Abstract
Both the availability and the quality of nutrients affect cellular functions by controlling gene activity. AreA, a member of the GATA family of transcription factors, globally activates expression of genes involved in nitrogen source utilization in Aspergillus nidulans. The quality of the nitrogen source determines the level and activation capacity of AreA through controls at the level of areA mRNA stability and by interaction of AreA with the corepressor NmrA. The availability of potential nitrogen sources also affects the activation capacity of AreA. We show that the complete absence of a nitrogen source results in an enhanced level of AreA-dependent gene expression and that this response is independent of mechanisms regulating AreA activity in response to nitrogen source quality. During nitrogen starvation AreA accumulates in the nucleus, but the presence of a potential nitrogen source or carbon starvation prevents this accumulation. Furthermore, accumulated AreA is rapidly lost from the nuclei of nitrogen-starved cells when a nitrogen source is supplied or when a carbon source is absent, and this accompanies arrest of the AreA-dependent nitrogen starvation response on regulated gene expression. By the generation of a leptomycin B-sensitive mutant, we have been able to show that nuclear exit occurs via the CrmA exportin. We conclude that sensing mechanisms discriminate between starvation and the presence of potential nutrients that can signal to the AreA transcription factor. Nitrogen source availability, but not quality, affects nuclear accumulation by regulating nuclear exit of AreA, providing a rapid response to changes in the supply of nutrients.
Filamentous fungi can use many compounds as sole nitrogen sources. Production of the enzymes and uptake systems required for utilization of nitrogen sources is regulated by a general control mechanism, termed nitrogen metabolite repression, in which expression of the relevant structural genes is low during growth on glutamine or ammonium and elevated under nitrogen-limiting conditions (31). In addition, pathway-specific transcription factors may activate sets of genes involved in catabolism of a particular compound in response to specific low-molecular-weight inducers.
The molecular genetics of nitrogen metabolite repression has been extensively studied in filamentous fungi (31). The key regulatory gene (areA in Aspergillus nidulans and nit-2 in Neurospora crassa) encodes a transcription activator containing a single GATA zinc finger DNA-binding domain (17, 24). Nitrogen catabolic gene expression requires transcription activation by AreA/NIT2, and nitrogen metabolite repression is signaled by nitrogen metabolites generated during growth on preferred nitrogen sources to prevent or diminish this activation. The major repressing metabolite is likely to be glutamine, as mutations blocking conversion of ammonium to glutamine result in loss of repression (30). Loss-of-function mutations in areA and nit-2 confer an inability to use nitrogen sources other than ammonium, and amino acid substitutions in the DNA-binding domain alter the pattern of nitrogen source utilization (24, 38, 42). AreA and NIT2 bind sequences containing the core GATAR sequence, and functional cis-acting sequences containing this motif are found in the 5′ regions of genes subject to nitrogen metabolite repression (7, 12, 19, 33, 38).
In A. nidulans, AreA activity is regulated in response to nitrogen sources. AreA levels are elevated under nitrogen-limiting conditions by autogenous transcription control via GATAR sequences in the areA 5′ region (28). Growth in the presence of ammonium, glutamate, or glutamine results in reduced areA mRNA stability compared with growth in the presence of limiting nitrogen sources (32, 36). This is mediated by sequences in the 3′ untranslated region (3′UTR) of areA, deletion of which results in increased mRNA stability and partial relief of nitrogen metabolite repression. AreA function is also controlled by interaction with the negative-acting regulator NmrA. Deletion of the nmrA gene or C-terminal truncation of AreA results in partial derepression (1, 36). NMR1, the product of the homologous gene in N. crassa, interacts specifically with NIT2 via C-terminal sequences and sequences within the GATA zinc finger domain (41). It is proposed that under nitrogen-sufficient conditions, NmrA interacts with AreA to prevent activation of nitrogen catabolic genes (1). Recent studies show that NmrA interacts with AreA in vitro and suggest that NmrA does not prevent DNA binding by AreA (26, 27). The two mechanisms controlling AreA activity are independent, as deletion of the AreA C-terminal sequence or the nmrA gene combined with deletion of the areA 3′UTR results in a high level of derepression (1, 36). These controls adjust levels of nitrogen catabolic gene expression according to nitrogen source quality.
Complete starvation for nitrogen may not be equivalent to nitrogen limitation. Some nitrogen catabolic enzymes such as acetamidase, histidase, and formamidase, which do not require addition of an inducer for expression, are produced at high levels in response to nitrogen starvation (14, 23, 37). It was thought that this response occurred by the normal mechanisms of AreA activation, with nitrogen starvation resulting in low levels of repressing metabolites. We show here, however, that nitrogen starvation results in an additional mechanism of AreA-dependent gene expression, independent of NmrA and the areA 3′UTR, and is rapidly reversed by addition of a wide variety of nitrogen compounds.
We have found that the response to nitrogen starvation is paralleled by accumulation of AreA in the nucleus and that its reversal by nitrogen compounds is paralleled by the rapid loss of accumulated AreA from the nucleus. The response to nitrogen starvation of enzymes involved in nitrogen source utilization does not occur during carbon starvation (14, 21, 22, 37). This effect is independent of creA-mediated carbon catabolite repression (14). We show here that accumulation of AreA in the nucleus does not occur when cells are starved for both carbon and nitrogen and that AreA is rapidly lost from the nucleus upon carbon starvation. Therefore, in addition to the previously described mechanisms for modulating AreA activity, control at the level of nuclear localization is important for regulating gene expression in response to starvation.
MATERIALS AND METHODS
Generation of areA gene replacement plasmids.
