionary change within a defined group of organisms, hence providing information about recombination, clonality, speciation, hybridization, and dispersal. Given a group of clonally reproducing organisms, the genealogies of multiple unrelated genes are expected to be the same since evolutionary changes arise mainly through random genetic drift. But given a group of organisms where sexual recombination has occurred to some degree, the genealogies of different genes may be expected to differ because of meiotic reassortment.
The relationships between isolates belonging to the same and different molecular types and subtypes from Vancouver Island are unclear, and additional studies are needed to examine aspects of clonality, dispersion, recombination, and hybridization for this population. To begin to explore these relationships, a comparative gene genealogy approach was used to assess the population structure of C. gattii isolates from B.C. The concordance of phylogenetic patterns (i.e., monophyly) and specific traits of the isolates, such as mating type, molecular type, host type, and geographic origin, were examined to further characterize the C. gattii lineages. Our other goals were to assess DNA sequence variation among B.C. isolates in the context of isolates from other areas of the world and to investigate potential epidemiological links between isolates.
MATERIALS AND METHODS
Yeast isolates.
Twenty-four Cryptococcus gattii isolates from human (n = 9), animal (n = 9), and environmental (n = 6) sources on Vancouver Island and from other parts of B.C. and Canada were selected for analysis; these strains were isolated between 2001 and 2004. Nineteen C. gattii isolates collected in other parts of the world were used for DNA sequence comparisons. Serotyping of isolates was performed using the CryptoCheck slide agglutination test (Iatron, Tokyo, Japan). Detailed information for each of the isolates is provided in Table 1.
TABLE 1.
Cryptococcal isolates used in this study and details of their isolation, molecular types, and designated sequence subtypes
| Isolate no.g | Serotype | Molecular type | Mating type | Sourcef | Year isolated | Geographical originf | Sequence subtype
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| URA5 | LAC | FTR1 | CAP1 | |||||||
| Cryptococcus gattii isolates from British Columbia | ||||||||||
| A2M R314A | B | VGI | MATα | Human, sputum | 2002 | Duncan, V.I. (travel history) | 1 | 1 | 1 | 1 |
| A2M R299A | B | VGI | MATα | Human, CSF | 2002 | Lantzville, V.I. (travel history) | 1 | 1 | 1 | 1 |
| A4M R64Aa | B | VGI | MATα | Human | 2004 | Vancouver (travel history) | 1 | 1 | 1 | 1 |
| KB7892Ba | B | VGI | MATα | Arbutus tree | 2004 | Saltspring Island, B.C. (environs of parrot isolate KB7091) | 3 | 2 | 2 | 1 |
| A1M R794A | B | VGI | MATαc | Human, CSF | 2001 | Vancouver, B.C. (travel history unknown) | 4 | 2 | 2 | 1 |
| A1M F2863C | B | VGI | MATαb | Dall's porpoise | 2002 | Washed up on shore of southern V.I. (assumed travel history) | 3 | 1 | 3 | 1 |
| A1M R265A | B | VGII ad | MATαb | Human, BAL | 2001 | Duncan, V.I. | 5 | 3 | 4 | 2 |
| A1M F3016C | B | VGII ad | MATαb | Dall's porpoise | 2002 | Washed up on shore of a gulf island, B.C. (assumed travel history) | 5 | 3 | 4 | 2 |
| MAC-9B | B | VGII ad | MATαb | Cedar | 2001 | Cathedral Grove, V.I. | 5 | 3 | 4 | 2 |
| AIM R272A | B | VGII bd | MATαc | Human, BAL | 2001 | Ladysmith, V.I. | 7 | 3 | 4 | 3 |
| KB2045Ba | B | VGII ad | MATα | Air sample | 2002 | Langley, B.C. (environs of tapir isolate KB1079) | 5 | 3 | 4 | 2 |
| RB28B | B | VGII bd | MATαb | Tree stump | 2002 | Parksville, V.I. | 7 | 3 | 4 | 3 |
| RB67B | B | VGII bd | MATαc | Douglas fir | 2002 | Parksville, V.I. | 7 | 3 | 4 | 3 |
| KB152A-6B | B | VGII bd | MATαc | Air sample | 2002 | Parksville, V.I. | 7 | 3 | 4 | 3 |
| A2M R282A | B | VGII | MATα | Human, sputum | 2002 | Victoria, V.I. | 5 | 3 | 4 | 2 |
| KB7091Da | B | VGII | MATα | Companion parrot | 2003 | Saltspring Island (travel history) | 5 | 3 | 4 | 3 |
| A3M R535A | B | VGII ad | MATα | Human, chest abscess | 2003 | Delta, B.C. | 5 | 3 | 4 | 2 |
| A3M R673A | B | VGII | MATα | Human, BAL | 2003 | Sidney, V.I. (travel history) | 5 | 3 | 4 | 2 |
| KB5746D | B | VGII | MATα | Horse | 2003 | Mill Bay, V.I. | 7 | 3 | 4 | 3 |
| KB7092Da | B | VGII | MATα | Llama | 2003 | Chilliwack, B.C. | 5 | 3 | 4 | 2 |
| KB4672D | B | VGII | MATα | Companion cat | 2003 | Nanaimo, V.I. | 5 | 3 | 4 | 2 |
