Abstract
Cryptococcus neoformans, a basidiomycetous fungal pathogen, infects hosts through inhalation and can cause fatal meningoencephalitis in individuals if untreated. This fungus undergoes a dimorphic transition from yeast to filamentous growth during mating and monokaryotic fruiting, which leads to the production of hyphae and airborne infectious basidiospores. Here we characterized a novel morphological feature associated with the filamentous stages of the life cycle of C. neoformans which resembles resting or survival structures known as chlamydospores in other fungi. The C. neoformans chlamydospore-like structure is rich in glycogen, suggesting that it might have a role as an energy store. However, characterization of mutants with decreased or increased levels of glycogen production showed that glycogen levels have little effect on filamentous growth, sporulation, or chlamydospore formation. These results suggest that the formation of chlamydospores is independent of glycogen accumulation level. We also show that chlamydospore formation does not require successful sporulation and that the presence of chlamydospores is not sufficient for sporulation. Although the biological functions of chlamydospores remain to be established for this pathogenic fungus, their formation appears to be an integral part of the filamentation process, suggesting that they could be necessary to support sexual sporulation under adverse conditions and thereby facilitate the production of infectious basidiospores or long-term survival propagules in harsh environments.
Chlamydospores are produced by many fungi and represent enlarged, thick-walled vegetative cells with varied forms and condensed cytoplasm that form within hyphae or at hyphal tips. Despite poor cytological descriptions or documentation of their mode of generation, chlamydospores have been observed in three major clades of the fungal kingdom. For example, the basidiomycete black ink mushroom Coprinus cinereus (28) and Cryptococcus laurentii (30), the ascomycete nematode-trapping fungus Duddingtonia flagrans (20, 41), and zygomycete mucorales, such as Rhizopus schipperae (4, 55), have all been shown to produce chlamydospores. Even the fungus-like oomycete plant pathogens Phytophthora cinnamomi and Phytophthora parasitica produce chlamydospores.
Biological functions ascribed to these chlamydospores differ between species. For example, desiccation-resistant chlamydospores of P. cinnamomi are produced within plant roots during drought and are transported in root fragments or soil, germinating to cause infections when warm, moist conditions are encountered. When chlamydospores of the nematode-trapping fungus D. flagrans are fed to domesticated animals, they can survive passage through the alimentary tract and reduce the number of parasitic nematode larvae that develop from eggs in the feces, thus preventing clinical disease (20, 41). In addition, the chlamydospore developmental phase of Aspergillus parasiticus has been associated with increased aflatoxin production, while chlamydospores of Fusarium species are the principal means of long-term survival during unfavorable periods in the soil and play an important role as the primary inocula infecting plants (1, 14). Chlamydospores have also been observed in human fungal pathogens such as Candida albicans, Paracoccidioides brasiliensis, Histoplasma capsulatum, and Blastomyces dermatitidis (19). Although the appearance of chlamydospores in C. albicans serves as a diagnostic test, these structures are rarely found in infected patients or animals and their pathological or biological functions are as yet unclear (11, 13, 36).
The environmental cues for chlamydospore formation for various fungi are usually species specific and include nutrients (6, 27, 40), osmolarity, light (25, 29), pH (44), temperature (40), air (6), drug treatment (13), and plant stimulants (26). Because chlamydospores are not known to be formed in the model yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, relatively less is known about the molecular mechanisms controlling chlamydospore formation, with the exception of information gleaned from certain strains of Candida species (2, 25, 39, 46, 47). Thus, although chlamydospores are produced by a broad assortment of fungi, their mechanisms of formation, biological functions, and molecular regulation remain enigmatic.
Here we describe the first identification of chlamydospores in the basidiomycetous pathogen Cryptococcus neoformans, the most common cause of fungal meningoencephalitis in humans and which is capable of infecting both immunocompromised and apparently healthy hosts (10). Although C. neoformans exists in a single-celled yeast form during infection and in culture, this dimorphic fungus can switch from yeast to filamentous growth during mating and monokaryotic fruiting (3, 22, 31, 34, 53), which eventually leads to production of airborne meiotic basidiospores, thought to be involved in dispersal and infection.
C. neoformans strains can be readily isolated from the environment, particularly from soil contaminated with aged pigeon guano (10). Due to their microscopic nature, structures that promote C. neoformans survival in the environment are not known and could be sexual basidiospores, mitotic yeast cells, filaments, or other unknown structures. While many soilborne fungi produce chlamydospores as long-lived survival structures under hostile environmental conditions (15, 44), there has been no previous report of such structures in C. neoformans. Here we describe the formation of a morphological structure associated with the filamentous growth of C. neoformans that is strikingly similar to chlamydospores produced by other fungi, providing the first documentation of such structures in this organism. These data reveal a novel growth option in the life cycle of C. neoformans and provide a robust and genetically tractable model for studying the morphogenesis and molecular basis of the development and function of these cellular structures.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. YPD medium contained 1% yeast extract, 2% Bacto peptone, and 2% dextrose. YNB with glucose medium (pH 6.0) was used as a minimal medium and contained 6.7 g of yeast nitrogen base without amino acids (Difco) and 20 g of glucose per liter. Filament agar (53) and V8 juice (32) agar were used for monokaryotic fruiting and mating. JEC21 cells were used for monokaryotic fruiting, and the coculture of JEC21and JEC20 was used for mating.
TABLE 1.