Sequences encoding three copies of the hemagglutinin (HA) epitope (YPYDVPDYA) were inserted in frame into areA at the PvuII site (position +156 relative to the start codon). The areA 5′ PvuII fragment (−895 to +156) was inserted into the EcoICRI site of the HA epitope vector pSM491 to give pJAF4688. The 3.1-kb PvuII/ScaI areA 3′ fragment (+157 to +3252) was then inserted into the SmaI site of pJAF4688 to give pJAF4689, which encodes AreA with three copies of the HA epitope inserted between residues 52 and 53. pJAF5198 was generated from pJAF4689 by deleting the 3′ end of areA after nucleotide +1475 as an ApaI fragment. The areA EcoICRI/BglII promoter fragment (−571 to −19) in pJAF5198 was replaced with an EcoRV/XhoI (end-filled) gpdA promoter fragment (nucleotides −1225 to −119 relative to the gpdA start codon) from pALX215 to give pJAF5200. The areA 5′ deletion construct was created by inserting a 2.2-kb BglII/SmaI riboB+ fragment from pPL1 (35) into pJAF5198 digested with BglII/SnaBI, replacing nucleotides −18 to +811 of areA to give pJAF5224.
Epitope tagging of areA derivatives.
MH8926 (yA1 areA217 pabaA1 fmdS-lacZ) was transformed with the areAHA gene on pJAF4689 and transformants selected on 10 mM nitrate and screened by Southern analysis. MH9641 (yA1 pabaA1 areAHA fmdS-lacZ [at argB]) was chosen for immunostaining and assays.
Simultaneous HA tagging of AreA variants and replacement of the areA promoter with the gpdA promoter involved two steps (Fig. 1A). (i) 5′ areA (−18 to + 811) was gene replaced with the riboB gene by transformation of MH764 (wA3 riboB2 facB101) with the areA::riboB(5′) construct pJAF5224 linearized with ApaI and selection for riboflavin prototrophs. An AreA− transformant (MH9922 [wA3 areA::riboB(5′) riboB2 facB101]) was identified by lack of growth on 10 mM nitrate, and the gene replacement was confirmed by Southern blotting. (ii) areA was reconstructed with modifications by transforming MH9922 with ApaI-linearized pJAF5200, containing gpdA(p)areAHA truncated at +1475, and selecting for growth on 10 mM nitrate. Transformants were tested for loss of riboB, and integration was confirmed by Southern blotting. Transformant MH9883 [wA3 gpdA(p)areAHA riboB2 facB101] was outcrossed, and the segregant MH9949 [biA1 gpdA(p)areAHA amdS-lacZ] was used in immunostaining, Western analysis, and assays. MH9927 [biA1 niiA4 amdS-lacZ gpdA(p)areAHA nmrA::BleR pyroA4] was isolated from progeny of a cross of MH9883 × MH8936 (biA1 niiA4 amdS-lacZ areA102 nmrA::BleR pyroA4).
FIG. 1.
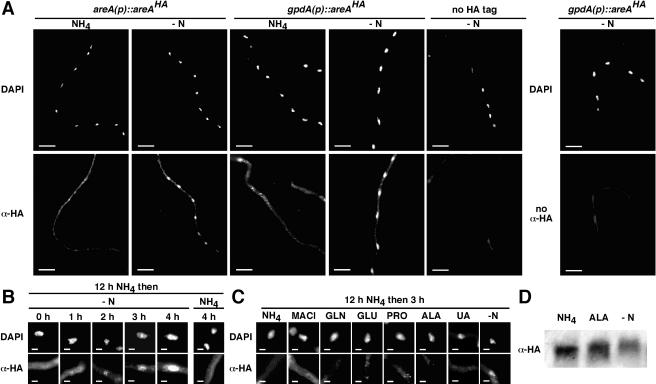
Regulation of expression of amdS-lacZ in response to different nitrogen sources. (A) AreA was HA tagged and the areA promoter replaced with the gpdA promoter in a two-step strategy (panel 1). 5′ areA (−18 to +811) was gene replaced with riboB by transformation of a riboflavin auxotroph with linearized pJAF5224 and selection for Ribo+ prototrophs (panel 2). areA was reconstructed with modifications by transformation of the areA::riboB(5′) strain from panel 1 with linearized pJAF5200 and selection on 10 mM nitrate. pJAF5200 contains gpdA(p)areAHA truncated at +1475 and confers AreA function only by homologous integration at the areA locus. (B) Strains of the indicated genotypes were grown for 16 h in 1% glucose medium with the nitrogen sources 10 mM ammonium tartrate (NH4), 10 mM l-glutamine (GLN), and 10 mM l-alanine (ALA) or subjected to nitrogen starvation (−N) by transferring mycelium pregrown in 1% glucose-10 mM ammonium tartrate for 16 h to 1% glucose medium lacking a nitrogen source for 4 h. Mycelium was harvested, extracted, and assayed for β-galactosidase. Specific activities with standard errors for at least three experiments are shown. (C) Wild-type and areA::riboB strains were grown for 16 h in 1% glucose-10 mM ammonium tartrate medium and then transferred to glucose medium lacking a nitrogen source, harvested at the indicated times, and assayed for β-galactosidase. (D) The indicated strains were grown for 16 h in glucose-ammonium medium (0 min), transferred to glucose medium containing various nitrogen sources (10 mM) for the indicated times, and then harvested and assayed for β-galactosidase. GLN, l-glutamine; NH4, ammonium tartrate; GLU, l-glutamate; ALA, l-alanine; PRO, l-proline; UREA, urea; ASN, l-asparagine; UA, uric acid; −N, no added nitrogen source.
gpd(p)areA-844HA was gene replaced using MH8421 (areA-844 tamA119 riboB2) as the recipient to generate MH9771 [areA-844::riboB(5′) tamA119 riboB2], in which 5′ areA-844 (−18 to +811) was gene replaced with riboB but the 3′ mutant areA coding sequences remained intact. pJAF5200 was then transformed into MH9771, and transformants were selected on 10 mM nitrate to generate MH9896 [gpdA(p)areAΔ844-876HA tamA119 riboB2]. MH9896 was outcrossed to give MH9895 [biA1 niiA4 amdS-lacZ gpdA(p)areAΔ844-876HA].