| KB1079Da | B | VGIII | MATα | Captive tapir | 2002 | Langley, B.C. (probably acquired infection in United States) | 8 | 5 | 6 | 6 |
| Cryptococcus gattii isolates from other parts of the world | ||||||||||
| A3M R29Aa | B | VGI | MATα | Koala | 2002 | Toronto Zoo, ON, Canada (previously from San Diego Zoo) | 1 | 1 | 1 | 1 |
| A2M R554Aa= UAMH9837 | B | VGI | MATα | Captive bottlenose dolphin | 2000 | San Diego, CA (no recent travel history) | 2 | 1 | 1 | 1 |
| WM179Ia | B | VGI | MATα | Human, CSF | 1993 | Sydney, NSW, Australia | 1 | 1 | 1 | 1 |
| WM276a,e | B | VGI | MATα | E. tereticornis | 1993 | Mt. Annan, NSW, Australia | 1 | 1 | 1 | 1 |
| KB10455Da | B | VGII | MATα | Companion cat | 2004 | Edmonton, AB, Canada (travel history to V.I.) | 5 | 3 | 4 | 2 |
| NIH444Fa= ATCC 32609 | B | VGII | MATα | Human, sputum | ca. 1971 | Seattle, WA | 5 | 3 | 4 | 2 |
| KB9944Ba | B | VGII | MATα | Unidentified tree species | 2004 | CA (environs of parrot isolate KB7091) | 5 | 3 | 4 | 2 |
| CBS 7750Ea | B | VGII | MATα | E. camaldulensis | 1990 | San Francisco, CA | 5 | 3 | 4 | 2 |
| LA55Ha= FOC417 | B | VGII | MATa | Human, CSF | 1995 | NE region of Piaui, Brazil | 5 | 3 | 4 | 5 |
| LA57Ha= FOC506 | B | VGII | MATα | Human, CSF | 1995 | NE region of Piaui, Brazil | 5 | 3 | 4 | 4 |
| LA61Ha= FOC557 | B | VGII | MATα | Human, CSF | 1997 | NE region of Piaui, Brazil | 7 | 3 | 4 | 4 |
| LA499Ga= HOO58 I-106 | B | VGII | MATa | Human, CSF | 1990 | Norte de Santander, Colombia | 6 | 3 | 4 | 5 |
| LA567Ga= HOO58 I-638 | B | VGII | MATa | Human, CSF | 1997 | Caqueta, Colombia | 6 | 3 | 4 | 5 |
| LA584Ga= HOO58 I-675 | B | VGII | MATa | Human, CSF | 1998 | Bolivar, Colombia | 6 | 3 | 4 | 5 |
| MC-S-115Ia | B | VGII | MATα | Human, CSF | 1993 | Thailand | 7 | 3 | 4 | 3 |
| WM178Ia | B | VGII | MATα | Human, lung | 1991 | Sydney, NSW, Australia | 5 | 4 | 5 | 3 |
| RAM005Ia | B | VGII | MATα | E. tetrodonta | 1999 | Arnhemland, NT, Australia | 7 | 3 | 4 | 3 |
| WM1008Ia | B | VGII | MATα | Insect frass | 2000 | Mt. Druitt, NSW, Australia | 7 | 3 | 4 | 3 |
| WM161Ia | B | VGIII | MATα | E. camaldulensis | 1992 | San Diego, CA | 8 | 5 | 6 | 6 |
| NIH191Fa | C | VGIII | MATab | Human, CSF | 1978 | CA | 9 | 6 | 7 | 7 |
| WM779Ia | C | VGIV | MATα | Cheetah | 1994 | Johannesburg, South Africa | 10 | 7 | 8 | 8 |
| Outgroups | ||||||||||
| H99a,e | A | VNI | MATαb | Human | 1978 | New York, NY | ||||
| B-3501Aa,e | D | VNIV | MATαb | Laboratory cross | NA | NA | ||||
Isolates not known to be connected to Vancouver Island.
Mating type of isolate was previously determined by laboratory crosses (15).
Isolate was previously found to be infertile (15).
Molecular subtype was determined from M13 PCR fingerprinting and/or AFLP analysis (15; S. E. Kidd, unpublished data).
URA5, LAC, FTR1, and CAP1 sequences were obtained from the following genome assemblies: H99, Duke Center for Genome Technology, NC, and Broad Institute, MA; B-3501A, Stanford Genome Technology Center, CA; WM276, British Columbia Genome Sciences Centre, Vancouver, B.C., Canada.
V.I., Vancouver Island; B.C., British Columbia; NA, not applicable; CSF, cerebrospinal fluid; BAL, bronchoalveolar lavage.
Isolates were donated by the following groups or individuals: A, British Columbia Centre for Disease Control, Vancouver, B.C., Canada; B, Karen Bartlett, School of Occupational and Environmental Hygiene, University of British Columbia, Vancouver, B.C., Canada; C, Stephen Raverty, Animal Health Centre, Abbotsford, B.C., Canada; D, Sally Lester, Central Laboratory for Veterinarians, Langley, B.C., Canada; E, Teun Boekhout, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; F, June Kwon-Chung, Laboratory of Clinical Investigation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.; G, Elizabeth Castañeda, Grupo de Microbiología, Instituto Nacional de Salud, Bogotá, Colombia; H, Ricardo Pereira Igreja, Doencas Infecciosas e Parasitarias, Hospital Universittario Clementino Fraga Filho, Rio de Janeiro, Brazil; and, I, Wieland Meyer, Molecular Mycology Laboratory, Westmead Hospital, Westmead, NSW, Australia.
DNA manipulations.
High-molecular-weight genomic DNA was isolated using previously described techniques (27, 39). The molecular types of all isolates were determined by URA5-RFLP analysis according to a previously described method (26). Some of the isolates in this study were included in a previous study, in which PCR fingerprinting and AFLP analysis revealed subtypes within the VGII molecular type (15).
