Abstract
The α-amino acid ester hydrolase from Acetobacter turbidans ATCC 9325 is capable of hydrolyzing and synthesizing β-lactam antibiotics, such as cephalexin and ampicillin. N-terminal amino acid sequencing of the purified α-amino acid ester hydrolase allowed cloning and genetic characterization of the corresponding gene from an A. turbidans genomic library. The gene, designated aehA, encodes a polypeptide with a molecular weight of 72,000. Comparison of the determined N-terminal sequence and the deduced amino acid sequence indicated the presence of an N-terminal leader sequence of 40 amino acids. The aehA gene was subcloned in the pET9 expression plasmid and expressed in Escherichia coli. The recombinant protein was purified and found to be dimeric with subunits of 70 kDa. A sequence similarity search revealed 26% identity with a glutaryl 7-ACA acylase precursor from Bacillus laterosporus, but no homology was found with other known penicillin or cephalosporin acylases. There was some similarity to serine proteases, including the conservation of the active site motif, GXSYXG. Together with database searches, this suggested that the α-amino acid ester hydrolase is a β-lactam antibiotic acylase that belongs to a class of hydrolases that is different from the Ntn hydrolase superfamily to which the well-characterized penicillin acylase from E. coli belongs. The α-amino acid ester hydrolase of A. turbidans represents a subclass of this new class of β-lactam antibiotic acylases.
In the search for microorganisms to be used in the biocatalytic production of semisynthetic antibiotics, Acetobacter turbidans ATCC 9325 was first described in 1972 by Takahashi et al. (35) as an organism able to synthesize cephalosporins. Since only α-amino acid derivatives could act as substrates and due to the preference for esters over amides, the enzyme involved was named α-amino acid ester hydrolase (AEH) (34).
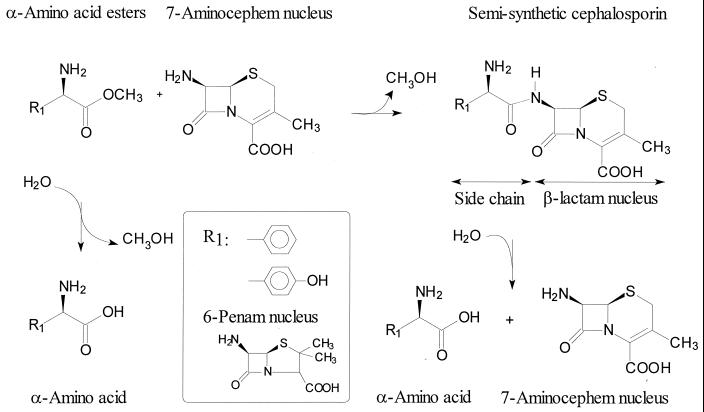
A similar AEH (EC 3.1.1.43) activity has been described for several other organisms. These enzymes catalyze the transfer of the acyl group from α-amino acid esters to amine nucleophiles such as 7-aminocephem and 6-penam compounds (synthesis) or to water (hydrolysis) (Fig. 1). Presumably, an acyl-enzyme intermediate is involved in this transfer reaction (9, 34). These AEHs show promising properties for the industrial enzymatic production of semisynthetic β-lactam antibiotics. Due to the preference for esters, it is conceivable that higher product (amide) accumulation can be reached in synthesis reactions using these enzymes than with the Escherichia coli penicillin G acylase (EC 3.5.1.11) (15, 28, 34). Moreover, the enzyme of A. turbidans showed high selectivity toward the d-form of phenylglycine methyl ester (d-PGM, the activated acyl donor). This enables an ampicillin synthesis starting from a racemic mixture of acyl donors, which is not feasible with the E. coli penicillin acylase (14). In contrast to penicillin acylase from E. coli, it was claimed that the AEHs are able to accept charged substrates (9). The low pH optimum of the α-amino acid ester hydrolases, i.e., pH 6, compared to a pH of 7.5 to 8 for penicillin G acylases (20), and the lack of inhibition by phenylacetic acid (9), a side product from the hydrolysis of penicillin G, are also interesting properties for biocatalytic applications.
FIG. 1.
Example of synthesis and hydrolysis of β-lactam antibiotics catalyzed by the AEH of A. turbidans. Shown as a 7-aminocephem nucleus is 7-aminodesacetoxycephalosporanic acid, and 6-aminopenicillanic acid is depicted as a 6-penam nucleus.
The structural characterization of the AEHs is limited to the determination of the subunit size and the quaternary structure. The AEHs from A. turbidans ATCC 9325 (29, 34), Xanthomonas citri IFO 3835 (16) and Pseudomonas melanogenum IFO 12020 (18) have been purified. All three enzymes have similar subunit sizes of either 70 or 72 kDa or both. However, there is some dissimilarity in the native subunit compositions, which were reported to be α2β2 for A. turbidans (29), α4 for X. citri (16), and α2 for P. melanogenum (18). Cloning of the gene which encodes AEH and using it for overproduction would be worthwhile since expression in the natural hosts is low, varying from 0.1 to 2% of the total cellular protein (16, 18, 29, 34; this study). In the past, effort was put into cloning an aehA gene, but this was unsuccessful (3, 25).
In this paper we describe the cloning and genetic characterization of the AEH of A. turbidans ATCC 9325. We succeeded in producing active AEH in E. coli and report some kinetic and structural properties of the purified recombinant protein. The sequence analysis showed that the α-amino acid ester hydrolase is a member of a new class of β-lactam antibiotic acylases.
MATERIALS AND METHODS
Materials.
d-2-Nitro-5-[(phenylglycyl)amino]benzoic acid (NIPGB) was obtained from Syncom (Groningen, The Netherlands). 6-Aminopenicillanic acid (6-APA), 7-aminodesacetoxycephalosporanic acid (7-ADCA), d-phenylglycine methyl ester (d-PGM), d-phenylglycine amide (d-PGA), d-4-hydroxyphenylglycine methyl ester (d-HPGM), amoxicillin, cefadroxil, glutaryl 7-aminocephalosporanic acid (glutaryl 7-ACA), adipoyl 7-ADCA, and cephalexin were provided by DSM Life Sciences (Delft, The Netherlands). The oligonucleotides for cloning of the aehA gene were provided by Eurosequence BV (Groningen, The Netherlands).