Cryptococcus neoformans strains
| Straina | Genotype |
|---|---|
| JEC21 | MATα |
| JEC20 | MATa |
| JEC169 | MATaade2 lys1 ura5 |
| XL467 | MATα gsy1::NAT |
| XL466 | MATagsy1::NAT |
| XL343 | MATagsy1::NAT ade2 |
| XL344 | MATα gsy1::NAT ura5 lys1 |
| XL468 | MATatps1::NAT |
| XL470 | MATα tps1::NAT |
| XL347 | MATatps1::NAT ade2 |
| XL349 | MATα tps1::NAT ura5 lys1 |
| XL832 | MATα tps1::NAT TPS1-NEO |
| XL835 | MATα gsy1::NAT GSY1-NEO |
| YSB139 | MATα hog1::NAT |
| YSB143 | MATahog1::NAT |
Congenic strains JEC21 and JEC20 were described previously (33). All other strains are their derivatives. XL466, XL343, and XL344 are progeny from the cross between XL467 and JEC169. XL468, XL347, and XL349 are progeny from the cross between XL470 and JEC169. YSB139 and YSB143 were provided by Yong-Sun Bahn (5).
Microscopy.
Cells were grown on V8 medium on the top of slides in the dark for 7 days. Hyphae were fixed in 3.7% formaldehyde in phosphate-buffered saline with 1% Triton X-100. Nuclei and septa were visualized by staining with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) and calcofluor (Sigma) as described previously (53). For FM4-64 and Nile Red staining of intracellular organelles and lipid bodies, live cells were used and were processed as described previously (35, 49).
Transformations.
Dominant selectable markers conferring resistance to nourseothricin (NAT1) and G418 (NEO) markers (18) were introduced biolistically using a Bio-Rad model PDS-1000/He biolistic particle delivery system as previously described (50). All the primers used in this study are listed in Table 2.
TABLE 2.
Primers used
| Primer name | Sequence |
|---|---|
| JOHE13917 | CTGGCCGTCGTTTTACGAGAGCTTCCTACCGCCAG |
| JOHE13918 | CTGGCCGTCGTTTTACCTTCCCTTGACGCCTCCCG |
| JOHE117342 | GGTGGAAGCGGAGACTAAGC |
| JOHE117343 | CTGGCCGTCGTTTTACGTGGGAGGATATATAGATGGGC |
| JOHE117344 | GTCATAGCTGTTTCCTGGCCCCGTAAACGCTCCGAC |
| JOHE117345 | GCCTCAGACAGCCTCCACTC |
| JOHE117360 | CGAAGAACAAGCGGCCTGC |
| JOHE117361 | CTGGCCGTCGTTTTACCGTCGCTGGACATTGTCGTC |
| JOHE117362 | GTCATAGCTGTTTCCTGGGACCCAAACTTAGGCTTCG |
| JOHE117363 | GGCCACAGACCGAACGGAG |
| M13 Forward | GTAAAACGACGGCCAG |
| M13 Reverse | CAGGAAACAGCTATGAC |
Gene disruption and complementation.
To disrupt the GSY1 gene, an overlap PCR product with the NAT marker amplified from plasmid pAI1 (23) and 5′ and 3′ flanking sequences of the GSY1 locus from strain JEC21 (967 bp and 859 bp, respectively) was generated using primers JOHE117342, JOHE117343, JOHE117344, JOHE117345, M13 Forward, and M13 Reverse. The PCR product was directly introduced into strain JEC21 (MATα) by biolistic transformation. Homologous replacement mutants were screened by PCR and confirmed by Southern blotting. Isogenic MATa strains with the GSY1 deletion were obtained by selecting NAT-resistant a progeny from a cross between the mutant α strains and the MATa strain JEC20.
For complementation of the gsy1 mutation, an overlap PCR product with the NEO marker from pJAF1 (18) linked to the wild-type GSY1 gene from strain JEC21 was generated using primers JOHE117342 and JOHE13917, M13 Forward, and M13 Reverse. The PCR product was directly introduced into gsy1 mutants by biolistic transformation.
To disrupt the TPS1 gene, an overlap PCR product with the NAT marker amplified from plasmid pAI1 (23) and 5′ and 3′ flanking sequences of the TPS1 locus from strain JEC21 (1,092 bp and 1,046 bp, respectively) was generated using primers JOHE117360, JOHE117361, JOHE117362, JOHE117363, M13 Forward, and M13 Reverse. The PCR product was directly introduced into strain JEC21 (MATα) by biolistic transformation. Homologous replacement mutants were screened by PCR and confirmed by Southern blot analysis. Isogenic MATa strains with the tps1 deletion were obtained by selecting NAT-resistant a progeny from a cross between the mutant α strains and the MATa strain JEC20.
For complementation of the tps1 mutation, an overlap PCR product with the NEO marker from pJAF1 (18) linked to the wild-type TPS1 gene from strain JEC21 was generated using primers JOHE117360, JOHE13918, M13 Forward, and M13 Reverse. The PCR product was directly introduced into tps1 mutants by biolistic transformation.
Glycogen level test.
For the filaments, glycogen was stained with I2KI solution (60 mg/ml KI and 10 mg/ml I2). For yeast colonies, glycogen was stained with iodine vapor as previously described (49).
Genetic manipulations and mating type assay.
Crosses were performed on V8 juice agar medium (pH 7.0) in the dark at 22°C. Individual basidiospores were dissected by micromanipulation and transferred to YPD medium. From these subcultures, progeny clones were isolated for analysis.
To determine the mating type, each isolate was grown on YPD medium for two days at 30°C and separately cocultured with the parental tester strains, JEC20 (MATa) and JEC21 (MATα), on V8 medium. The isolate and tester strains alone were always cultured on the same plate as controls. The mating reactions were examined daily for mating hypha formation. To measure mating efficiency quantitatively, cell fusion efficiency was determined. Five microliters of the MATα ura5 lys1 strains were cocultured with the MATa ade2 strains at a concentration of 107 cells/ml on V8 medium supplemented with uracil, lysine, and adenine. At 24 and 48 h, cells were removed, washed, and plated on YNB medium to select prototrophic colonies at room temperature. CFU were counted to measure the efficiency of cell fusion.