Fragments of four unrelated genes were studied, namely, LAC (encodes diphenol oxidase/laccase) (36), URA5 (encodes orotidine monophosphate pyrophosphorylase) (4), FTR1 (encodes a high-affinity iron permease) (21), and CAP1 (encodes a capsule-associated protein), located at the mating locus (11). To verify that these genes were physically unlinked, the positions of these loci in the serotype B genome were determined (www.bcgsc.ca). LAC and URA5 both lie on chromosome 7 separated by 688 kb and the centromere, such that these loci are expected to undergo independent reassortment; FTR1 lies on chromosome 3; and CAP1 lies within the mating locus on chromosome 9. Primers designed to amplify and sequence the PCR products of each of the gene fragments were as follows (5′-3′): for LAC (565-bp product) (36), GGCGATACTATTATCGTA (forward) and TTCTGGAGTGGCTAGAGC (reverse); for URA5 (744-bp product) (4), ACGCCTGCCTGTTACTTAA (forward) and GGACATGATGATTGGAGT (reverse); for FTR1 (865-bp product), GTTCTCGGTCACCATCTTC (forward) and TCTCAGGCTCGCCATCTTC (reverse); and for CAP1 (815-bp product), CGCCATAGAGAGAGGATGAC (forward) and CCGCCTTACCTTCACAGTCG (reverse). PCR products were gel purified using a Qiaquick gel purification kit (QIAGEN, Mississauga, Ontario, Canada) or Wizard spin columns (Promega, Madison, Wis.). Sequences for these genes from WM276 (VGI) were obtained from the genome assembly from the B.C. Genome Sciences Centre (www.bcgsc.ca). Orthologous sequences were obtained from genome assemblies of H99 (serotype A) (Duke Center for Genome Technology, NC [http://cneo.genetics.duke.edu/], and Broad Institute, MA [http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans/]) and B-3501 (serotype D) (Stanford Genome Technology Center, CA [http://altoid.stanford.edu/sgtc/group/C.neoformans/index.html])(24) for use as outgroups in the analyses.
The accuracy of the nucleotide sequences was assessed by a BLASTn search of the A1M R265 genome assembly that recently became available for this isolate (http://www.broad.mit.edu), using the nucleotide sequences obtained for all four loci for this strain.
The mating type was determined by using BLAST 2 sequences (34) to align the CAP1 sequence from each isolate in our study to the recently available sequences of the C. gattii VGI MATα (GenBank accession no. AY710430.1) and MATa loci (AY710429.1) (11), which share 87% nucleotide identity at the CAP1 gene. Classification of the mating type in this way was consistent with previously determined mating types for some of the isolates used in this study (16). For those strains in which the CAP1 sequence matched that of the MATa locus, classical mating tests were used for confirmation by previously described methods (15).
Data analyses.
Phylogenetic analyses of the four individual gene fragments as well as the combined data (2,717 nucleotides) were performed using PAUP* v 4.0b10 software (D. L. Swofford, Sinauer Associates, Massachusetts), where transitions and transversions were weighted equally. The most parsimonious trees were obtained by a heuristic search based upon 500 random sequence additions. Statistical support for each clade was assessed using bootstrap analysis with 500 replicate samples of phylogenetically informative characters. Sequences from serotype A and D strains were used to root each tree. The sequence divergence between and within isolates grouped according to molecular types and geographical regions was estimated using mean pairwise Kimura 2 parameter distances (18).
The congruence of gene genealogies was assessed using the partition homogeneity test as well as pairwise comparisons of the tree topologies for the four genes (10). These tests were applied to the following six subsets of isolates: (i) all isolates used in the study (n = 45), (ii) MATα isolates only (n = 40), (iii) VGI isolates only (n = 10), (iv) VGII isolates only (n = 29), (v) VGII MATα isolates only (n = 25), and (vi) B.C. isolates only (n = 22).
To test whether there were significant phylogenetic patterns based upon mating types, molecular types (as determined by URA5-RFLP analysis), isolation host types, and geographic origins, we used the topology-dependent permutation tail probability test (T-PTP) (9). This test compares the lengths of maximum parsimony (MP) trees with and without the monophyletic constraint defined by each of the described traits. If the constrained trees are significantly longer than the MP tree without any constraint, the results would suggest a lack of a phylogenetic pattern based upon the specific traits. For each constraint analysis, the statistical significance was derived from permutation of the combined sequence data under the assumption of nonmonophyly to generate a null distribution of tree lengths. Statistical support for nonmonophyly is achieved when >95% of all permuted data sets have tree lengths shorter than the MP tree generated with the constraint of monophyly (9). The T-PTP test was implemented in PAUP* v. 4.0b10. One thousand permuted data sets were generated and analyzed for each of the constraint tests. The serotype A and D outgroup isolates were not included in these analyses.
Nucleotide sequence accession numbers.
All sequences obtained in this study were submitted to GenBank under the following accession numbers: LAC sequences, AY973072 to AY973113; URA5 sequences, AY973114 to AY973155; FTR1 sequences, AY972002 to AY972043; CAP1 sequences, AY971960 to AY972001.
RESULTS AND DISCUSSION
Nucleotide sequence diversity and lineages within C. gattii.