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (11), using the following strains: MH3408 (biA1 niiA4 amdS-lacZ), MH9949 [biA1 gpd(p)areAHA amdS-lacZ], MH8882 (biA1 niiA4 amdS-lacZ nmrA::BleR), MH9102 (biA1 areA-3′UTRΔ amdS-lacZ nmrA::BleR pyroA4), MH9058 (biA1 areA-3′UTRΔ amdS-lacZ pyroA4 riboB2), MH9939 (biA1 areAHA amdS-lacZ), and MH10384 (biA1 areA::riboB amdS-lacZ).
Microscopy.
Hyphae were grown in liquid medium on coverslips, fixed, and immunostained as described previously (39) except that 10 mg/ml Glucanex (Novozymes) replaced Novozyme, the anti-HA rat monoclonal antibody 3F10 (Boehringer Mannheim) was used at 1:100 as the primary antibody, and Alexa-488 goat anti-rat (Molecular Probes) was used at 1:1,000 as the secondary antibody. Indirect immunofluorescence microscopy was performed using a Reichardt-Jung Polyvar microscope equipped with Nomarski difference interference contrast and fluorescence optics and a Plan Apo Oel Iris 100× objective with a numerical aperture of 1.32. Alexa-488 immunofluorescence was detected using a standard fluorescein isothiocyanate filter set (excitation wavelength band pass, 450 to 495 nm; dichroic mirror, 510 nm; long-pass barrier filter, 520 nm). DAPI (4′,6′-diamidino-2-phenylindole) fluorescence was detected using a DAPI filter set (excitation wavelength band pass, 330 to 380 nm; dichroic mirror, 420 nm; long-pass barrier filter, 418 nm). At least 30 nuclei from each of two independent experiments were analyzed for each growth condition. Photomicrographs were captured using a Spot camera (Diagnostic Instruments) and Adobe Photoshop 6.0 (Adobe Systems). Images were manipulated similarly within and between experiments by using Adobe Photoshop 6.0. Images were cropped, and the tonal range was increased by adjusting highlights and shadows without altering the color balance and converted to gray scale.
Western blot analysis.
One hundred milligrams of mycelia, frozen at harvest in liquid nitrogen, were lysed in 1 ml of denaturing buffer (10 mM Tris-HCl, pH 8.0, 25 mM NH4 acetate, 1 mM EDTA, 10% trichloroacetic acid), using FastPrep FP120 at speed 6.5 four times for 15 s with cooling intervals. The lysate was centrifuged (16,100 × g) at 4°C for 10 min. The pellet was rinsed with 1 M Tris-HCl, pH 11.0, resuspended in 400 μl resuspension buffer (0.1 M Tris-HCl, pH 11.0, 3% sodium dodecyl sulfate), and boiled for 3 min and particulate matter was removed by centrifugation (16,100 × g) twice for 2 min. Fifty micrograms of total protein extract (determined using the DC protein assay kit [Bio-Rad]) was separated by 6% SDS-polyacrylamide gel electrophoresis and immunoblotted to a polyvinylidene difluoride membrane (Millipore). AreAHA was detected using 1:4,000 anti-HA 12CA5 antibodies (Roche), 1:5,000 horseradish peroxidase-conjugated goat anti-mouse antibodies (Promega), and chemiluminescence with the ECL Plus Western blotting detection system (Amersham).
Generation of a leptomycin B (LMB)-sensitive strain.
Expressed sequence tag sequences with high identity to Schizosaccharomyces pombe CRM1 were identified by BLAST search of the A. nidulans expressed sequence tag database (http://www.genome.ou.edu/asper.html). Primers Crm1a (5′-ATGAACGAAGAGACCGAGAAGC-3′ [+1803 to +1823 relative to the ATG]) and Crm1b (5′-TAGCGGCACTTTATCATCCATC-3′ [+2920 to +2899]) were used to PCR amplify a 1.1-kb product from A. nidulans MH1 (biA1) genomic DNA. The product was cloned (pJAF5248), and a bacterial artificial chromosome library (R. Dean, Clemson University, South Carolina) was probed with the insert. The crmA gene was subcloned as a 5.5-kb XhoI fragment (into SalI of pBluescript-SK+ [Stratagene] to give pJAF5256) and partially sequenced. The crmA sequence (also called kapK [GenBank accession number AY555733]) corresponds to the A. nidulans “Crm1” sequence described by Kudo et al. (25) (GenBank Accession Number AA966051) and hypothetical protein AN1401.2 in the A. nidulans genome sequence (Aspergillus Sequencing Project, Broad Institute of the Massachusetts Institute of Technology and Harvard [http://www.broad.mit.edu]).
The crmAT525C mutation was generated by in vitro mutagenesis of codon 525 (ACG to TGT). A silent C-to-A substitution at +1859, destroying the StuI site, was also introduced. PCR from the crmA plasmid with Crm1b and the mutagenic primer Crm1c (5′-ACTCTGTGAAATGAAGAGAGGG-3′ [+1859 to +1880]; changes are underlined) using Pfu Turbo DNA polymerase generated a 1.0-kb PCR product, which was digested with BamHI, and the fragment (+1859 to +2355), with one blunt end and one BamHI end, was inserted into pJAF5256 digested with BamHI (partial) and StuI to reconstruct the 3′ portion of crmA in pJAF5164. The pyrG+ gene was inserted 3′ of the crmA coding region: the 2.2-kb EcoRV fragment from pJAF5256 was cloned into the SmaI site of the pyrG plasmid pAB4626 to give pJAF5265, and then the crmA(3′)-pyrG EcoRV-SacII fragment was cloned into EcoRV/SacII-digested pJAF5164 to give pJAF5294. MH10077 [biA1 pyrG89 pabaB22 gpd(p) areAHA] was transformed with selection for PyrG+, and Southern hybridization identified 18 homologous gene replacements. Two lacked the StuI site at +1856 and therefore were generated by crossover 5′ of the mutation. The mutation was confirmed by PCR and direct sequence analysis. MH10635 [biA1 pyrG89 pabaB22 gpd(p)areAHA crmAT525C pyrG+ (at crmA)] was chosen for further analysis. MH10654 [biA1 pyrG89 pabaB22 gpd(p)areAHA crmAT525 pyrG+ (at crmA)], containing a single-copy integration at crmA but retaining the StuI site (due to crossover 3′ of the mutation and retaining the wild-type crmA sequence), was chosen as a control.