RESULTS
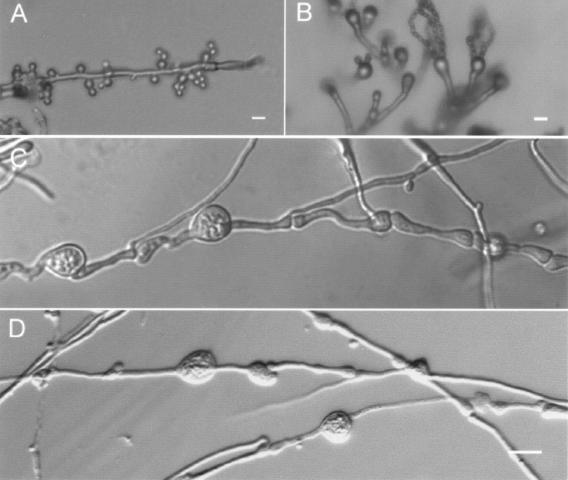
Chlamydospore formation during hyphal growth in Cryptococcus neoformans. In response to nutrient limitation, desiccation, darkness, and pheromone stimulation, Cryptococcus neoformans can switch from a single-celled yeast form to a multicellular filamentous form. During hyphal growth of this fungus, round mitotic blastospores are formed by mitotic budding from the edges of hyphal cells and oval meiotic basidiospores are produced in long chains on the surface of the basidium by basipetal budding (Fig. 1A and B). Close microscopic analysis of the sexual cycle, however, revealed that another type of structure could form either at the intercalary or terminal hyphae, and this structure resembled chlamydospores formed by other fungi (Fig. 1). These structures vary in size, but most are greater than 10 μm in diameter, significantly larger than the almost uniform 2- to 5-μm round blastospores or 1.5- to 3-μm oval basidiospores.
FIG. 1.
Chlamydospore structures associated with the filamentous growth of C. neoformans. (A) Blastospores bud off the hyphae. (B) Basidiospores on the basidium at the top of aerial hyphae. (C) Chlamydospores formed during monokaryotic fruiting. (D) Chlamydospores formed during mating. Cells of JEC21 (monokaryotic fruiting) or a JEC21 and JEC20 (mating) coculture were grown on V8 medium for 2 weeks at 22°C in the dark. Scale bars = 10 μm.
We were quite intrigued by these novel round structures observed during filamentous differentiation and considered three hypotheses. First, they might be water droplets whose role is to survive the desiccating conditions normally required to trigger filamentous growth. Second, they might be enlarged hyphal cells that have responded to pheromone, similar to the dramatically enlarged MATa cells observed during confrontation assays in which MATa cells have undergone isotropic expansion, likely to enhance fusion with conjugation tubes produced by α cells. Third, they might represent a novel cell type, namely, chlamydospores. The structures were not dispersed by micromanipulation, indicating that the structures had a solid surface and thus were not water drops. Since we observed these structures associated with hyphal growth during both mating (produced by a cross between a and α cells) and monokaryotic fruiting (produced by α cells alone) (Fig. 1C and D), this suggests that these structures in C. neoformans are not likely to be swollen hyphal cells in response to opposite mating pheromones, in contrast to what occurs with the swollen MATa cells induced by α pheromone prior to cell fusion during mating. Manipulating or isolating these chlamydospores, however, presents a technical challenge due to their large size and physical connection with the hyphae. Unlike blastospores or basidiospores, these structures are not formed by budding; instead, these unusual structures appear to be formed by the conversion of hyphal compartments themselves, consistent with observations of chlamydospore generation in other fungi.
We find it quite remarkable that, despite nearly a century of investigation, including an appreciation of the sexual cycle for three decades and of the monokaryotic fruiting cycle for a decade, the chlamydospores of C. neoformans have not been previously reported. Yet these structures are readily apparent under a variety of conditions, including during mating, monokaryotic fruiting, and filamentous differentiation of diploid cells. These structures were produced during hyphal growth in both C. neoformans var. neoformans and var. grubii strains, two predominant varieties found in clinical isolates (10). These findings reveal that further detailed morphological analysis of these processes is clearly warranted, as well as studies to understand the formation and function of these unique specialized cells, including their possible roles in energy storage and orchestration and support of the differentiation cascades that are likely involved in the generation of infectious basidiospores.
Chlamydospores could be independent entities.
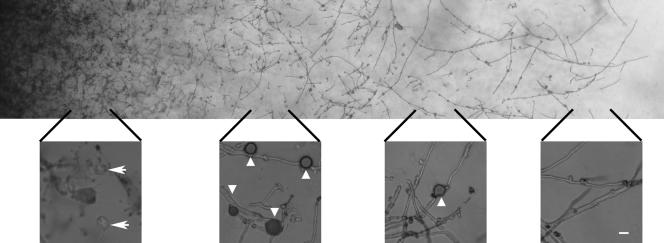
Indeed, the development of chlamydospores appears to be temporally and/or spatially regulated, as they first appear in the mycelial zone behind the actively growing hyphae during both mating and monokaryotic fruiting. Chlamydospores are predominantly produced before the production of abundant aerial hyphae (specialized hyphae bearing basidia), and become deflated (devoid of cytoplasm) in the center of an aged mycelium, where large numbers of basidiospores are produced and mature (Fig. 2).
FIG. 2.
Chlamydospore formation appears to be temporally controlled. Morphology of the filamentous mycelium of C. neoformans from the cross of strains JEC21 and JEC20 on V8 medium, with enlargement of the mycelial zones above. Triangles point to chlamydospores, and the arrowheads indicate deflated chlamydospores. Scale bars = 10 μm.