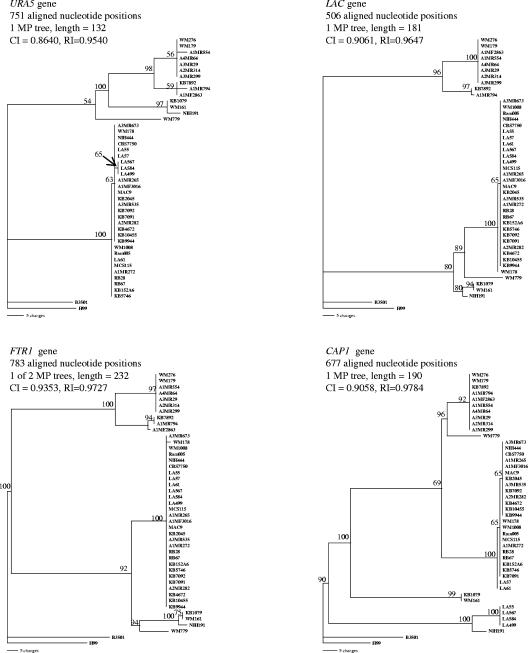
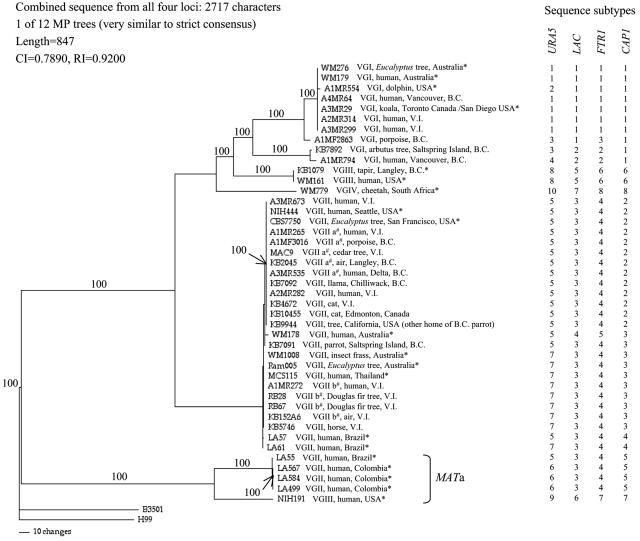
To begin an analysis of C. gattii lineages, we examined the genealogy of each of the four genes using maximum parsimony and found that the genes possessed various levels of divergence (Fig. 1). URA5 revealed the greatest diversity, with 10 alleles observed among the 43 C. gattii isolates in this study. There were eight FTR1 alleles and seven LAC alleles. CAP1 also revealed eight alleles, but this gene contained mating type-specific sequence variation due to its location at the mating locus (11); six CAP1 alleles were observed among C. gattii MATα isolates. These allele distributions were compiled to generate a multilocus sequence type (MLST) profile for each of the isolates in the study. The combined sequence data for the four gene fragments resulted in 2,717 aligned nucleotide sites. Of these, 614 sites were variable, including 388 that were phylogenetically informative. Figure 2 shows one of the 12 maximum parsimony trees generated from the combined sequence data that closely resembles the strict consensus tree.
FIG. 1.
Maximum parsimony trees for 43 isolates of Cryptococcus gattii from each of four gene regions sequenced. MP, maximum parsimony; CI, consistency index; RI, retention index. Values above branches indicate bootstrap support of >50% from 500 replicates.
FIG. 2.
One of 12 maximum parsimony trees from the combined DNA sequences of the LAC, URA5, FTR1, and CAP1 genes. Numbers (100) above branches denote strict consensus branches. V.I., Vancouver Island; B.C., British Columbia. *, isolates not known to be related to the emergence of C. gattii in B.C.; #, previously determined VGII subtypes (15; S. E. Kidd, unpublished data). An artificial clade of MATa strains was observed, arising due to the mating type-specific sequences of the CAP1 gene that outweighed the sequence variation due to molecular type divergence at all other loci.
The constraint of isolates belonging to the same mating type as a monophyletic group (for MATα, n = 38; for MATa, n = 5) revealed little change in the tree length compared to the tree without constraint (P = 0.999), indicating significant support for isolates of each mating type representing two monophyletic groups. At the CAP1 gene, sequence variation due to the mating type (104 and 110 of 672 sites within VGIII and VGII molecular types, respectively) was greater than that due to molecular type divergence (12 and 27 of 672 sites among MATa and MATα isolates, respectively). This mating type-specific sequence variation at one locus outweighed variation due to the molecular type at all four loci in the combined sequence data set, with the creation of an artificial clade corresponding to MATa isolates (Fig. 2). These data support an ancient divergence of the cryptococcal mating locus (11) that predates the divergence of molecular types. Since the nucleotide sequences of genes at the bipolar cryptococcal mating locus are known to be mating type specific (11, 28, 34), all further analyses were performed with exclusion of the MATa isolates (for VGII, n = 4; for VGIII, n = 1) to avoid distortion of the data.
The MATα isolates were constrained in monophyletic groups according to their molecular type (for VGI, n = 10; for VGII, n = 25; for VGIII, n = 2; for VGIV, n = 1), with little change in tree length compared to that without constraint (P = 0.999). This indicates significant support for isolates of different molecular types forming monophyletic groups representing potentially ancient lineages within the species, which is consistent with the findings of previous studies (14, 15). In addition, this reinforces the usefulness of molecular typing techniques for approximating sequence divergence and evolutionary distance between strains of C. gattii.
With the exception of the CAP1 gene, most of the sequence loci that we investigated revealed greater diversity among the 10 VGI isolates used in this study than that observed among the 25 VGII MATα isolates. The mean pairwise Kimura 2 parameter distance (Table 2) within VGI was significantly greater than that observed within the MATα VGII isolates (P = 0.000), and the mean divergence between VGI and VGII MATα isolates was >10 times greater than that within either the VGI or VGII isolates (P = 0.000). Other comparisons between and within molecular types showed a similar pattern of greater divergence between the molecular types than within them. A previous study also demonstrated greater nucleotide sequence diversity within VGI than within the other molecular types, using the internal transcribed spacer regions of the rRNA gene (14). Such a significant divergence between molecular types suggests the existence of several phylogenetic species within C. gattii.
TABLE 2.