Bacterial strains, plasmids, and growth conditions.
A. turbidans ATCC 9325 was grown at 30°C in a 10-liter fermentor on the medium described by Takahashi et al. (34) without the addition of antifoam. Bacto Peptone was purchased from Difco (Sparks, Nev.), and the meat extract was obtained from Fluka (Buchs, Switzerland). E. coli strains were grown in shake flasks at 30°C on Luria-Bertani medium. Antibiotics were added to the media at the following final concentrations: tetracycline (Tc), 12.5 μg/ml; kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 34 μg/ml; and ampicillin (Ap), 50 μg/ml. When required, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 to 1 mM. E. coli HB101 (10) was used for cloning derivatives of pLAFR3 (32) and pEC (DSM Life Sciences). E. coli strains BL21(DE3)pLysS (Promega) and TOP10F’ (Invitrogen, Leek, The Netherlands) were used for cloning derivatives of pET9 (Promega Corporation, Madison, Wis.) and pCR-TOPO (Invitrogen), respectively.
DNA manipulation and sequencing.
All chemicals used in DNA manipulation procedures were purchased from Roche Diagnostics GmbH (Mannheim, Germany) and used as recommended by the manufacturer. The DNA sequences were determined at the Department of Medical Biology of the University of Groningen.
Isolation of α-amino acid ester hydrolase from A. turbidans.
Cells of A. turbidans were harvested in the stationary phase by continuous centrifugation at 6,000 × g, washed twice with 10 mM potassium phosphate buffer (pH 6.2), and resuspended in this buffer. All further steps were carried out at 4°C. A cell extract was made by sonification, and cell debris was removed by centrifugation at 13,000 × g for 40 min. To the supernatant DNase and RNase (final concentration, 6 mg/liter each) were added in the presence of 5 mM MgSO4. The solution was incubated for 3 h under mild stirring and centrifuged at 50,000 rpm in a type 50 Ti rotor (Beckman) for 60 min and then applied to a CM Sepharose fast-flow column (5- by 15-cm column; Amersham Pharmacia Biotech Ltd., Hertfordshire, United Kingdom) preequilibrated with 10 mM K2HPO4-KH2PO4, pH 6.2. Prior to elution the nonbinding proteins were washed from the column with equilibration buffer. The retained protein eluted in a linear gradient of 0 to 1 M KCl (30 ml/min) at 0.2 M. Activity-containing fractions were pooled and (NH4)2SO4 was added to a final concentration of 1.5 M, after which the pool was loaded on a hydrophobic-interaction column (Resource Phenyl, 2.6 by 7.5 cm; Amersham Pharmacia) pre-equilibrated with 1.5 M (NH4)2SO4-50 mM sodium phosphate buffer, pH 6.2. After being washed with the equilibration buffer the AEH eluted at 0.8 to 0.68 M (NH4)2SO4 in a decreasing linear gradient from 1.5 to 0 M (NH4)2SO4 in 50 mM sodium phosphate buffer (pH 6.2) at 5 ml/min. Fractions that contained AEH were pooled and concentrated by ultrafiltration (YM30, Amicon bioseparations; Millipore, Bedford, Mass.) and loaded on a Superdex 200 HR 10/30 column (24-ml bed volume; Amersham Pharmacia). AEH was eluted at 1 ml/min in a 50 mM sodium phosphate buffer (pH 6.2) with 0.15 M NaCl.
Isolation of recombinant α-amino acid ester hydrolase from E. coli.
The recombinant AEH was purified from E. coli BL21(DE3)pLysS (Cmr) cells carrying the pETAT (Kmr) construct. The cells were harvested from two 2.5-liter cultures by centrifugation, and the crude extract was prepared as described above. The extract was loaded onto a DEAE Sepharose column (5- by 13-cm column; Amersham Pharmacia) pre-equilibrated with 50 mM sodium phosphate buffer, pH 6.2. The AEH activity was eluted from the column in the nonbinding fraction in the equilibration buffer at 30 ml/min. The activity was then applied to a ceramic hydroxyapatite column (2.6- by 11-cm column; Bio-Rad Laboratories, Hercules, Calif.), which was equilibrated with 50 mM sodium phosphate, pH 6.2. After being washed with the equilibration buffer, the AEH activity was eluted from the column at 275 mM sodium phosphate in a linear gradient of 50 to 500 mM sodium phosphate (pH 6.2) at 10 ml/min. The AEH was purified further to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) homogeneity by hydrophobic interaction and gel filtration chromatography as described above.
Preparation and screening of the A. turbidans genomic library.
Genomic DNA was isolated as described by Poelarends et al. (27). An incubation of 30 min at 37°C with proteinase K (0.10 mg/ml) after the first hour of incubation with SDS was added to the procedure. DNA of the cosmid pLAFR3 used for the construction of the gene library was isolated from E. coli HB101 according to the alkaline lysis method and purified by ultracentrifugation using a CsCl gradient (30). The chromosomal DNA of A. turbidans was partially digested with Sau3A to yield fragments with an average size of 30 to 50 kb. These fragments were ligated in the cosmid pLAFR3 (Tcr) which had been completely digested with BamHI and dephosphorylated with alkaline phosphatase. In vitro packaging and infection of E. coli HB101 was carried out according to the recommendations of the manufacturer (Roche). Recombinant clones were stored at −80°C in microtiter plates.
Colony hybridization was carried out essentially as described by Van Hylckama Vlieg et al. (38) with an AEH-specific probe. An incubation of the membrane with proteinase K for 30 min at room temperature after fixation of the DNA was included in the procedure. After hybridization at 68°C the membrane was washed with 2 × SSC (10× SSC is 1.5 M NaCl with 0.15 M sodium citrate)-0.1% SDS at room temperature and with 0.5× SSC-0.1% SDS for 15 min at 68°C. The digoxigenin (DIG)-labeled DNA was visualized with alkaline phosphatase and a chemiluminescence substrate, CPSD (C18H20ClO7PNa2; Roche) following the recommendations of the manufacturer.
PCR amplification of the DIG-labeled aehA probe.