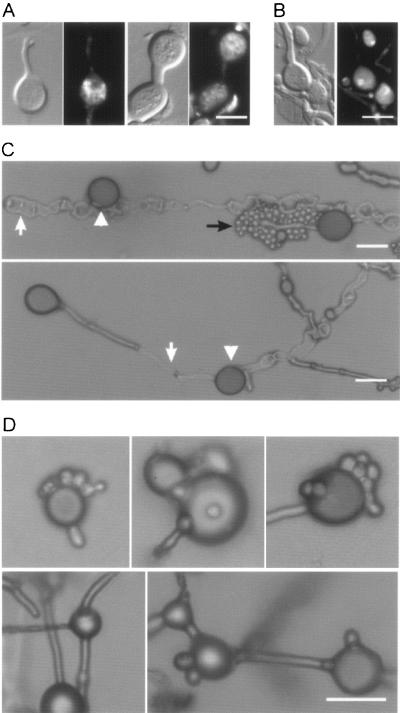
Because in S. cerevisiae, G0 arrest (a stage during which the cell cycle is arrested for an indefinite period) leads to the production of enlarged yeast cells filled with an enlarged vacuole, we hypothesized that the chlamydospore-like structures that we observed during filamentous growth could in fact represent G0-arrested hyphal compartments filled with an enlarged vacuole. Intracellular organelles can be visualized using FM4-64, a membrane-selective dye that is internalized via endocytosis. As shown in Fig. 3, these chlamydospore-like structures contain numerous small vesicles or internal organelles, suggesting that these structures are not G0-arrested enlarged hyphal compartments but are, on the contrary, metabolically active cells (Fig. 3A). To establish whether these structures contain genetic material, we stained the hyphae with DAPI and found that chlamydospores indeed contain nuclei (Fig. 3B), raising the possibility that these structures could be independently surviving cells. Interestingly, with prolonged incubation, these intercalary and terminal structures can be released from the hyphae after hyphal autolysis, again supporting the idea that these structures can be independent entities (Fig. 3C) and might survive longer than their producing hyphae. Furthermore, we observed that the chlamydospores could also give rise to yeast cells by budding or generate new hyphal branches, indicating that they are viable and capable of further cell division and reproduction (Fig. 3D). These observations raise the important question of their biological functions in the long-term survival for this pathogenic fungus in harsh environments.
FIG. 3.
Chlamydospores are metabolically active and could be independent structures. Chlamydospores were obtained from monokaryotic fruiting on V8 medium. (A) FM4-64 staining revealing internal organelles. (Left) DIC. (Right) Fluorescent images of the same field. (B) DAPI staining revealing nuclei. (Left) DIC. (Right) Fluorescent images of the same field. (C) Examples of released chlamydospores after hyphal autolysis. (D) Chlamydospores can generate yeast cells and new branches. (Upper panel) Yeast cells formed by budding from chlamydospores. (Lower panel) Branches generated from chlamydospores. Triangles indicate chlamydospores, white arrows indicate lysed hyphae, and black arrows indicate blastospores. Scale bars = 10 μm.
Chlamydospores are enriched in glycogen and may serve as energy stores.
As described above, chlamydospore formation in C. neoformans is first initiated near the periphery of the mycelium prior to the production of basidiospores. The chlamydospores become deflated in the center of the mycelium from which basidiospores have matured. Therefore, we hypothesized that chlamydospores may serve as an energy storage structure for basidiospore production and/or maturation. Nutrient limitation is one of the critical factors triggering the mating and monokaryotic fruiting in C. neoformans that lead to filamentous growth and fruiting body formation. Since nutrient transport in the long vegetative mycelium becomes inefficient as the mycelium expands, there may exist some stages of basidiospore formation or maturation that rely on stored carbon sources rather than their transport over longer distances. It is possible that the formation of energy-storing chlamydospores might serve to provide this type of stable energy source for sporulation through regulated translocation and degradation of its stored nutrients.
In order to begin answering these questions, we addressed whether chlamydospores of C. neoformans are energy storage structures. Because fungi can store energy in the form of lipids, we stained the mycelium with the lipid body dye Nile Red and found that the lipid accumulation level in chlamydospores is comparable with that in other sections of the hyphae (data not shown). We then examined whether these structures are rich in complex carbohydrates by staining the mycelium with iodine, which stains glycogen and starch. We found that the chlamydospores are highly enriched in iodine-stainable substances compared with other regions of the hyphae (Fig. 4A). Since fungi, like animals, store energy primarily in the form of glycogen, whereas plants store energy in the form of starch, this result suggests that the chlamydospore may serve as an energy store by accumulating glycogen.
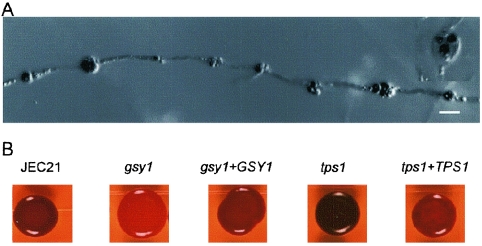
FIG. 4.
(A) Chlamydospores are enriched in glycogen. Filaments from monokaryotic fruiting of strain JEC21 were stained with an I2-KI solution. (B) Altered glycogen levels in the gsy1 and tps1 mutant strains. XL467 and XL470 were grown on YPD medium for 2 days at 22°C and then stained with iodine vapor. Scale bar = 10 μm.
What is the purpose of this energy storage? One possibility is that the fungus may use this specialized energy storage structure for basidiospore production and/or their maturation, as hypothesized above. Alternatively, energy stored in the chlamydospores could act purely as a carbon reserve for chlamydospores to promote their own long-term survival or reproduction. In many fungi, chlamydospores are long-term survival structures produced in response to harsh environments and a sufficient endogenous energy supply is essential for chlamydospores to conduct their survival functions. Moreover, it is also possible that energy stored in chlamydospores produced by C. neoformans during filamentous growth could serve both purposes, and which function they play may depend on the appropriate environmental cues. Characterization of mutants with altered glycogen levels can further elucidate the relationships between energy storage, formation of chlamydospores, and sporulation in C. neoformans.