Mean pairwise Kimura 2 parameter distances among C. gattii isolates within and between molecular types (or predetermined VGII subtypes [15])a
| Molecular type | Kimura 2 parameter distance (mean ± SD)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
MATα isolates only (n = 40)
|
MATa isolates only (n = 5)
|
Subtyped isolates only (n = 9)
|
||||||||
| VGI (n = 10) | VGII (n = 23) | VGIII (n = 2) | VGIV (n = 1) | VNI (n = 1) | VNIV (n = 1) | VGII (n = 4) | VGIII (n = 1) | VGII a (n = 5) | VGII b (n = 4) | |
| VGI | 0.00328 ± 0.00322 | 0.04895 ± 0.00438 | 0.03847 ± 0.00070 | 0.04874 ± 0.00070 | 0.13075 ± 0.00063 | 0.13560 ± 0.00079 | ||||
| VGII | 0.00052 ± 0.00046 | 0.04766 ± 0.00048 | 0.04888 ± 0.00043 | 0.13888 ± 0.00053 | 0.13998 ± 0.00025 | 0.00019 ± 0.00021 | 0.04514 ± 0.00020 | |||
| VGIII | 0.00000 | 0.04625 ± 0.00000 | 0.13045 ± 0.00000 | 0.13712 ± 0.00000 | ||||||
| VGIV | 0.13742 | 0.13703 | ||||||||
| VNI | 0.08896 | |||||||||
| VNIV | ||||||||||
| VGII a | 0.00000 | 0.00075 ± 0.00000 | ||||||||
| VGII b | 0.00000 | |||||||||
Bold type indicates a mean genetic distance among isolates of a single molecular type or subtype.
Other T-PTP tests of geography- and host type-based phylogenetic patterns indicated significantly longer trees in the presence of the monophyletic constraints than those in the absence of such constraints. These results are consistent with the hypotheses of extensive strain dispersal among geographic areas and the lack of host specificity among the analyzed strains. Specifically, of the strains from each of the three host types (for humans, n = 15; for animals, n = 11; for the environment, n = 12), none formed a statistically robust monophyletic group (P = 0.000). Although the sample size is small, this indicates that the host type cannot be used to predict evolutionary groups among isolates and is consistent with previous studies that found clinical and environmental isolates to be genotypically indistinguishable (7, 23, 31).
Epidemiological links between B.C. isolates and those from other parts of the world.
One of the goals of this study was to investigate the possibility of an epidemiological link between the VGII isolates from Vancouver Island and the NIH444 strain, isolated in Seattle circa 1971, and recently found to belong to the VGII molecular type (1, 15). The MLST profile for NIH444 was identical to those of many isolates from Vancouver Island (A1M R265, A1M R282, A3M R673, MAC-9, and KB4672), other parts of B.C. (A1M F3016, A3M R535, KB2045, KB7091, and KB7092), and other parts of North America (KB10455, CBS7750, and KB9944), some of which were previously subtyped as VGIIa/AFLP6A (15). It is also possible that NIH444 could share sequence identity with isolates from areas other than North America which were not included in this study. A recent study of the mini-intein sequence from the cryptococcal PRP8 gene (2) revealed that NIH444 differed from seven Vancouver Island environmental VGII isolates at 1 of 510 nucleotide sites. We independently sequenced the PRP8 mini-intein sequence for this strain and found that it was identical across all 510 nucleotides to the isolates from Vancouver Island. Therefore, it is possible that NIH444 may be related to isolates from B.C., but knowledge of the travel history of the patient from which NIH444 was isolated is lacking. On the other hand, the NIH444 URA5 and CAP1 sequences described in this study differ from those of the VGIIb isolates from Vancouver Island (RB28, RB67, KB152A-6, A1M R272, and KB5746). Rather, these VGIIb isolates share identical MLST profiles with VGII environmental isolates from Australia (Ram005 and WM1008) and a clinical isolate from Thailand (MC-S-115; travel history unknown). Thus, isolates from B.C. share similar or identical genotypes with isolates from several different areas of the world. However, there is insufficient evidence from these studies to conclude that C. gattii strains were introduced to B.C. from sources from any specific part of the world. Indeed, it is not implausible that strains from B.C. may have dispersed to other parts of North America or the world, thus accounting for the shared genotypes.
Based on their MLST profiles, four of the five VGI sequence variants observed in this study are associated with B.C. isolates, and three B.C. VGI isolates represent unique strains in this study. One of these unique strains, KB7892 (from an arbutus tree on Saltspring Island), represents the only VGI environmental strain isolated in B.C. at the time of these analyses. Therefore, there appears to be no epidemiological link between the B.C. VGI environmental isolate and any of the clinical VGI isolates from B.C. This is supported by preliminary studies of a second environmental VGI isolate, recently obtained from a different tree on Saltspring Island, which indicate that it is genotypically identical to KB7892 (S. E. Kidd, unpublished data). Clinical VGI isolates from B.C. (A2M R314, A2M R299, and A4M R64) possessed identical MLST profiles to those of an isolate from a captive koala (A3M R29) residing at Toronto Zoo, Ontario, Canada (and at San Diego Zoo, CA, 6 months prior to cryptococcal isolation) and environmental isolates from Australia (WM276 and WM179). As mentioned above, many of the clinical VGI cases from B.C. are associated with a known travel history (wild porpoises were assumed to have a travel history since their geographical ranges were not known), and it is possible that some or all of the VGI infections were acquired outside of B.C., particularly since there is limited evidence at this time for a truly colonized source of VGI in the B.C. environment. Further investigation is under way to determine whether the environmental VGI isolates from Saltspring Island represent colonization or a transient presence. Overall, these data indicate that the genetic variation among VGI strains isolated from sources in B.C. is roughly equivalent to that among VGI isolates from other parts of the world, although this conclusion is in the context of the limited number of isolates available for use in this study and the possibility that the VGI infections were acquired outside of B.C.