Part of the aehA gene was initially cloned by PCR amplification from chromosomal DNA using the LA PCR in vitro cloning kit (TaKaRa Biomedicals, Takara Shuzo Co., Ltd., Otsu, Shiga, Japan) and a degenerated primer (pNTd) based on the N-terminal sequence of purified AEH, 5′-ATGGCNCCNGCNGCNGAYGCNGCNCARGCNCAYGA-3′ (Y is T or C; R is A or G; N is any base). The PCR products were isolated from gel (Qiaquick kit; Qiagen, GmbH, Hilden, Germany), cloned, and sequenced. A gene probe for the AEH gene was made with primers based on the DNA sequence of the PCR fragment that encoded the N terminus of AEH. The matching forward primer was 5′-CCGCTAAGCGTGCAGACCGGCAGC-3′ (upstream of pNTd), and the reverse primer was 5′-CATGCATACCGTGCCAGAACG-3′. These primers were used to amplify a 696-bp fragment (NTaehA) with Taq polymerase and the PCR DIG probe synthesis mix from Roche.
Cloning of aehA into an expression host.
For expression of the aehA gene in E. coli the vector pETAT (aehA cloned in pET9) was constructed. The aehA gene was cloned in the NdeI and BamHI site of pET9 using a forward primer based on the N-terminal sequence including the leader sequence in which an AsnI site is incorporated, 5′-CCGCCGCCGATTAATGGTGGGACAGATTACCCTTT-3′ (AsnI site underlined, start codon in bold) and a reverse primer in which a BamHI site was incorporated (underlined), 5′-ACCCATACTGGATCCTTACTGTTTCACAACCGGGAG-3′. The gene was also cloned without the N-terminal leader sequence, which was replaced by a methionine, using 5′-GGTCGCGCATTAATGGCTCCGGCAGCGGATGC-3′ (AsnI site underlined, start codon in bold) as a primer. After denaturation of the DNA (pLAFR3 (aehA)) the amplifications were established in 30 cycles of 30 s at 94°C, 1 min at 58°C, and 1.5 min at 72°C. Products were digested with AsnI and BamHI and ligated into pET9 cut with NdeI and BamHI. The ligation mixture was used to transform CaCl2-competent E. coli BL21(DE3)pLysS. The constructs were confirmed by sequence analysis. For cloning in the NdeI/HindIII site of pEC, the gene was amplified with the forward primers as described above and the reverse primer 5′-CATACTGGCAAGCTTTTACTGTTTCACAACCGGGAGCAG-3′ (HindIII site underlined, stop codon in bold).
N-terminal sequence determination and protein analysis.
For N-terminal sequence analysis, approximately 15 μg of protein was sliced from an SDS-PAGE gel. Eurosequence BV carried out further preparation of the sample and automated Edman degradation (Model 477A; Applied Biosystems).
Subunit composition determination.
The subunit composition was determined via dynamic light scattering using a DynaPro MS 80 Tc (Protein Solutions, Charlottesville, Va.) in combination with the Dynamics V4.0 software from Protein Solutions. A pure protein solution of 1.2 mg/ml in 50 mM sodium phosphate buffer, pH 6.2, was placed in the laser bundle at 20°C, and data were collected for 3 min.
Inactivation.
Stock solutions of the inhibitors phenylmethylsulfonyl fluoride (PMSF), 4-(2-aminoethyl) benzenesulfonyl fluoride (Pefabloc SC), and p-nitrophenyl p′-guanidinobenzoate · HCl (p-NPGB) of 100 mM were made in acetonitrile, 50 mM sodium phosphate buffer (pH 6.2), and dimethylformamide, respectively. The enzyme, at a final concentration of 2.5 μM (molecular size, 140 kDa), was incubated with the inhibitor for 15 min at 30°C. Inhibitors and their concentrations were as follows: PMSF, 2 mM; Pefabloc SC, 5 mM; and NPGB, 1 mM. The degree of inactivation was determined by measuring the remaining initial hydrolysis activity on NIPGB.
Enzyme assays and determination of kinetic constants.
Activity of AEH was routinely assayed at 30°C by following the hydrolysis of 15 mM NIPGB in a spectrophotometer at 405 nm in 50 mM phosphate buffer, pH 6.2 (2).
The synthesis of cephalexin was followed by high-performance liquid chromatography (HPLC) (2). Incubations were done at 30°C and contained 30 mM 7-ADCA and 15 mM D-PGM at pH 6.2 (50 mM sodium phosphate buffer). One cexU is defined as the amount of enzyme needed to produce 1 μmol of cephalexin per min. Before analysis the samples were quenched and diluted 50-fold by the addition of HPLC eluent (20 mM phosphate buffer [pH 3], 30% acetonitrile).
The initial rates (<10% conversion) of hydrolysis of all the substrates were determined by measuring product formation by HPLC (2) except for NIPGB, which was monitored as described above. The enzyme was incubated with varying concentrations in the range of 0 to 25 mM for cephalexin, ampicillin, HPGM, and cefadroxil, or 0 to 50 mM for D-PGM and NIPGB, or 0 to 10 mM for amoxicillin. Reactions were done at 30°C in 50 mM phosphate buffer, pH 6.2. The calculations involved nonlinear regression fitting (Scientist, Micromath) using Michaelis-Menten and substrate inhibition kinetics, and the calculated kinetic parameters are given with their standard deviations. The hydrolysis of PGA was measured at 5 and 50 mM and the kcat/Km was calculated from the initial linear slope of the Michaelis-Menten curve. Hydrolysis of glutaryl 7-ACA and adipoyl 7-ADCA was measured at 5 and 25 mM.
Nucleotide sequence accession number.
The nucleotide sequence from the AEH-has been submitted to GenBank and assigned accession no. AF439262.
RESULTS
Cloning of the gene (aehA) encoding the AEH of A. turbidans.