The gsy1 and tps1 mutants show altered levels of cellular glycogen.
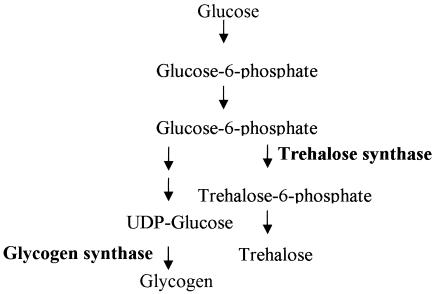
Glycogen metabolism has been shown to be linked to sporulation in some fungi (7, 8, 12, 17, 21, 24, 42, 51). Because both glycogen and trehalose are products of branched glycogenesis, they share some common precursors (Fig. 5) and there are possible links between glycogen and trehalose metabolism during growth and differentiation (7, 16, 37, 38, 45, 48, 51, 52). We therefore decided to isolate both glycogen synthase and trehalose synthase mutants to study the effects of altered glycogen levels on the development of chlamydospores in C. neoformans.
FIG. 5.
Schematic pathway of glycogen and trehalose synthesis (16). Both trehalose synthase and glycogen synthase were mutated to test the role of glycogen in chlamydospore formation.
The glycogen synthase gene GSY1 and the trehalose synthase gene TPS1 of C. neoformans were identified by BLAST searches with the S. cerevisiae orthologous GSY1 and TPS1 genes against the C. neoformans genome (http://www.tigr.org/tdb/e2k1/cna1/). Both genes showed high homology with the yeast counterparts (57.3% identity for Gsy1 and 46.1% identity for Tps1). The two genes were replaced with a drug resistance marker through biolistic transformation and homologous recombination. Disruption mutants were confirmed by PCR and Southern blot analysis (data not shown). As expected, the gsy1 mutant produces much less glycogen than the wild type (Fig. 4B). The tps1 mutant, however, accumulates more glycogen than the wild type. Reintroduction of the wild-type copy of either the GSY1 or the TPS1 gene restored each respective mutant to the wild-type phenotype. These mutant phenotypes are similar to those observed in S. cerevisiae in which tps1 mutants hyperaccumulate glycogen and have elevated levels of glycogen synthase activity (9). This could be due to increased substrate availability for glycogen synthesis when the trehalose synthesis pathway is blocked. The TPS1 gene has also been shown to be essential for virulence in C. neoformans var. grubii strains (54). Our findings reveal that the gsy1 and tps1 mutations have opposing effects on the accumulation of cellular glycogen levels.
The gsy1 and tps1 mutants still form chlamydospores during mating yet have differing effects on filamentation and sporulation.
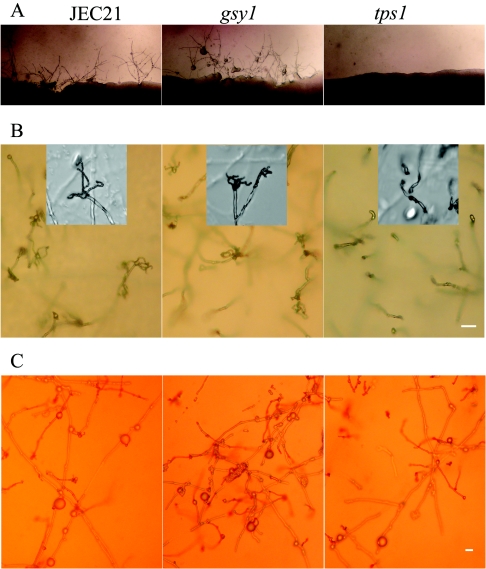
Deletions of the GSY1 and TPS1 genes conferred opposing phenotypes in many respects. As mentioned above, the gsy1 mutant accumulates low levels of glycogen, while the tps1 mutant contains high glycogen levels. Furthermore, the tps1 mutation blocks monokaryotic fruiting, while the gsy1 mutation does not (Fig. 6A). Although both mutants do form filaments during bilateral mating (mutant α cells crossed with mutant a cells), the tps1 mutant displayed a reduction in filamentation. This is in part due to the fact that cell fusion in the tps1 mutant is less efficient (10% of the wild-type level [data not shown]). The gsy1 mutant mated and sporulated normally, while sporulation in the tps1 mutant was blocked and development was arrested at the stage of basidium formation (Fig. 6B). Surprisingly, both mutants formed chlamydospores during mating (Fig. 6C), indicating that although glycogen is stored in chlamydospores, altered glycogen levels do not affect the formation of chlamydospores under these conditions; thus, events beyond simple glycogen accumulation must be responsible for the formation of these structures. These data also indicate that chlamydospore formation itself is not sufficient to support sporulation in C. neoformans given that the tps1 mutant forms chlamydospores but not basidiospores.
FIG. 6.
Effects of the gsy1 and tps1 mutations on filamentation, chlamydospore formation, and sporulation. (A) The tps1 mutant is blocked in filamentation during monokaryotic fruiting. From left to right are wild-type JEC21 (α), XL467 (α gsy1), and XL470 (α tps1). Cells were cultured on V8 medium for 1 week. (B) The tps1 mutant is blocked in sporulation during bilateral mating. From left to right are crosses between JEC21 and JEC20, XL467 (α gsy1) and XL466 (a gsy1), and XL470 (α tps1) and XL468 (a tps1). Insets are enlarged images of basidia of corresponding mating cultures. Cells were cultured on V8 medium for 2 weeks. (C) Chlamydospore formation in a gsy1 and tps1 mutant during bilateral mating is normal. The order is the same as in panel B. Scale bars = 10 μm.