Population structure of C. gattii in B.C.
The congruence of gene genealogies was tested to assess the population structure among the C. gattii isolates in this study. When all 45 isolates in the study were considered and when only the MATα isolates (n = 40) were considered, the gene genealogies were found to be highly incongruent (P = 0.000). However, when the isolates from B.C. were considered alone, the four gene genealogies were highly congruent (P = 0.450), and when only the VGII isolates were considered, the four genes had highly congruent genealogies (P = 0.734), with increased congruence observed when the four VGII MATa isolates were excluded from this analysis (P = 0.810). When VGI isolates were considered alone, the four genes had marginally congruent genealogies (P = 0.053) which, in the absence of providing evidence for recombination, was interpreted as an indication of clonality. These data suggest that sexual recombination has occurred between C. gattii isolates of different molecular types on a global scale but that the C. gattii population in B.C. has a predominantly clonal mode of reproduction in nature. Furthermore, it appears that there is a clonal propagation of isolates within each of the VGI and VGII molecular types, both among isolates from B.C. and among the global isolates used in this study. This is consistent with the hypothesis presented in a previous study, in which all fertile C. gattii isolates tested from B.C. belonged to the alpha mating type and an association was observed between mating incompetence and the VGIIb/AFLP6B molecular subtype (15). In contrast, a recent study of C. gattii VGII isolates from Sydney and the Northern Territory of Australia found evidence for recombination among the isolates from each geographical area and genetic connectivity between the two populations (3). Studies of the population structure among environmental C. gattii VGI isolates from Australia and Papua New Guinea found no evidence of sexual recombination, and all isolates were infertile (3, 13).
Studies of fertility among C. gattii isolates from Vancouver Island (obtained between 2001 and 2002) observed that all fertile isolates from clinical and environmental sources were of the alpha mating type (12, 15). Among the models presented in one of these studies (12), it was suggested that mating and meiotic recombination in a fertile clade may have given rise to a strain with increased virulence, resulting in the comparatively high rate of infection reported for Vancouver Island. A previously reported disparity in the fertility of VGIIa and VGIIb isolates (15) may provide a useful basis upon which to investigate this model further. While we observed no evidence for recombination specifically relating to the VGII isolates or the group of VGI and VGII isolates from B.C., the relationship between the VGIIa and VGIIb genotypes was unclear from our data set and should be further investigated. These VGII variants appear to be clonally propagated at present, but this does not preclude the possibility of a more ancient recombination. Other models invoking an increased ability of Vancouver Island strains to undergo haploid fruiting or a contribution of fertility to the formation of infectious propagules remain to be investigated. The recent demonstration that C. neoformans cells of the same (alpha) mating type can undergo sexual reproduction may be relevant in this context (22). Specifically, the fertile MATα cells of the VGII molecular type found on Vancouver Island may be particularly well adapted for alpha-alpha sexual development leading to spore formation; this type of interaction may not be revealed by our methods if the strains were closely related. In general, it is clear that more detailed experiments are needed to understand the fertility properties of the Vancouver Island isolates in the context of their ecological niche and their virulence. The availability of sequenced genomes for strains of the VGI and VGII molecular types (see Materials and Methods) will facilitate such experiments.
Geographical relatedness of C. gattii isolates.
To examine the phylogeographic patterns of the strains in this study, MATα isolates were constrained within monophyletic groups according to their geographical origins (for Vancouver Island, n = 12; for other parts of B.C., n = 10; for other parts of North America, n = 7; for South America, n = 2; for Australia, n = 5; for Africa, n = 1; and for Asia, n = 1). These constraints led to significant increases in tree length (P = 0.000). To assess whether there was any bias introduced as a result of arbitrary limits of the geographical regions, we varied the geographical boundaries into which isolates were constrained, e.g., Canada (n = 24), the United States (n = 5), Brazil (n = 2), South Africa (n = 1), Australia (n = 5), and Thailand (n = 1) (P = 0.000), or North America (n = 29), South America (n = 2), Australia (n = 5), Africa (n = 1), and Asia (n = 1) (P = 0.000). Therefore, among the isolates used in this study, there was no correlation between sequence divergence among isolates and their geographic distance. Varying the limits of geographical regions that were constrained as monophyletic groups made no difference to the probability of constrained data, and all analyses yielded trees with significantly greater lengths than that of the tree with no geographic constraints. The mean Kimura 2 parameter (Table 3) distances between isolates from different geographical regions indicated that there is no significant difference in the level of sequence diversity among isolates from Vancouver Island (or from all of B.C.) compared to that among isolates from other regions examined in this study. In addition, the global genetic diversity of the isolates used in this study is not subdivided according to geographical borders. Under the assumption that the dispersal of strains increases the heterogeneity of a given population at a greater rate than the accumulation of mutations among a clonal lineage (random genetic drift), these data indicate a significant migration of C. gattii strains between different regions of the world and are consistent with the findings of a previous study (38). However, since we cannot be certain that a given isolate is truly representative of the geographical region from which it was isolated (particularly for clinical isolates, which may have been acquired outside of the host's residential area) and since each geographic population appears to be as genetically diverse as the others, it was not possible to unambiguously infer the center(s) of origin for individual strains or populations. Indeed, the extent of cryptococcal strain migration throughout the world seems to be such that searching for specific sources of particular strains may be irrelevant.
TABLE 3.