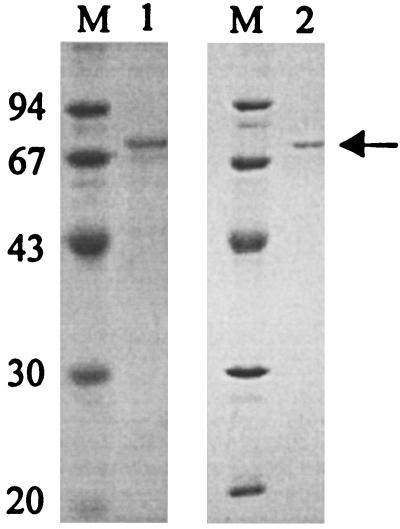
To obtain an N-terminal amino acid sequence, the AEH from A. turbidans was purified by ion-exchange, hydrophobic interaction, and gel filtration chromatography (Table 1). The native enzyme was found to be a multimer, as determined by gel filtration, varying from a dimer to a multiple of dimers, which is in agreement with earlier observations (29). Although the yield was rather low, a small amount of pure protein of 70 kDa, in agreement with the activity peak, was obtained, which was sufficient for SDS-PAGE and amino acid sequencing (Fig. 2, lane 1).
TABLE 1.
Purification of the AEH from A. turbidans ATCC 9325
| Purification step | Total vol (ml) | Total protein (mg) | Total activitya (cexU) | Sp acta (cexU/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| Cell extract | 165 | 461 | 599 | 1.3 | 1 | 100 |
| CM-Sepharose | 32 | 31 | 477 | 15 | 12 | 80 |
| Hydrophobic interaction | 5.8 | 1.1 | 68 | 62 | 48 | 11 |
| Gel filtration | 2.0 | 0.033 | 22 | 667 | 513 | 3.7 |
Cephalexin synthesis activity.
FIG. 2.
SDS-PAGE of AEH. The enzyme was purified from A. turbidans and E. coli as described in Materials and Methods. The pooled Superdex 200 fractions were analyzed by SDS-PAGE (12.5% separating and 2.5% stacking gel) and stained with Coomassie brilliant blue. The purified AEHs from A. turbidans (lane 1) and E. coli BL21(DE3)pLysS(pETAT) (lane 2) are shown. The band corresponding to AEH is indicated with an arrow. Molecular mass markers were loaded in lanes labeled M, and their masses (in kilodaltons) are shown at the left of the gels.
The N-terminal sequence of the 70-kDa subunit was determined to be Ala-Pro-Ala-Ala-Asp-Ala-Ala-Gln-Ala-His-Asp-Pro-Leu-Ser-Val-Gln-Thr-Gly-Ser-Asp-Ile-Pro. Based on the first 12 amino acids, and adding a starting methionine, a degenerated oligonucleotide primer (pNTd) was designed. From total DNA of A. turbidans a PCR product of 2.6 kb was obtained with the LA PCR in vitro cloning kit and pNTd. Sequence analysis of the 2.6-kb fragment indicated that the fragment contained the correct gene since downstream of the primer sequence, the DNA sequence encoded the remaining 10 amino acids of the determined N terminus of the protein.
Sequence analysis of the aehA gene and its region.
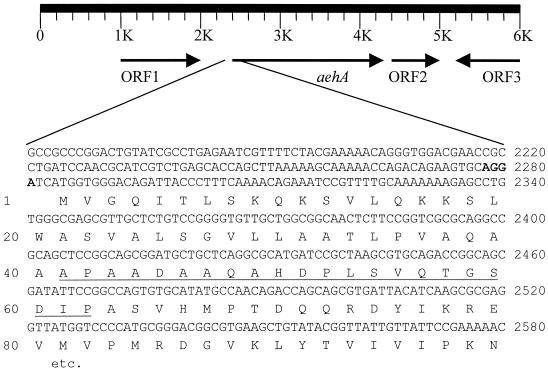
To ensure the completeness of the gene and to be able to study the surroundings of the aehA gene, a cosmid library was constructed and transduced to E. coli. A bank of 5,670 clones with 99.9% completeness was obtained and screened with a 696-bp DIG-labeled probe (NTaehA) based on part of the gene found in the 2.6-kb PCR product mentioned above. Of the 1,248 colonies screened, 2 hybridized with the probe. From one of these clones the cosmid was isolated, and 6 kb of its insert was sequenced (Fig. 3). Four open reading frames (ORFs) were identified, of which one harbored the determined N-terminal amino acid sequence and the sequence downstream of it found on the 2.6-kb PCR fragment. This gene (aehA) encoded a polypeptide with a molecular weight of 74,060, corresponding to a polypeptide of 667 amino acids. The determined N-terminal amino acid sequence was found at positions 41 to 62 (Fig. 3), indicating that the first 40 amino acids of the protein were cleaved off during maturation in A. turbidans.
FIG. 3.
Genetic organization around aehA and the N-terminal part of the protein and its nucleotide sequence. The deduced amino acid sequence is shown below the nucleotide sequence. The N-terminal sequence found by amino acid sequencing of the mature wild-type protein is underlined; the putative ribosome binding site is shown in bold. The ORFs are described in the text.
The genetic organization of the DNA region in which the aehA gene is situated could give indications about the biological function of the AEH. Identification of ORFs surrounding the ORF of the aehA gene was possible (Fig. 3) after a search for sequence similarity in the nonredundant database at the National Center for Biotechnology Information using BLASTX (4). Upstream of the aehA gene, ORF1, which has 56% identity to a phosphoserine aminotransferase from Methanosarcina barkeri (22), was detected. Downstream of the aehA gene a putative protein of 30 kDa (ORF2), which shows 22% identity with part of the creatinine amido hydrolase of Bacillus halodurans (protein ID no. BAB03945) (36), was found. Further downstream, in the opposite direction, the C-terminal part of a protein (ORF3), which has significant identity (50%) to a succinic semialdehyde dehydrogenase from Deinococcus radiodurans (protein ID no. BAA21377), was found.
Database searches using BLAST (4) indicated that the deduced amino acid sequence of aehA showed homology with several (putative) proteins, most of which originated from genome-sequencing projects and have unknown functions (Table 2; Fig. 4). The most closely related protein for which the activity is described is the intracellular cocaine esterase from the gram-positive strain Rhodococcus sp. strain MB1 (12). This enzyme hydrolyses the ester bond in cocaine resulting in benzoate and ecgonine methyl ester. The next-most-related studied enzyme is the glutaryl 7-ACA acid acylase of Bacillus laterosporus that hydrolyzes glutaryl 7-ACA acid to 7-amino-cephalosporanic acid. Activity of these enzymes for α-amino esters or β-lactam antibiotics carrying an α-amino acid acyl side chain has not been reported. The glutaryl-7-ACA acid acylase is composed of a polypeptide with a molecular size of 70 kDa, which corresponds to the size of the subunits found for AEH from A. turbidans (this study).