Chlamydospore formation in C. neoformans is species specific.
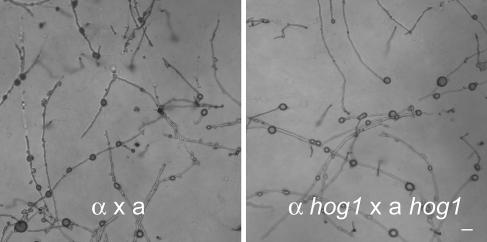
In the dimorphic fungal pathogen C. albicans, where some molecular insights into chlamydospore formation are available, the mitogen-activated protein kinase Hog1p is required for chlamydospore formation, as are genes involved in morphogenesis (2, 46). Although Hog1 has also been shown to be a key regulator of stress responses in C. neoformans as in other fungi (5), the hog1 mutation surprisingly did not affect chlamydospore formation in C. neoformans (Fig. 7) and neither did mutations in the cyclic AMP signaling pathway (data not shown). In a recent study, a panel of 217 C. albicans insertional mutants were screened for defects in chlamydospore formation and several genes were identified (39), suggesting that chlamydospore development in Candida is a complex process regulated by multiple genes. In contrast, we screened 2,500 random insertional mutants produced by Agrobacterium transformation and did not find any single mutation that blocks chlamydospore formation specifically; that is, all of the insertional mutants that produced monokaryotic hyphae also produced chlamydospores (data not shown). We did, on the other hand, find mutations that block filamentation and, by extension, chlamydospore formation, which supports the hypothesis that chlamydospore formation is intimately associated with filamentous growth. The failure to find genes specifically controlling the production of chlamydospores suggests that their formation is an integral part of filamentous growth in C. neoformans or that redundancy in gene function associated with chlamydospore formation may mask developmental defects in strains with only single gene disruptions, as occurs in Agrobacterium-mediated mutagenesis. Moreover, there may be far fewer genes that regulate this process in C. neoformans and a larger screen may be needed to identify them. Our results therefore suggest that the regulation of chlamydospore development in C. neoformans may be fundamentally different from the process in C. albicans, necessitating species-specific research into the regulation of their production.
FIG. 7.
The mitogen-activated protein kinase Hog1 does not control chlamydospore formation in C. neoformans during mating. Chlamydospores on bilateral mating filaments of wild-type strains JEC21 and JEC20 (A) and the hog1 mutant strains YSB139 and YSB143 (B) on V8 medium. Scale bar = 10 μm.
Besides this difference in genetic regulation, the chlamydospores of C. neoformans are very different from those of C. albicans in that the chlamydospores observed in C. neoformans are likely to play a more dynamic role in cell physiology and differentiation. First, chlamydospores of C. neoformans are more commonly observed to be intercalary instead of terminal, in contrast to the terminal chlamydospores produced by C. albicans. Second, C. albicans chlamydospores may have only very transient viability, and the germination of C. albicans chlamydospores is very rarely observed (43). By contrast, chlamydospores of C. neoformans are viable and capable of further cell divisions (Fig. 3D). These findings suggest that the roles of chlamydospores are likely to be distinct in different fungi and in different pathogenic fungi, and thus their functions and the pathways that give rise to them will require direct identification and analysis in each system.
DISCUSSION
A clearer understanding of the reproductive modes and survival structures of fungal pathogens in the environment is of great importance with regard to their ecology and epidemiology. Budding, conidiation, sporulation, fragmentation of hyphae, and conversion of hyphal elements into chlamydospores are common modes of reproduction. In some soilborne fungi, chlamydospores have been documented to have a role as survival structures. This study is the first report of C. neoformans chlamydospores, which are produced behind the active hyphal growth zone during filamentous growth, and it elucidates a novel stage of the life cycle for this pathogen. Although chlamydospores have been observed in the related fungus Cryptococcus laurentii (30), it is, however, dangerous to extrapolate to other Cryptococcus species, as in the case in Candida species. Among all the Candida species, only the two most prevalent pathogens, Candida albicans and Candida dubliniensis, have been observed to produce chlamydospores, while other Candida species, such as Candida tropicalis, Candida glabrata, Candida parapsilosis, Candida krusei, Candida lusitaniae, Candida kefyr, and Candida guilliermondii, do not.
Filaments of C. neoformans produce both intercalary and terminal chlamydospores, which are potentially fully functional (independent from the mycelium) and physiologically active, as they are capable of generating new branches and yeast cells. Although we found that chlamydospores in C. neoformans are enriched in glycogen, mutants with altered glycogen levels still form chlamydospores, suggesting the possibility for the storage of other materials and regulatory mechanisms independent of glycogen accumulation. Whether the energy stored in the chlamydospores is for their own survival and reproduction or to support proficient basidiospore production and/or maturation is an important unanswered question. Since we were unable to identify any single mutation that specifically blocks chlamydospore formation, the exact nature of the process that leads to chlamydospore production and the biological function of the structures remains to be defined. The inability of our genetic screen to separate the formation of chlamydospores from filamentous growth suggests that chlamydospore production could be an integral part of hyphal growth in C. neoformans in response to harsh environments.
Basidiospores are proposed to be the propagules for C. neoformans dispersal and infection, and it is likely that basidiospores are also long-term survival structures in nature. However, in many other fungi, chlamydospores serve this role. Our identification of chlamydospores in C. neoformans suggests the interesting possibility of an overlooked role for these structures in the popular model of C. neoformans survival and propagation. It is possible that these two different reproduction modes of C. neoformans coexist in nature and serve independent biological roles, or alternatively, there may be a key connection between the formation of chlamydospores and the production of basidiospores. While we have shown that the formation of chlamydospores is apparently independent of the production of basidiospores, given that the tps1 mutant is blocked in sporulation during mating but can still form chlamydospores, this does not mean that sporulation is independent of chlamydospore formation. The availability of large-scale screens of insertional mutants or of a genome-wide deletion mutation collection in C. neoformans may yield insight into the relationship between these two processes and provide a model for chlamydospore production in related fungi.