RESULTS
AreA-dependent gene expression in response to nitrogen starvation.
The amdS gene is controlled by AreA-mediated nitrogen metabolite repression (23). Expression of a gene-replaced amdS-lacZ reporter gene (11) increased in response to growth on the limiting nitrogen source alanine compared with the repressing nitrogen sources ammonium and glutamine and was further elevated in response to complete nitrogen starvation (Fig. 1B). The elevated response of amdS-lacZ levels to nitrogen starvation was areA dependent (Fig. 1C) and was retained in the nmrAΔ mutant and the areA 3′UTR element deletion mutant, each of which showed partial derepression on ammonium and glutamine compared with the limiting nitrogen source alanine (Fig. 1B). Furthermore, simultaneous deletion of nmrA and the areA 3′UTR element resulted in a high level of derepression of amdS-lacZ expression on ammonium compared with alanine but did not prevent the response to nitrogen starvation (Fig. 1B). Regulation of an fmdS-lacZ fusion gene present in single copy at the argB locus showed a similar pattern of areA-dependent expression, with an elevated response to nitrogen starvation compared with levels in the presence of alanine (14) (data not shown). Overall, these data suggest that nitrogen starvation results in an additional areA-dependent mechanism different from that effected by NmrA or by areA mRNA stability.
AreA accumulates in the nucleus in response to nitrogen starvation.
We modified the areA genomic locus to encode AreA with the HA epitope inserted between residues 52 and 53 (AreAHA), within a region dispensable for AreA regulation (5, 28) (see Materials and Methods). We also uncoupled transcriptional regulation of areA by replacing the native areA promoter with the constitutive gpdA promoter (Fig. 1A). The pattern of nitrogen metabolite repression and nitrogen starvation response of amdS-lacZ (Fig. 1B) and fmdS-lacZ (data not shown) was not affected by either the presence of the HA tag or expression of AreA from the gpdA promoter. However, overall levels were slightly lower than wild type in the areAHA strain and were increased in the presence of the gpdA promoter.
Immunofluorescence microscopy was used to examine the subcellular distribution of AreAHA. Nuclear accumulation of AreAHA occurred within 1 hour of transfer to medium lacking a nitrogen source, and the amount of AreAHA accumulated in the nucleus increased over the course of 4 h of nitrogen starvation. This regulated distribution of AreAHA was apparent whether expression was from the areA or gpdA promoter (Fig. 2A and B). Thus, nuclear accumulation of AreAHA paralleled the increase in AreA-dependent gene expression in response to nitrogen starvation. Furthermore, nuclear accumulation of AreAHA is a specific consequence of nitrogen starvation. AreAHA nuclear accumulation was not observed when mycelium was transferred for 3 h to medium containing the nitrogen source ammonium, glutamine, glutamate, proline, alanine, or uric acid or the ammonium analog methylammonium (Fig. 2C) or when mycelium was grown for 12 h on medium containing ammonium, glutamine, glutamate, proline, alanine, formamide, gamma-aminobutyric acid, urea, or uric acid (data not shown).
FIG. 2.
Subcellular localization of AreAHA. (A) AreAHA accumulates in the nucleus upon nitrogen starvation. areA(p)areAHA and gpd(p)areAHA strains were grown at 25°C in 1% glucose minimal medium containing 10 mM ammonium tartrate for 12 h and then transferred to medium containing 10 mM ammonium tartrate (NH4) or nitrogen-free (−N) medium for a further 4 h. Hyphae were fixed and immunostained with rat monoclonal anti-HA primary antibody and Alexa-488-conjugated goat anti-rat secondary antibody. Nuclei were stained with DAPI. Hyphae were viewed using a DAPI filter set (DAPI) and a fluorescein isothiocyanate filter set (α-HA). (B) Time course of AreAHA nuclear accumulation. gpd(p)areAHA hyphae were fixed and immunostained after 12 h of growth in glucose minimal medium plus 10 mM ammonium tartrate (0 h); transfer to nitrogen-free medium (-N) for 1, 2, 3, and 4 h; and transfer to ammonium tartrate-containing medium (NH4) for 4 h. Representative DAPI-stained nuclei (DAPI) and AreAHA fluorescence (α-HA) in the corresponding region are shown. (C) AreAHA does not accumulate in the nucleus in response to nitrogen sources. gpd(p)areAHA hyphae grown for 12 h in glucose minimal medium containing 10 mM ammonium tartrate were fixed and immunostained after transfer for 3 h to medium containing 10 mM ammonium tartrate (NH4), 10 mM methylammonium chloride (MACl), 10 mM glutamine (GLN), 10 mM glutamate (GLU), 10 mM proline (PRO), 10 mM alanine (ALA), or 10 mM uric acid (UA) or lacking nitrogen (−N). Scale bars represent 20 μm (A) and 2 μm (B and C). (D) AreAHA is differentially modified under different nitrogen conditions. Western blot analysis of total protein extracts (50 μg) from gpd(p)areAHA mycelia grown for 16 h in glucose minimal medium containing 10 mM ammonium tartrate (NH4) or 10 mM alanine (ALA) or for 16 h in glucose minimal medium containing 10 mM ammonium tartrate and then transferred for 4 h to medium lacking a nitrogen source (−N) is shown. AreAHA was detected with anti-HA antibody.