TABLE 2.
Amino acid sequence similarities of the AEH of A. turbidans ATCC 9325
| Protein IDa | Organism | Function (reference) | Identityb (%) | Size (kDa) | Motifc |
|---|---|---|---|---|---|
| AF439262 | Acetobacter turbidans | AEH precursor (this study) | 100 | 74.0 | GsSYeG |
| AAD29644 | Zymomonas mobilis | NDc | 60 | 73.4 | GsSYeG |
| AAF83839 | Xylella fastidiosa | ND | 61 | 75.9 | GsSYeG |
| AAF42807 | Rhodococcus sp. | Cocaine esterase (12) | 29 | 62.1 | GvSYlG |
| AAK23601 | Caulobacter crescentus | ND | 27 | 68.0 | GlSYga |
| AAK22924 | Caulobacter crescentus | ND | 28 | 69.1 | GcSssa |
| BAA10148 | Bacillus laterosporus | Glutaryl 7-ACA acylase precursor (6) | 26 | 72.2 | GlSYma |
| BAA17073 | Synechocystis sp. | ND | 25 | 60.0 | GfSYoG |
| CAB67720 | Streptomyces rochei | ND | 24 | 58.1 | GrSYea |
| AAC26133 | Streptomyces griseus | ND | 26 | 61.6 | GtSYoa |
| CAB03646 | Mycobacterium tuberculosis | ND | 26 | 60.3 | GlpYlG |
| CAB53331 | Streptomyces coelicolor | ND | 25 | 69.9 | GkSYda |
| CAB07817 | Mycobacterium tuberculosis | ND | 28 | 62.6 | GsSYla |
| CAB01467 | Mycobacterium tuberculosis | ND | 23 | 69.9 | GnSYdG |
| BAB53185 | Rhizobium loti | ND | 22 | 74.0 | GiSwgG |
| BAB51629 | Rhizobium loti | ND | 22 | 73.6 | GiSwgG |
| AAK05333 | Lactococcus lactis | ND | 27 | 64.9 | GtSYla |
| BAB43685 | Staphylococcus aureus | ND | 26 | 64.4 | GvSYla |
| AAF82828 | Xylella fastidiosa | ND | 38d | 91.1 | GgSYgG |
Accession number of the protein database of the National Center for Biotechnology Information.
Determined using the pairwise alignment option of BLAST with default settings of the parameters.
ND, not described.
This identity was found in a stretch of 100 residues.
Bold, conserved amino acids.
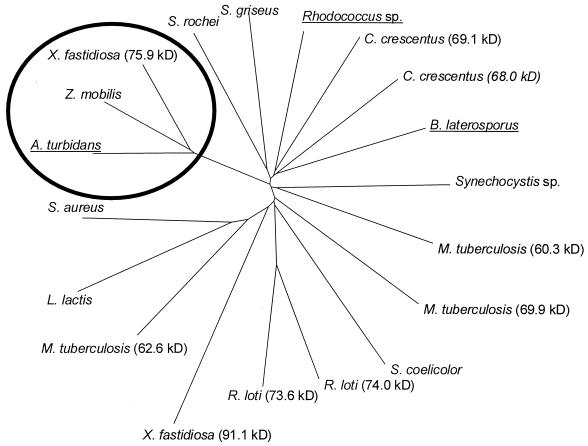
FIG. 4.
A dendrogram of proteins that are homologous to the AEH of A. turbidans as found by BLAST search. The distance can be read as the number of nucleotide substitutions per site. The proteins that share more than 60% sequence identity with AEH are circled. The proteins with described activity are underlined. The tree was constructed using Clustal W and TreeView.
An extended search for homologous proteins using the position-specific iterated PSI-BLAST program (5) indicated low identity (average 14%) to X-prolyl dipeptidyl aminopeptidases from Lactococcus and Lactobacillus strains. The X-prolyl dipeptidyl aminopeptidases belong to the peptidase_S15 family as defined by the Pfam database (8). These enzymes are serine proteases with the active-site serine located in the consensus sequence GxSYxG, where x is a nonconserved amino acid (13). The AEH shows conservation of this motif and its direct surroundings (Fig. 5). However, there is no significant overall similarity, which makes it impossible to judge if the proteins are structurally related.
FIG. 5.
Partial alignment of AEH with other (putative) serine hydrolases. The sequences shown are glutaryl-7-ACA-acylase precursor from B. laterosporus (protein ID no. I40217), cocaine esterase from Rhodococcus sp. (protein ID no. AAF42807), X-prolyl dipeptidyl aminopeptidases from Lactococcus lactis (protein ID no. AAA25207), Lactobacillus delbrueckii (protein ID no. CAB38074), and Lactobacillus helveticus (protein ID no. CAA88273). The alignment was performed with the pattern-induced (local) multiple alignment (PIMA 1.4) facilitated by the BCM search launcher on the World Wide Web. The region of relatively high identity among all seven proteins is boxed. The bold line indicates the GxSYxG motif.
Expression in E. coli and properties of the recombinant protein.
The cosmid clones that harbor the aehA gene and about 20 kb of surrounding DNA did not show any AEH activity, indicating that the enzyme was not expressed properly. The complete aehA gene of 2,004 bp was subcloned in the expression vectors pEC and pET9, which resulted in active constructs that were designated pAT and pETAT, respectively. This confirms that AEH is encoded by aehA and indicates that probably the bad positioning of the ribosome binding site (Fig. 3) or an unrecognized promoter caused the lack of expression from the cosmids in E. coli. Although overexpression was limited, due to the increased culture densities and the improved purification method, 11 times greater quantities of pure protein (Fig. 2, lane 2) per liter of culture were obtained from E. coli BL21(DE3)pLysS(pETAT) (Table 3) than from the natural host.
TABLE 3.