Mean pairwise Kimura 2 parameter distances among C. gattii isolates within and between different geographical regions
| Region | Kimura 2 parameter distance (mean ± SD)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Vancouver Island (n = 12) | B.C., Excluding Vancouver Island (n = 10) | B.C., Including Vancouver Island (n = 22) | Rest of North America (n = 8) | South America (n = 6) | Australia (n = 5) | Thailand (n = 1) | South Africa (n = 1) | |
| Vancouver Island | 0.01518 ± 0.02258 | 0.02570 ± 0.02412 | NAb | 0.02589 ± 0.02813 | 0.02175 ± 0.02258 | 0.02175 ± 0.02433 | 0.00844 ± 0.01883 | 0.04883 ± 0.00039 |
| B.C., excluding Vancouver Island | 0.03288 ± 0.02216 | NAb | 0.03132 ± 0.02619 | 0.04551 ± 0.02551 | 0.02721 ± 0.02350 | 0.02471 ± 0.02535 | 0.04853 ± 0.00103 | |
| B.C., including Vancouver Island | 0.02383 ± 0.02403 | 0.02840 ± 0.02732 | 0.03809 ± 0.02481 | 0.02423 ± 0.02400 | 0.01583 ± 0.02302 | 0.04870 ± 0.00074 | ||
| Rest of North America | 0.03911 ± 0.02839 | 0.04395 ± 0.02456 | 0.03320 ± 0.02709 | 0.02791 ± 0.03050 | 0.05207 ± 0.00915 | |||
| South America | 0.01967 ± 0.01884 | 0.04162 ± 0.02566 | 0.02504 ± 0.01897 | 0.06490 ± 0.01292 | ||||
| Australia | 0.02998 ± 0.02517 | 0.01987 ± 0.02654 | 0.04874 ± 0.00033 | |||||
| Thailand | 0.048430 | |||||||
| South Africa | ||||||||
Bold type indicates a mean genetic distance among isolates from within a defined geographical area.
NA, not applicable.
Given that the B.C. C. gattii population appears to be clonal, there are a number of possible explanations for the recent emergence of C. gattii infection on Vancouver Island. These possibilities include (i) C. gattii on Vancouver Island represents an ancient population, where heterogeneity has arisen through random genetic drift, strain dispersal, or recombination that was not detectable within our data set; (ii) the Vancouver Island C. gattii population was recently established by introductions of both VGI and VGII strains from one or more parts of the world, which could have occurred independently or at the same time; and (iii) the Vancouver Island C. gattii population was recently established by the introduction, colonization, and rapid expansion of a VGII strain from another part of the world, where genetic drift, strain dispersal, or undetected recombination has effected at least two distinct strains within the VGII molecular type, and the presence of VGI isolates is due to strain dispersal.
Even though the emergence of C. gattii infection in B.C. is recent, no geographical area represented by isolates in this study revealed any more genetic diversity than other areas (Table 3), and the hypothesis of an ancient C. gattii population in B.C. cannot be rejected. Our data indicate that the B.C. C. gattii population possesses as much genetic diversity as those from other geographical areas and shares many identical or similar genotypes. Therefore, either the ancient population or the recent introduction hypothesis can explain the observed emergence of C. gattii in B.C. However, the evidence for clonal propagation of the isolates from B.C. investigated here suggests that the C. gattii isolates from B.C. are unlikely to represent an ancient population.
An ongoing environmental sampling study of C. gattii has amassed strong evidence for the colonization and propagation of VGII isolates on Vancouver Island (K. H. Bartlett and S. E. Kidd, unpublished results). However, despite the isolation of several VGI strains from clinical cases, there is little evidence at this time for permanent colonization of VGI in the B.C. environment, aside from two isolates from Saltspring Island. A comparison of MLST profiles revealed no similarity of the VGI environmental strain included in this study to any of the clinical strains. In the absence of a confirmed environmental reservoir for the VGI infections, we suggest that some or all of these cases may have been acquired through travel to other parts of the world; indeed, many of the patients had a travel history within the 12 months prior to diagnosis. Continued investigation of B.C. environmental isolates may make clear whether VGI is transient or has a colonized source in B.C.
Summary.
The unprecedented emergence of Cryptococcus gattii as a saprophyte and infectious agent in the temperate climate of B.C. since 1999 (15, 33; Bartlett et al., Abstr. 16th Biometeorol. Aerobiol. Meet. 2004, abstr. 5.5, 2004 [http://ams.confex.com/ams/pdfpapers/80027.pdf]), together with evidence for extensive strain dispersion, indicates a dynamic distribution of strains throughout the world. However, despite their unforeseen colonization in a temperate region, the C. gattii isolates from Vancouver Island and B.C. do not appear to be exceptional in terms of their phylogeny compared to those from other areas of the world. These isolates are clonal, possess a level of nucleotide sequence diversity that is equivalent to that of the global population, and share identical MLST profiles with many isolates collected in other parts of the world.
Acknowledgments
We thank the following groups and individuals for kindly providing cryptococcal isolates: British Columbia Centre for Disease Control, Vancouver, B.C., Canada; Sally Lester of the Central Laboratory for Veterinarians, Langley, B.C., Canada; Stephen Raverty of the Animal Health Centre, Abbotsford, B.C., Canada; Teun Boekhout of the Centraalbureau voor Schimmelcutures, Utrecht, The Netherlands; June Kwon-Chung of the Laboratory of Clinical Investigation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.; Elizabeth Castañeda of the Grupo de Microbiología, Instituto Nacional de Salud, Bogotá, Colombia; Ricardo Pereira Igreja of the Doencas Infecciosas e Parasitarias, Hospital Universittario Clementino Fraga Filho, Rio de Janeiro, Brazil; and Wieland Meyer of the Molecular Mycology Laboratory, Westmead Hospital, Westmead, NSW, Australia. In addition, we thank Mary Berbee, Department of Botany, University of British Columbia, for her critical review of the manuscript.