Western blot analysis showed that the relative levels of AreAHA in the gpd(p)areAHA strain did not differ significantly when mycelia were nitrogen starved or grown on ammonium or alanine (Fig. 2D). However, the mobility of AreAHA was reduced in mycelia grown on alanine compared with those grown on ammonium. A further reduction in AreAHA mobility was observed in nitrogen-starved mycelia. These data indicate that AreA accumulates in the nucleus during nitrogen starvation and is subject to differential posttranslational modification according to both nitrogen source quality and availability.
Nuclear accumulated AreA is rapidly lost in response to nitrogen sources.
The effect of addition of nitrogen sources to nitrogen-starved hyphae was examined. Loss of accumulated AreAHA from the nucleus was observed within 1 minute after transfer of nitrogen-starved cells to ammonium-containing medium (Fig. 3A). Western analysis showed that the AreAHA protein was stable for up to 60 min after transfer to ammonium (Fig. 3B), indicating that rapid loss of AreAHA from the nucleus reflected increased nuclear export and not AreAHA degradation. Furthermore, the reduced mobility of AreAHA on nitrogen starvation medium (Fig. 2D) was maintained for more than 15 min after transfer to ammonium (Fig. 3B). Transfer to glutamine, methylammonium, proline, alanine, or uric acid also resulted in rapid loss of nuclear accumulated AreAHA (Fig. 3C). Therefore, the presence of a nitrogen source is sufficient for enhanced export of AreA from the nucleus. This correlates with the observed effects on gene expression of nitrogen source addition to nitrogen-starved mycelium. The response of amdS-lacZ to nitrogen starvation ceased rapidly after addition of various nitrogen sources, and this was not affected by deletion of nmrA or the areA 3′UTR element (Fig. 1D). Deletion of nmrA or truncation of the C terminus of AreA at residue 844 did not alter the distribution of AreAHA in hyphae transferred to ammonium or nitrogen starvation medium or when ammonium was reintroduced to nitrogen-starved hyphae (data not shown).
FIG. 3.
Accumulated AreAHA is rapidly lost from the nucleus upon addition of a nitrogen source. (A and C) The gpd(p)areAHA strain was grown for 12 h in glucose-10 mM ammonium tartrate minimal medium and transferred to nitrogen-free medium (−N) for 4 h (0 min). Hyphae were then transferred to 10 mM ammonium tartrate medium for 1, 2, 5, 10, and 15 min (A). Control hyphae were transferred to nitrogen-free (−N) medium for 15 m (A). Hyphae were transferred to glucose minimal medium containing 0.2 mM glutamine (GLN), 1 mM methylammonium chloride (MACl), 10 mM alanine (ALA), 10 mM proline (PRO), and 10 mM uric acid (UA) for 1 and 15 min (C). The hyphae were fixed and immunostained. Representative DAPI-stained nuclei (DAPI) and AreAHA fluorescence (α-HA) in the corresponding region are shown. Scale bars represent 2 μm. (B) AreAHA is stable after transfer to ammonium. Western blot analysis with anti-HA antibody of total protein extracts (50 μg) from areAHA mycelia grown for 16 h in glucose minimal medium containing 10 mM ammonium tartrate, transferred for 4 h to medium lacking a nitrogen source (0 min), and then transferred to medium containing 10 mM ammonium tartrate for 1, 5, 15, 30, and 60 min is shown.
Carbon starvation prevents nuclear accumulation of AreA.
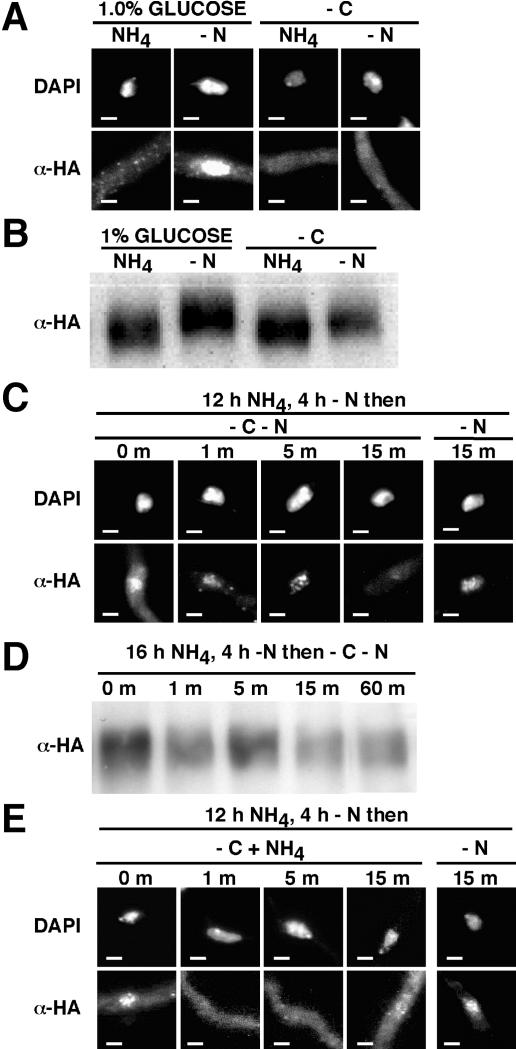
Enzymes involved solely in nitrogen source utilization, including formamidase, histidase, and a general amidase, show an areA-dependent response to nitrogen starvation but not during simultaneous carbon starvation (14, 16, 37). Furthermore, transfer of nitrogen-starved mycelium to medium lacking a carbon source results in rapid loss of areA-dependent expression of an fmdS-lacZ fusion gene (14). Carbon starvation was found to prevent accumulation of AreAHA in the nucleus in response to nitrogen starvation (Fig. 4A). Western analysis showed that AreAHA was present under carbon starvation conditions and that AreAHA mobility was reduced during nitrogen starvation but not carbon starvation (Fig. 4B). Transfer of nitrogen-starved mycelium to simultaneous nitrogen and carbon starvation conditions resulted in loss of accumulated AreAHA from the nucleus within 15 minutes (Fig. 4C). The AreAHA protein was stable for up to 60 min after transfer (Fig. 4D). Therefore, rapid loss of AreAHA from the nucleus upon carbon starvation paralleled the response of expression of areA-regulated genes to carbon starvation. Transfer of nitrogen-starved mycelium to carbon starvation conditions in the presence of ammonium resulted in loss of accumulated AreAHA from the nucleus within 1 minute (Fig. 4E).