Purification of AEH from E. coli BL21(DE3)pLysS(pETAT)
| Purification step | Total vol (ml) | Total protein (mg) | Total activity (cexU) | Sp acta (cexU/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| Cell extract | 184 | 1174 | 2348 | 2 | 1 | 100 |
| DEAE | 195 | 60 | 2050 | 34 | 17 | 87 |
| CM-Hapb | 80 | 5.8 | 1109 | 191 | 96 | 47 |
| Hydrophobic interaction | 32 | 0.96 | 633 | 605 | 303 | 27 |
| Gel filtration | 6.6 | 0.18 | 153 | 859 | 430 | 7 |
Cephalexin synthesis activity.
CM-Hap, ceramic hydroxyapatite.
As described above, the aehA gene codes for a precursor protein with an N-terminal leader sequence of 40 amino acids and the 70-kDa subunit of AEH. The sequence of the first 40 amino acids has features typical for signal peptides, like positively charged residues at the N terminus followed by a stretch of hydrophobic residues (40). In the carboxy-terminal segment of the leader sequence, a consensus pattern specific for the cleavage by signal peptidase I (AXA) is present (23, 24, 26). This, however, predicts the cleavage site at positions 39 to 40 (AQA-AAP), which is one position earlier than what was found by N-terminal amino acid sequencing of the enzyme (positions 40 to 41, AQAA-AP). To obtain insight in the function of the leader sequence and localization of the enzyme, the osmotic shock procedure was performed on the wild-type organism and the clones. Moreover, constructs without the leader sequence were made. The procedure developed for E. coli (1) was used with A. turbidans, but no released proteins were detected. In contrast, the same procedure released DNA from E. coli BL21(DE3)pLysS(pETAT), indicating that the cell wall of this strain is not rigid enough, which is ascribed to the presence of lysozyme, coded by pLysS. A periplasmic extract from E. coli HB101(pAT) was made, but no AEH activity was detected. A control experiment using E. coli HB101(pEC), expressing the periplasmic penicillin acylases from E. coli ATCC 11105, showed that the procedure worked. Cloning of the aehA gene in E. coli without the leader sequence, starting with M-41-APAAD, resulted in inactive clones, using two different expression systems (pET9, pEC).
To determine the way of processing in E. coli BL21(DE3)pLysS(pETAT), the first five N-terminal amino acid residues of the recombinant enzyme were determined. Multiple signals were present, but the major sequence found was 40-AAPXAD, which is in agreement with the predicted cleavage site between residues 39 and 40. This indicates that the leader sequence is processed in a similar way in E. coli and in A. turbidans. The other signals present indicate some N-terminal heterogeneity, which might be caused by cleavage at the other potential peptidase I cleavage sites (Fig. 3, positions 41 to 49) or by nonspecific protease activity.
Characterization and kinetic properties of recombinant AEH.
The subunit composition of the recombinant purified enzyme was determined via dynamic light scattering. Averaging two measurements yielded a molecular size of 150 kDa, which is in good agreement with a dimeric form of the enzyme (140 kDa).
To study the substrate specificity of the recombinant AEH, steady-state kinetic parameters were determined for a range of substrates (Table 4). For most compounds Michaelis-Menten kinetics was observed. Substrate inhibition was confirmed in the hydrolysis of ampicillin (34) and also observed for the chromogenic substrate NIPGB. The V versus [S] curves could be fitted according to the common equations given for substrate inhibition. The Km values for cephalexin and D-PGM obtained with the recombinant AEH were in reasonable agreement with values given in the literature for the enzyme from A. turbidans, 1.5 and 4.9 mM, respectively (28) (Table 4). The AEH did not display detectable activity towards glutaryl 7-ACA, which is a substrate for the related glutaryl 7-ACA acylase from B. laterosporus. Adipoyl 7-ADCA was not hydrolyzed by AEH either. In general we see for esters a high kcat in combination with a high Km. The addition of an OH group on the aromatic moiety of the acyl group reduces the activity. This shows that the nature of the acyl group has a large influence on the activity.
TABLE 4.
Kinetic parameters of AEH
| Substratea | Mean ± SE of:
|
||
|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | |
| NIPGBb | 1.1 ± 0.5 | 0.4 ± 0.07 | 0.4 ± 0.2 |
| Cephalexin | 0.34 ± 0.03 | 347 ± 10 | 1021 ± 95 |
| Ampicillinb | 1.0 ± 0.6 | 162 ± 61 | 162 ± 115 |
| D-PGM | 7 ± 2 | 1035 ± 123 | 148 ± 46 |
| D-PGA | >13 | >43 | 3.3c |
| Cefadroxild | 1.7 ± 0.3 | 9.6 ± 0.4 | 6 ± 1 |
| Amoxicillind | 2.6 ± 0.2 | 10 ± 0.3 | 3.9 ± 0.3 |
| HPGM | 11 ± 3 | 263 ± 30 | 24 ± 7 |
For glutaryl 7-ACA and adipoyl 7-ADCA the hydrolysis was less than 300 nmol · s−1 · mg−1 of AEH at 25 mM substrate.
Substrate inhibition was observed with Ki of 200 mM (NIPGB) and Ki of 3 mM (ampicillin).
Calculated from the initial slope of the Michaelis-Menten curve; the error is 30%.
Data were measured using His-tagged protein with the same catalytic properties as the wild type (unpublished data).
Inhibition.
The conservation of the GxSYxG motif suggests that AEH is a serine hydrolase. To explore this possibility some known serine protease and/or hydrolase inhibitors, Pefabloc SC, PMSF, and p-NPGB, were tested. Incubation with Pefabloc SC resulted in a 10% loss in activity, and PMSF did not give inactivation at all. Significant reduction (75%) of the initial enzyme activity was observed using p-NPGB. This suggests that a serine is involved in the catalytic mechanism.
DISCUSSION
Almost 30 years ago the AEHs were first described, and they have since been repeatedly explored for use in biocatalysis, but no amino acid sequence information has been reported so far. We cloned the gene encoding the AEH from A. turbidans out of a cosmid library via Southern blotting. From the literature it was expected that the gene for AEH would code for two different subunits, one of 70 and one of 72 kDa (29, 34). However, only one gene (aehA) coding for a protein of 74 kDa was found. Since the determined N-terminal sequence was found at position 41, it was concluded that aehA encodes a precursor of AEH that undergoes processing to yield an active enzyme of 70 kDa. No evidence for a second subunit of 72 kDa was obtained, neither by purification (Fig. 2) nor by sequence analysis (Fig. 3). However, incomplete processing of the leader sequence might have caused the presence of two apparently different subunits of 70 and 74 kDa, which could easily have been interpreted as subunits of 70 and 72 kDa.