During our small-scale screen of insertion mutants, we did notice an inverse relationship between blastospore and chlamydospore production by vegetative hyphae, suggesting that there may be a balance between the formation of these two reproductive forms. We also observed that robust blastospore formation is usually associated with suppressed hyphae, poor aerial hyphal production, and sporulation, while chlamydospore formation is associated with better aerial hyphae production and sporulation. The different developmental pathways to produce blastospores or chlamydospores might reflect the choice that the hyphae make, either to maintain vegetative growth and multiply rapidly or to enter terminal growth, leading to the production of aerial hyphae and basidiospores. The balance between the three reproduction forms (blastospores, chlamydospores, and basidiospores) may be dependent on genetic background, developmental stages, and environmental cues that require further clarification and may yield new clues to the nature and formation of infectious C. neoformans propagules.
Acknowledgments
This work was supported by NIAID R01 grant AI50113 to J.H. J.H. was a Burroughs Welcome Fund Scholar in Molecular Pathogenic Mycology and an Investigator of the Howard Hughes Medical Institute.
We thank Yong-Sun Bahn for strains; John Perfect, Alex Idnurm, James Fraser, Weihua Fan, and Felicia Walton for critical reading; and Arturo Casadevall, June Kwon-Chung, and Andrew Alspaugh for comments.
REFERENCES
- 1.Abou-Gabal, M., and J. Fagerland. 1981. Ultrastructure of the chlamydospore growth phase of Aspergillus parasiticus associated with higher production of aflatoxins. Mykosen 24:307-311. [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., F. Navarro-Garcia, E. Roman, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5:217-238. [DOI] [PubMed] [Google Scholar]
- 4.Anstead, G. M., D. A. Sutton, E. H. Thompson, I. Weitzman, R. A. Otto, and S. K. Ahuja. 1999. Disseminated zygomycosis due to Rhizopus schipperae after heatstroke. J. Clin. Microbiol. 37:2656-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barran, L. R., E. F. Schneider, and W. L. Seaman. 1977. Requirements for the rapid conversion of macroconidia of Fusarium sulphureum to chlamydospores. Can. J. Microbiol. 23:148-151. [DOI] [PubMed] [Google Scholar]
- 7.Brana, A. F., C. Mendez, L. A. Diaz, M. B. Manzanal, and C. Hardisson. 1986. Glycogen and trehalose accumulation during colony development in Streptomyces antibioticus. J. Gen. Microbiol. 132:1319-1326. [DOI] [PubMed] [Google Scholar]
- 8.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 9.Cannon, J. F., J. R. Pringle, A. Fiechter, and M. Khalil. 1994. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics 136:485-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 11.Chabasse, D., J. P. Bouchara, L. de Gentile, and J. M. Chennebault. 1988. Candida albicans chlamydospores observed in vivo in a patient with AIDS. Ann. Biol. Clin. (Paris) 46:817-818. [PubMed] [Google Scholar]
- 12.Chakraborty, T. K., N. Das, and M. Mukherjee. 2003. Evidences of high carbon catabolic enzyme activities during sporulation of Pleurotus ostreatus (Florida). J. Basic Microbiol. 43:462-467. [DOI] [PubMed] [Google Scholar]
- 13.Cole, G. T., K. R. Seshan, M. Phaneuf, and K. T. Lynn. 1991. Chlamydospore-like cells of Candida albicans in the gastrointestinal tract of infected, immunocompromised mice. Can. J. Microbiol. 37:637-646. [DOI] [PubMed] [Google Scholar]
- 14.Couteaudier, Y., and C. Alabouvette. 1990. Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f. sp. lini in soil. Can. J. Microbiol. 36:551-556. [DOI] [PubMed] [Google Scholar]
- 15.Croan, S. C., H. H. Burdsall, Jr., and R. M. Rentmeester. 1999. Preservation of tropical wood-inhabiting basidiomycetes. Mycologia 91:908-916. [Google Scholar]
- 16.De Silva-Udawatta, M. N., and J. F. Cannon. 2001. Roles of trehalose phosphate synthase in yeast glycogen metabolism and sporulation. Mol. Microbiol. 40:1345-1356. [DOI] [PubMed] [Google Scholar]
- 17.Fonzi, W. A., M. Shanley, and D. J. Opheim. 1979. Relationship of glycolytic intermediates, glycolytic enzymes, and ammonia to glycogen metabolism during sporulation in the yeast Saccharomyces cerevisiae. J. Bacteriol. 137:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrison, R. G., and K. S. Boyd. 1975. Aspects of the dimorphism of Histoplasma farciminosum: a light and electron microscopic study. Sabouraudia 13:174-184. [DOI] [PubMed] [Google Scholar]
- 20.Gronvold, J., P. Nansen, S. A. Henriksen, M. Larsen, J. Wolstrup, J. Bresciani, H. Rawat, and L. Fribert. 1996. Induction of traps by Ostertagia ostertagi larvae, chlamydospore production and growth rate in the nematode-trapping fungus Duddingtonia flagrans. J. Helminthol. 70:291-297. [DOI] [PubMed] [Google Scholar]
- 21.Hopper, A. K., P. T. Magee, S. K. Welch, M. Friedman, and B. D. Hall. 1974. Macromolecule synthesis and breakdown in relation to sporulation and meiosis in yeast. J. Bacteriol. 119:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 23.Idnurm, A., J. L. Reedy, J. C. Nussbaum, and J. Heitman. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3:420-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane, S. M., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kertesz-Chaloupkova, K., P. J. Walser, J. D. Granado, M. Aebi, and U. Kues. 1998. Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet. Biol. 23:95-109. [DOI] [PubMed] [Google Scholar]