FIG. 4.
Carbon starvation prevents AreAHA nuclear accumulation. (A) The gpd(p)areAHA strain was grown for 12 h in glucose minimal medium plus 10 mM ammonium tartrate and transferred to 1% glucose or carbon-free (−C) minimal medium containing (NH4) or lacking (−N) 10 mM ammonium tartrate for 4 h. (B) Analysis of AreAHA during carbon starvation. Western blot analysis with anti-HA antibody of total protein extracts (50 μg) from gpd(p)areAHA mycelia grown for 16 h in glucose-10 mM ammonium tartrate medium and transferred to 1% glucose or carbon-free (−C) minimal medium containing (NH4) or lacking (−N) 10 mM ammonium tartrate for 2 h. (C) Loss of accumulated AreAHA in response to carbon starvation. The gpd(p)areAHA strain was grown for 12 h in glucose-10 mM ammonium tartrate minimal medium, transferred to 1% glucose nitrogen-free minimal medium for 4 h (0 min), and then transferred to minimal medium lacking both a carbon and a nitrogen source (−C−N) for 1, 5, and 15 min. A control 15-min transfer to 1% glucose nitrogen-free medium (−N) was included. (D) AreAHA is stable after transfer to carbon starvation medium. Western blot analysis with anti-HA antibody of total protein extracts (50 μg) from gpd(p)areAHA mycelia grown for 16 h in glucose-10 mM ammonium tartrate medium, transferred for 4 h to minimal medium lacking a nitrogen source (0 min), and then transferred to medium lacking both a carbon and nitrogen source (−C−N) for 1, 5, 15, and 60 min. (E) Effects of ammonium predominate over effects of carbon starvation. The gpd(p)areAHA strain was grown for 12 h in glucose minimal medium plus 10 mM ammonium tartrate, transferred to glucose nitrogen-free minimal medium for 4 h (0 min), and then transferred to minimal medium lacking a carbon source (−C) but containing 10 mM ammonium chloride (NH4) as a nitrogen source for 1, 5, and 15 min. A control 15-min transfer to 1% glucose nitrogen-free medium (−N) was included. For panels A, C, and E, hyphae were fixed and immunostained, representative DAPI-stained nuclei (DAPI) and AreAHA fluorescence (α-HA) in the corresponding region are shown, and the scale bars represent 2 μm.
AreA nuclear export occurs via the CrmA exportin.
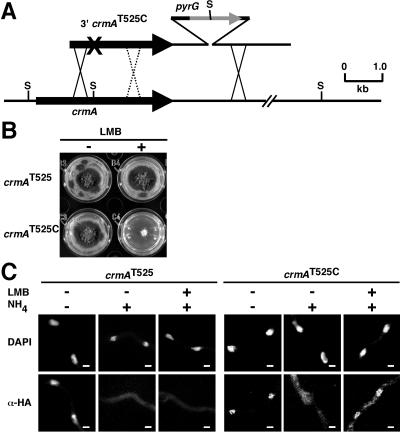
The slow nuclear accumulation and rapid loss of AreA suggests that nuclear export is the major control of AreA nuclear accumulation. The AreA protein contains a putative leucine-rich nuclear export signal at residues 703 to 712 (LHGVVRPLSL), conforming to the consensus for nuclear export signals bound by CRM1 exportins (13, 18). S. pombe CRM1 is inhibited by the drug LMB, which binds to a specific cysteine residue (C529), resulting in growth sensitivity and inhibition of CRM1-dependent nuclear export (25). Saccharomyces cerevisiae is LMB resistant, and the equivalent CRM1 residue is threonine, which when mutated to cysteine causes LMB sensitivity (34). Wild-type A. nidulans is LMB resistant, and the A. nidulans crmA gene specifies a threonine at residue 525, corresponding to S. pombe CRM1 C529 (25). We isolated the full-length crmA gene (see Materials and Methods) and altered codon 525 to specify cysteine (Fig. 5A). The crmAT525C mutant was sensitive to the presence of 100 ng/ml LMB (Fig. 5B). The crmAT525C mutation had no effect on AreA nuclear export in the absence of LMB (Fig. 5C). We examined the effect of LMB on export of AreAHA in the presence and absence of the crmAT525C mutation. Rapid nuclear export of AreAHA in the presence of LMB occurred in response to ammonium within 1 minute in the control strain, but AreAHA was retained in the nucleus in the crmAT525C mutant (Fig. 5C). Therefore, CrmA is the major export route from the nucleus for AreA.
FIG. 5.