Expression of this single gene in E. coli, including the identified leader sequence, produces active dimeric AEH. Processing of the leader sequence in E. coli implies a periplasmic localization, since signal peptidase I is active on the periplasmic face of the cytoplasmic membrane (7). However, the enzyme was not released from the E. coli HB101(pAT) cells by a standard osmotic shock procedure. It is therefore assumed that the protein sticks to cell envelope. Since no activity was detected when the enzyme was expressed without the leader sequence, it is proposed that the 40-amino-acid N-terminal part facilitates proper folding of the enzyme. Overall, it can be concluded that the N-terminal sequence is needed for production of active enzyme and probably serves for transport to the periplasm.
The deduced amino acid composition of aehA from A. turbidans is similar to the experimental data published in the literature, except for the number of methionines and cysteines, which we find to be significantly lower (29). Nevertheless, the deduced values for these amino acids are in reasonable agreement with the experimental data found for both X. citri and P. melanogenum (16, 18). This suggests a high similarity between the different AEHs, as expected from their comparable catalytic and structural properties.
Database searches with the aehA-encoded protein revealed no homology with any other known penicillin or cephalosporin acylase, apart from the glutaryl 7-ACA acylase of B. laterosporus. However, glutaryl 7-ACA was not hydrolyzed by the AEH of A. turbidans, which is probably due to the absence of an amino group on the Cα position (28, 34). The conservation of the GxSYxG motif in AEH, cocaine esterase (12), and other putative acylases (Table 2) suggests that AEH is a serine hydrolase, which was further indicated by the inactivation by p-NPGB. In glutaryl 7-ACA acylase the GxSYxG motif is not fully conserved, the last glycine being replaced by an alanine (GlSYma, Table 2). This might indicate that the second glycine influences the substrate range.
The physiological role of the β-lactam antibiotic acylases has not been elucidated yet. It has been suggested that penicillin G acylase from E. coli is involved in the degradation of phenylacetylated compounds for the generation of phenylacetic acid as a carbon source (37). Although the aehA gene appears to be located in an area where genes involved in the metabolism of amino compounds are situated, the real function of the AEH remains unclear. Further investigation of the substrate range of the AEH might reveal a relation to the surrounding enzymes.
Comparison of some kinetic values of the cloned AEH to literature data showed that the recombinant AEH has kinetic properties similar to those of the wild-type enzyme (28, 34). Remarkable features are the esterase activity being better than amidase activity with related substrates (D-PGM compared to D-PGA) and the need for an α-amino group. The higher specificity for the acyl donor than for the corresponding antibiotic in the cases of cefadroxil and amoxicillin is favorable for high product accumulation in a synthesis reaction. Even though esters are generally preferred, the specificity constant for cephalexin is higher than for D-PGM. This is in contradiction with the classification of AEH as an esterase. Therefore, a broader exploration of the substrate range is needed.
The AEH of A. turbidans was classified, based on the preferred antibiotic substrate, as an ampicillin acylase (31, 39). However, cephalexin is the preferred β-lactam antibiotic for AEH, as described for other AEHs as well (9, 17, 18, 28), and the enzyme should therefore be designated a cephalexin acylase.
X-ray analysis and mutational studies have shown that β-lactam antibiotic acylases from different substrate specificity classes all belong to the Ntn hydrolase superfamily (11, 19, 21, 33). Since both AEH and the glutaryl 7-ACA acylase from B. laterosporus (6) (i) do not have an N-terminal serine, threonine, or cysteine, (ii) contain the serine protease motif, and (iii) show no homology with any other known penicillin acylases (Fig. 4), it is very unlikely that these enzymes belong to the superfamily of Ntn hydrolases. We could not identify homologous proteins with a known X-ray structure that might reveal a structural relation of AEH to other protein families. Therefore we conclude that AEH, together with the glutaryl 7-ACA acylase from B. laterosporus, represents a new class of β-lactam antibiotic acylases, each representing a different subclass.
The expression of the AEH from A. turbidans ATCC 9325 in E. coli makes it possible to study the enzyme in more detail. The catalytic mechanism and the structural features that together determine the biocatalytic performance will be investigated further in order to gain more insight in the structure-function relationship of this new class of acylases.
Acknowledgments
We thank Peter Terpstra for sequencing the material presented in this paper. We are also grateful to P. Wietzes and T. Pijning for their technical assistance.
This work was financially supported by the Dutch Ministry of Economic Affairs.