This work was funded in part by grants from the Canadian Institutes of Health Research (to J.W.K.) and the National Sciences and Engineering Research Council of Canada (to J.X.). S.E.K. and K.H.B. are supported by the Michael Smith Foundation for Health Research. J.W.K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology. DNA sequencing was performed by the Nucleic Acids and Protein Service (NAPS) Unit at the University of British Columbia and by the Mobix Laboratory at McMaster University.
REFERENCES
- 1.Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, W. C. J. Hop, E. C. A. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 2.Butler, M. I., and R. T. Poulter. 2005. The PRP8 inteins in Cryptococcus are a source of phylogenetic and epidemiological information. Fungal Genet. Biol. 42:452-463. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, L. T., B. J. Currie, M. Krockenberger, R. Malik, W. Meyer, J. Heitman, and D. A. Carter. 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell 4:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall, A., L. F. Freundlich, L. Marsh, and M. D. Scharff. 1992. Extensive allelic variation in Cryptococcus neoformans. J. Clin. Microbiol. 30:1080-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, and K. Byth. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 6.Chuck, S. L., and M. A. Sande. 1989. Infections with Cryptococcus neoformans in the acquired immune deficiency syndrome. N. Engl. J. Med. 321:794-799. [DOI] [PubMed] [Google Scholar]
- 7.Delgado, A. C., H. Taguchi, Y. Mikami, M. Myiajy, M. C. Villares, and M. L. Moretti. 2005. Human cryptococcosis: relationship of environmental and clinical strains of Cryptococcus neoformans var. neoformans from urban and rural areas. Mycopathologia 159:7-11. [DOI] [PubMed] [Google Scholar]
- 8.Evans, E. E. 1950. The antigenic composition of Cryptococcus neoformans. I. A serological classification by means of the capsular and agglutination reactions. J. Immunol. 64:423-440. [PubMed] [Google Scholar]
- 9.Faith, D. L. 1991. Cladistic permutation tests for monophyly and nonmonophyly. Syst. Zool. 40:366-375. [Google Scholar]
- 10.Farris, J. S., M. Kallersjo, A. G. Kluge, and C. Bult. 1995. Testing significance of incongruence. Cladistics 10:315-319. [Google Scholar]
- 11.Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler, F. S. Dietrich, and J. Heitman. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:2243-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halliday, C. L., and D. A. Carter. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsu, M., S. Kidd, A. Ando, M. Moretti-Branchini, Y. Mikami, K. Nishimura, and W. Meyer. 2004. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans complex. FEMS Yeast Res. 4:377-388. [DOI] [PubMed] [Google Scholar]
- 15.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. MacDougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidd, S. E., T. C. Sorrell, and W. Meyer. 2003. Isolation of two molecular types of Cryptococcus neoformans var. gattii from insect frass. Med. Mycol. 41:171-176. [DOI] [PubMed] [Google Scholar]
- 17.Kidd, S. E. 2003. Ph.D. thesis. University of Sydney, Sydney, Australia.
- 18.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 20.Kwon-Chung, K. J., and J. E. Bennett. 1984. Epidemiological differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123-130. [DOI] [PubMed] [Google Scholar]
- 21.Lian, T., M. I. Simmer, C. A. D'Souza, B. R. Steen, S. D. Zuyderduyn, S. J. M. Jones, M. A. Marra, and J. W. Kronstad. 2005. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 55:1452-1472. [DOI] [PubMed] [Google Scholar]
- 22.Lin, X., C. M. Hull, and J. Heitman. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017-1021. [DOI] [PubMed] [Google Scholar]
- 23.Litvintseva, A. P., L. Kestenbaum, R. Vilgalys, and T. G. Mitchell. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 43:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, K. J. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome and transcriptome of Cryptococcus neoformans, a basidiomycetous fungal pathogen of humans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madrenys, N., C. De Vroey, C. Raes-Wuytack, and J. M. Torres-Rodriguez. 1993. Identification of the perfect state of Cryptococcus neoformans from 195 clinical isolates including 84 from AIDS patients. Mycopathologia 123:65-68. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, W., A. Castañeda, S. Jackson, M. Huynh, E. Castañeda, and the Ibero-American Cryptococcal Study Group. 2003. Molecular typing of Ibero-American Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by PCR-fingerprinting and RAPD—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 28.Moore, T. D., and J. C. Edman. 1993. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruma, P., S. C. A. Chen, T. C. Sorrell, and A. G. Brownlee. 1996. Characterization of Cryptococcus neoformans by random DNA amplification. Lett. Appl. Microbiol. 23:312-316. [DOI] [PubMed] [Google Scholar]
- 30.Sorrell, T. C. 2001. Cryptococcus neoformans variety gattii. Med. Mycol. 39:155-168. [PubMed] [Google Scholar]
- 31.Sorrell, T. C., S. C. A. Chen, P. Ruma, W. Meyer, T. J. Pfeiffer, D. H. Ellis, and A. G. Brownlee. 1996. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J. Clin. Microbiol. 34:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speed, B., and D. Dunt. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28-34. [DOI] [PubMed] [Google Scholar]
- 33.Stephen, C., S. Lester, W. Black, M. Fyfe, and S. Raverty. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43:792-794. [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. (Erratum, 177:187-188.) [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, D. E., J. E. Bennett, and J. W. Bailey. 1968. Serological grouping of Cryptococcus neoformans. Proc. Soc. Exp. Biol. Med. 127:820-823. [DOI] [PubMed] [Google Scholar]
- 38.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
- 39.Xu, J., A. R. Ramos, R. Vilgalys, and T. G. Mitchell. 2000. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J. Clin. Microbiol. 38:1214-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, Z., X. Li, and J. Xu. 2002. Geographic distributions of mating type alleles of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 40:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]