AreA export from the nucleus occurs via the CrmA exportin. (A) Construction of the LMB-sensitive crmAT525C mutant. Transformants were selected for pyrimidine prototrophy due to insertion of pyrG 3′ of the crmA gene and screened by Southern analysis for loss of a genomic StuI site (S) associated with the introduced crmAT525C mutation (×). Crossovers 5′ of the mutation resulted in gene replacements carrying the crmAT525C mutation and lacking the StuI site, whereas crossovers 3′ of the mutation generated gene replacements lacking the desired mutation but retaining the StuI site. (B) The A. nidulans crmAT525C mutant is LMB sensitive. The crmAT525C and control gene replacement strains were tested for growth on 1% glucose-10 mM ammonium tartrate minimal medium with 100 ng/ml LMB dissolved in 100% ethanol (+LMB) or lacking LMB (−LMB) and grown for 3 days at 37°C. (C) AreA nuclear export occurs via the CrmA exportin. The gpd(p)areAHA crmAT525C and control gpd(p)areAHA crmAT525 strains were grown in 1% glucose-10 mM ammonium tartrate minimal medium for 12 h and then transferred to medium lacking a nitrogen source (−N) for 4 h. LMB (100 ng/ml) or ethanol (control) was then added and left for 5 m prior to treatment with 10 mM ammonium tartrate for 1 min. Samples were fixed and immunostained. Representative DAPI-stained nuclei (DAPI) and AreAHA fluorescence (α-HA) in the corresponding regions are shown. Scale bars represent 2 μm.
DISCUSSION
We have shown that A. nidulans can distinguish between conditions of nitrogen sufficiency, nitrogen limitation, and nitrogen starvation and adjust accordingly the expression levels of nitrogen catabolic genes. These changes are AreA dependent and reflect alterations in the level and activity of AreA. Furthermore, we have shown that A. nidulans can respond to nitrogen starvation by altering the dynamics of nuclear entry and exit of AreA, using regulated nuclear exit to provide a rapid response when nutritional conditions change.
We favor a model in which, in the presence of nitrogen sources, AreA is present in the nucleus at a relatively low level due to a balance between nuclear entry and exit. When repressing metabolites are at high levels (for example, when ammonium or glutamine is the source of nitrogen), the level of transcriptionally active AreA is low due to accelerated areA mRNA turnover and NmrA inhibition of AreA activation ability (1, 32). When poorer (limiting) nitrogen sources are present and glutamine levels are reduced, the increased stability of areA mRNA and diminished NmrA inhibition of AreA activity result in increased expression of AreA-dependent genes (28, 32, 36). Together these controls are sufficient to account for the difference in AreA activation under nitrogen-sufficient and nitrogen-limiting conditions. However, neither of these controls accounts for the elevated level of AreA-dependent gene expression observed under conditions of complete nitrogen starvation. Increased activation of AreA target genes correlates with progressive accumulation of AreA in the nuclei of nitrogen-starved cells. Increased concentrations of the AreA transcription factor inside the nucleus may result in high-level expression of target genes, although we cannot exclude the possibility that additional changes to AreA upon nitrogen starvation act to enhance its activity. Addition of a nitrogen source, whether limiting or nonlimiting, results in rapid loss of accumulated AreA and a loss of the response to nitrogen starvation. Furthermore, carbon starvation of nitrogen starved cells also causes rapid loss of nuclear accumulated AreA and loss of AreA-dependent gene expression. We have shown that these rapid responses, which occur within minutes, involve a redistribution of AreA from the nucleus rather than degradation of the protein. AreA is stable for at least 1 h after the addition of nitrogen sources to nitrogen-starved cells or transfer to carbon starvation conditions. We have shown that nuclear export of AreA via the CrmA exportin is likely to be the main control point for regulation of AreA nuclear accumulation.
Our model predicts that nitrogen starvation leads to modification of AreA resulting in its retention within the nucleus and that exposure to a nitrogen source leads to rapid CrmA-mediated nuclear exit of AreA. We have shown that AreA is posttranslationally modified according to nitrogen source quality and additionally modified during nitrogen starvation. Preliminary studies indicate that AreA is highly phosphorylated under all conditions (unpublished data). We observed no clear difference in AreAHA mobility accompanying rapid loss from the nucleus. The rapid loss of AreA from the nucleus could result from a minor posttranslational modification of AreA, which would not have been resolved in our analysis, or from modification of a protein that interacts with AreA. The AreA-dependent response of gene expression to nitrogen starvation is prevented by carbon starvation (14, 20, 37). This is paralleled by a lack of accumulation of AreA in the nucleus, which may be due to energy-dependent nuclear import being sensitive to carbon starvation. Carbon starvation also results in loss of nuclear accumulated AreA, and this could be by a mechanism different from the nitrogen source mechanism.
In S. cerevisiae control of nuclear localization of the GATA DNA-binding activator proteins Gln3 and Gat1/Nil1 is the major mechanism for the response of genes involved in nitrogen acquisition to changes in nitrogen source availability (2, 3, 10, 29). However, the molecular basis of this regulation differs from that in A. nidulans. Unlike the situation in A. nidulans, there is no reported difference in response to nitrogen starvation compared to limitation, and Gln3 nuclear export via the CrmA homolog Crm1 is not subject to regulation (6). Instead, under conditions of nitrogen excess the product of the URE2 gene, defined by mutations resulting in insensitivity of gene expression to the nitrogen source, prevents nuclear entry of Gln3 and Gat1. In A. nidulans, the closest homolog of URE2 encodes a glutathione S-transferase with no detectable role in nitrogen metabolite repression (15). In the presence of a good nitrogen source, the rapamycin-sensitive Tor kinase complex is thought to phosphorylate Gln3, which complexes with Ure2 in the cytoplasm (2, 4, 10). Nitrogen limitation or starvation leads to dephosphorylation of Gln3, allowing release from Ure2 and nuclear entry. However, recent data indicate that the phosphorylation state of Gln3 is complex and not necessarily correlated with nuclear localization and the nitrogen source present, raising the possibility that Tor activity and nutrient signaling affect Gln3 by different mechanisms (8, 40). The role, if any, of the Tor complex in controlling AreA activity in A. nidulans is unknown. In complete contrast to A. nidulans, glucose starvation in the presence of ammonium results in nuclear localization of Gln3 (4), but not when glutamine is present (9). These observations have been interpreted as resulting from carbon starvation causing reduced levels of ammonium assimilation into glutamine, the signaling molecule for control of Gln3 localization (9).