REFERENCES
- 1.Alkema, W. B. L., R. Floris, and D. B. Janssen. 1999. The use of chromogenic reference substrates for the kinetic analysis of penicillin acylases. Anal. Biochem. 275:47–53. [DOI] [PubMed] [Google Scholar]
- 2.Alkema, W. B. L., C. M. H. Hensgens, E. H. Kroezinga, E. de Vries, R. Floris, J.-M. van der Laan, B. W. Dijkstra, and D. B. Janssen. 2000. Characterization of the β-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies. Protein Eng. 13:857–868. [DOI] [PubMed] [Google Scholar]
- 3.Alonso, J., and J. L. García. 1996. Proline iminopeptidase gene from Xanthomonas campestris pv. citri. Microbiology 142:2951–2957. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aramori, I., M. Fukagawa, M. Tsumura, M. Iwami, H. Ono, H. Kojo, M. Kohsaka, Y. Ueda, and H. Imanaka. 1991. Cloning and nucleotide sequencing of a novel 7β-(4-carboxybutanamido)cephalosporanic acid acylase gene of Bacillus laterosporus and its expression in Escherichia coli and Bacillus subtilis. J. Bacteriol. 173:7848–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkowitz, R., and M. Bassilana. 1994. Protein translocation in Escherichia coli. Biochim. Biophys. Acta 1197:311–343. [DOI] [PubMed] [Google Scholar]
- 8.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam contribution to the annual NAR database issue. Nucleic Acids Res. 28:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blinkovsky, A. M., and A. N. Markaryan. 1993. Synthesis of β-lactam antibiotics containing α-aminophenylacetyl group in the acyl moiety catalyzed by D-(−)-phenylglycyl-β-lactamide amidohydrolase. Enzyme Microb. Technol. 15:965–973. [DOI] [PubMed] [Google Scholar]
- 10.Boyer, H. W., and D. Roulland-Dussiox. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472. [DOI] [PubMed] [Google Scholar]
- 11.Brannigan, J. A., G. Dodson, H. J. Duggleby, P. C. E. Moody, J. L. Smith, D. R. Tomchick, and A. G. Murzin. 1995. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378:416–419. [DOI] [PubMed] [Google Scholar]
- 12.Bresler, M. M., S. J. Rosser, A. Basran, and N. C. Bruce. 2000. Gene cloning and nucleotide sequencing and properties of a cocaine esterase from Rhodococcus sp. strain MB1. Appl. Environ. Microbiol. 66:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chich, J.-F., M.-P. Chapot-Chartier, B. Ribadeau-Dumas, and J.-C. Gripon. 1992. Identification of the active site serine of X-prolyl dipeptidyl aminopeptidase from Lactococcus lactis. FEBS Lett. 314:139–142. [DOI] [PubMed] [Google Scholar]
- 14.Fernández-Lafuente, R., O. Hernández-Jústiz, C. Mateo, M. Terreni, J. Alonso, J. L. García-López, M. A. Moreno, and J. M. Guisán. 2001. Stabilization of a tetrameric enzyme (α-amino acid ester hydrolase from Acetobacter turbidans) enables a very improved performance of ampicillin synthesis. J. Mol. Catal. 11:633–638. [Google Scholar]
- 15.Hernández-Jústiz, O., M. Terreni, G. Pagani, J. L. García, J. M. Guisán, and R. Fernández-Lafuente. 1999. Evaluation of different enzymes as catalysts for the production of β-lactam antibiotics following a kinetically controlled strategy. Enzyme Microb. Technol. 25:336–343. [Google Scholar]
- 16.Kato, K., K. Kawahara, T. Takahashi, and A. Kakinuma. 1980. Purification of α-amino acid ester hydrolase from Xanthomonas citri. Agric. Biol. Chem. 44:1069–1074. [Google Scholar]
- 17.Kato, K., K. Kawahara, T. Takahashi, and A. Kakinuma. 1980. Substrate specificity of α-amino acid ester hydrolase from Xanthomonas citri. Agric. Biol. Chem. 44:1075–1081. [Google Scholar]
- 18.Kim, D. J., and S. M. Byun. 1990. Purification and properties of ampicillin acylase from Pseudomonas melanogenum. Biochim. Biophys. Acta. 1040:12–18. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y., K.-H. Yoon, Y. Khang, S. Turley, and W. G. J. Hol. 2000. The 2.0 Å crystal structure of cephalosporin acylase. Structure 8:1059–1068. [DOI] [PubMed] [Google Scholar]
- 20.Kutzbach, C., and E. Rauenbusch. 1974. Preparation and general properties of crystalline penicillin acylase from Escherichia coli ATCC 11105. Hoppe-Seyler’s Z. Physiol. Chem. 355:45–53. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y. S., H. W. Kim, and S. S. Park. 2000. The role of α-amino group of the N-terminal serine of β subunit for enzyme catalysis and autoproteolytic activation of glutaryl 7-aminocephalosporanic acid acylase. J. Biol. Chem. 275:39200–39206. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf, W. W., J. K. Zhang, X. Shi, and R. S. Wolfe. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 178:5797–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakai, K. 2000. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54:277–344. [DOI] [PubMed] [Google Scholar]
- 24.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34–35. [DOI] [PubMed] [Google Scholar]
- 25.Nam, D. H., and D. D. Y. Ryu. 1988. Molecular cloning of the gene for α-acylamino-β-lactam acylhydrolase from Acetobacter turbidans by immunochemical detection method. Kor. J. Appl. Microbiol. Bioeng. 16:363–368. [Google Scholar]
- 26.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6. [DOI] [PubMed] [Google Scholar]
- 27.Poelarends, G. J., M. Wilkens, M. J. Larkin, J. D. van Elsas, and D. B. Janssen. 1998. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl. Environ. Microbiol. 64:2931–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu, Y. W., and D. D. Y. Ryu. 1988. Semisynthetic β-lactam antibiotic synthesizing enzyme from Acetobacter turbidans: catalytic properties. Enzyme Microb. Technol. 10:239–245. [Google Scholar]
- 29.Ryu, Y. W., and D. D. Y. Ryu. 1987. Semisynthetic β-lactam antibiotics synthesizing enzyme from Acetobacter turbidans: purification and properties. Enzyme Microb. Technol. 9:339–344. [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Savidge, T. A. 1984. Enzymatic conversions used in the production of penicillins and cephalosporins. Drugs Pharm. Sci. 22:171–224. [Google Scholar]
- 32.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suresh, C. G., A. V. Pundle, H. SivaRaman, K. N. Rao, J. A. Brannigan, C. E. McVey, C. S. Verma, Z. Dauter, E. J. Dodson, and G. G. Dodson. 1999. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 6:414–416. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, T., Y. Yamazaki, and K. Kato. 1974. Substrate specificity of an α-amino acid ester hydrolase produced by Acetobacter turbidans A.T.C.C. 9325. Biochem. J. 137:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, T., Y. Yamazaki, K. Kato, and M. Isona. 1972. Enzymatic synthesis of cephalosporins. J. Am. Chem. Soc. 94:4035–4037. [DOI] [PubMed] [Google Scholar]
- 36.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle, F., P. Balbas, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 16:36–40. [DOI] [PubMed] [Google Scholar]
- 38.Van Hylckama Vlieg, J. E. T., H. Leemhuis, J. H. Lutje Spelberg, and D. B. Janssen. 2000. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 182:1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandamme, E. J., and J. P. Voets. 1974. Microbial penicillin acylases. Adv. Appl. Microbiol. 17:311–369. [DOI] [PubMed] [Google Scholar]
- 40.von Heijne, G. 1985. Signal sequence, the limits of variation. J. Mol. Biol. 184:99–105. [DOI] [PubMed] [Google Scholar]