- 26.Khan, Z. U., S. Ahmad, E. Mokaddas, and R. Chandy. 2004. Tobacco agar, a new medium for differentiating Candida dubliniensis from Candida albicans. J. Clin. Microbiol. 42:4796-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kono, Y., H. Yamamoto, M. Takeuchi, and H. Komada. 1995. Alterations in superoxide dismutase and catalase in Fusarium oxysporum during starvation-induced differentiation. Biochim. Biophys. Acta 1268:35-40. [DOI] [PubMed] [Google Scholar]
- 28.Kües, U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64:316-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kües, U., J. D. Granado, R. Hermann, R. P. Boulianne, K. Kertesz-Chaloupkova, and M. Aebi. 1998. The A mating type and blue light regulate all known differentiation processes in the basidiomycete Coprinus cinereus. Mol. Gen. Genet. 260:81-91. [DOI] [PubMed] [Google Scholar]
- 30.Kurtzman, C. P. 1973. Formation of hyphae and chlamydospores by Cryptococcus laurentii. Mycologia 65:388-395. [PubMed] [Google Scholar]
- 31.Kwon-Chung, K. J. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197-1200. [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., J. E. Bennett, and J. C. Rhodes. 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek 48:25-38. [DOI] [PubMed] [Google Scholar]
- 33.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, X., C. M. Hull, and J. Heitman. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017-1021. [DOI] [PubMed] [Google Scholar]
- 35.Lin, X., and M. Momany. 2003. The Aspergillus nidulans swoC1 mutant shows defects in growth and development. Genetics 165:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, S. W., L. M. Douglas, and J. B. Konopka. 2005. Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot. Cell 4:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride, M. J., and D. R. Zusman. 1989. Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J. Bacteriol. 171:6383-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neves, M. J., S. Hohmann, W. Bell, F. Dumortier, K. Luyten, J. Ramos, P. Cobbaert, W. de Koning, Z. Kaneva, and J. M. Thevelein. 1995. Control of glucose influx into glycolysis and pleiotropic effects studied in different isogenic sets of Saccharomyces cerevisiae mutants in trehalose biosynthesis. Curr. Genet. 27:110-122. [DOI] [PubMed] [Google Scholar]
- 39.Nobile, C. J., V. M. Bruno, M. L. Richard, D. A. Davis, and A. P. Mitchell. 2003. Genetic control of chlamydospore formation in Candida albicans. Microbiology 149:3629-3637. [DOI] [PubMed] [Google Scholar]
- 40.Ohara, T., and T. Tsuge. 2004. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot. Cell 3:1412-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paraud, C., H. Hoste, Y. Lefrileux, A. Pommaret, V. Paolini, I. Pors, and C. Chartier. 2005. Administration of Duddingtonia flagrans chlamydospores to goats to control gastro-intestinal nematodes: dose trials. Vet. Res. 36:157-166. [DOI] [PubMed] [Google Scholar]
- 42.Ramaswamy, N. T., L. Li, M. Khalil, and J. F. Cannon. 1998. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics 149:57-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raudonis, B. M., and A. G. Smith. 1982. Germination of the chlamydospores of Candida albicans. Mycopathologia 78:87-91. [DOI] [PubMed] [Google Scholar]
- 44.Regulez, P., J. Ponton, J. B. Dominguez, F. M. Goni, and F. Uruburu. 1980. Lipid composition and the transition from yeast-like to chlamydospore cells of Pullularia pullulans. Can. J. Microbiol. 26:1428-1437. [DOI] [PubMed] [Google Scholar]
- 45.Rueda, B., E. M. Miguelez, C. Hardisson, and M. B. Manzanal. 2001. Changes in glycogen and trehalose content of Streptomyces brasiliensis hyphae during growth in liquid cultures under sporulating and non-sporulating conditions. FEMS Microbiol. Lett. 194:181-185. [DOI] [PubMed] [Google Scholar]
- 46.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staib, P., and J. Mörschhauser. 2005. Differential expression of the NRG1 repressor controls species-specific regulation of chlamydospore development in Candida albicans and Candida dubliniensis. Mol. Microbiol. 55:637-652. [DOI] [PubMed] [Google Scholar]
- 48.Teusink, B., J. Passarge, C. A. Reijenga, E. Esgalhado, C. C. van der Weijden, M. Schepper, M. C. Walsh, B. M. Bakker, K. van Dam, H. V. Westerhoff, and J. L. Snoep. 2000. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267:5313-5329. [DOI] [PubMed] [Google Scholar]
- 49.Thines, E., R. W. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandercammen, A., J. M. Francois, B. B. Torres, J. C. Maia, and H. G. Hers. 1990. Fructose 2,6-bisphosphate and carbohydrate metabolism during the life cycle of the aquatic fungus Blastocladiella emersonii. J. Gen. Microbiol. 136:137-146. [DOI] [PubMed] [Google Scholar]
- 52.Wannet, W. J., E. M. Aben, C. van der Drift, L. J. Van Griensven, G. D. Vogels, and H. J. Op den Camp. 1999. Trehalose phosphorylase activity and carbohydrate levels during axenic fruiting in three Agaricus bisporus strains. Curr. Microbiol. 39:205-210. [DOI] [PubMed] [Google Scholar]
- 53.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills, E. A., U. Himmelreich, E. Mylonakis, and J. R. Perfect. 2003. Abstr. 15th Congr. International Society of Human and Animal Mycology, p. 126.
- 55.Wolf, J. C., and S. A. Smith. 1999. Systemic zygomycosis in farmed tilapia fish. J. Comp. Pathol. 121:301-306. [DOI] [PubMed] [Google Scholar]