The pattern of regulation of genes by AreA would seem to be adapted to the ecology of filamentous fungi, which scavenge a wide variety of nitrogen sources derived from growth in soil and on decaying organic matter. Hyphae are likely to experience nutrient starvation during growth in microenvironments completely lacking particular nutrients and as substrates are exhausted. Increased expression of genes involved in nitrogen source acquisition during nitrogen starvation would allow a rapid response when new nutrients are encountered. However, in the absence of a carbon or energy source, derepression of nitrogen catabolism would not provide the nutrients necessary for growth. The ability of A. nidulans to adjust the nucleocytoplasmic distribution of AreA in response to both the carbon and nitrogen status of its environment highlights a previously undiscovered level of regulation of nitrogen catabolic gene expression.
Acknowledgments
This work was supported by Australian Research Council (ARC) grants to M.J.H. and M.A.D. and by a University of Melbourne Early Career Researcher Grant to R.B.T. J.A.F. was supported by an Australian Postgraduate Award (APA), and K.H.W. was supported by an International Postgraduate Research Scholarship (IPRS) and a Melbourne International Research Scholarship (MIRS).
We acknowledge assistance of K. Nguyen, S. Delimitrou, S. Murray, M. Wallis, and K. Smith. We are indebted to Minoru Yoshida (Riken Institute) for generous provision of leptomycin B.
REFERENCES
- 1.Andrianopoulos, A., S. Kourambas, J. A. Sharp, M. A. Davis, and M. J. Hynes. 1998. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180:1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, P. G., J. H. Choi, J. Carvalho, T. F. Chan, W. Ai, and X. F. Zheng. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caddick, M. X., and H. N. Arst, Jr. 1998. Deletion of the 389 N-terminal residues of the transcriptional activator AREA does not result in nitrogen metabolite derepression in Aspergillus nidulans. J. Bacteriol. 180:5762-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho, J., and X. F. Zheng. 2003. Domains of Gln3p interacting with karyopherins, Ure2p, and the target of rapamycin protein. J. Biol. Chem. 278:16878-16886. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, T. Y., and G. A. Marzluf. 1995. Binding affinity and functional significance of NIT2 and NIT4 binding sites in the promoter of the highly regulated nit-3 gene, which encodes nitrate reductase in Neurospora crassa. J. Bacteriol. 177:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, K. H., A. Kulkarni, J. J. Tate, and T. G. Cooper. 2004. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J. Biol. Chem. 279:10270-10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, K. H., J. J. Tate, and T. G. Cooper. 2002. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 277:37559-37566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, M. A., C. S. Cobbett, and M. J. Hynes. 1988. An amdS-lacZ fusion for studying gene regulation in Aspergillus. Gene 63:199-212. [DOI] [PubMed] [Google Scholar]
- 12.Feng, B., X. Xiao, and G. A. Marzluf. 1993. Recognition of specific nucleotide bases and cooperative DNA binding by the trans-acting nitrogen regulatory protein NIT2 of Neurospora crassa. Nucleic Acids Res. 21:3989-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, J. A., M. A. Davis, and M. J. Hynes. 2001. The formamidase gene of Aspergillus nidulans: regulation by nitrogen metabolite repression and transcriptional interference by an overlapping upstream gene. Genetics 157:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, J. A., M. A. Davis, and M. J. Hynes. 2002. A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance. Appl. Environ. Microbiol. 68:2802-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, J. A., M. A. Davis, and M. J. Hynes. 2002. The genes gmdA, encoding an amidase, and bzuA, encoding a cytochrome P450, are required for benzamide utilization in Aspergillus nidulans. Fungal Genet. Biol. 35:135-146. [DOI] [PubMed] [Google Scholar]
- 17.Fu, Y. H., and G. A. Marzluf. 1990. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 10:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 19.Gomez, D., I. Garcia, C. Scazzocchio, and B. Cubero. 2003. Multiple GATA sites: protein binding and physiological relevance for the regulation of the proline transporter gene of Aspergillus nidulans. Mol. Microbiol. 50:277-289. [DOI] [PubMed] [Google Scholar]
- 20.Hynes, M. J. 1975. Amide utilization in Aspergillus nidulans: evidence for a third amidase enzyme. J. Gen. Microbiol. 91:99-109. [DOI] [PubMed] [Google Scholar]
- 21.Hynes, M. J. 1973. The effect of lack of a carbon source on nitrate-reductase activity in Aspergillus nidulans. J. Gen. Microbiol. 79:155-157. [DOI] [PubMed] [Google Scholar]
- 22.Hynes, M. J. 1974. The effects of carbon source on glutamate dehydrogenase activities in Aspergillus nidulans. J. Gen. Microbiol. 81:165-170. [DOI] [PubMed] [Google Scholar]
- 23.Hynes, M. J. 1975. Studies on the role of the areA gene in the regulation of nitrogen catabolism in Aspergillus nidulans. Aust. J. Biol. Sci. 28:301-313. [DOI] [PubMed] [Google Scholar]
- 24.Kudla, B., M. X. Caddick, T. Langdon, N. M. Martinez-Rossi, C. F. Bennett, S. Sibley, R. W. Davies, and H. N. Arst, Jr. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb, H. K., K. Leslie, A. L. Dodds, M. Nutley, A. Cooper, C. Johnson, P. Thompson, D. K. Stammers, and A. R. Hawkins. 2003. The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J. Biol. Chem. 278:32107-32114. [DOI] [PubMed] [Google Scholar]
- 27.Lamb, H. K., J. Ren, A. Park, C. Johnson, K. Leslie, S. Cocklin, P. Thompson, C. Mee, A. Cooper, D. K. Stammers, and A. R. Hawkins. 2004. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 13:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langdon, T., A. Sheerins, A. Ravagnani, M. Gielkens, M. X. Caddick, and H. N. Arst, Jr. 1995. Mutational analysis reveals dispensability of the N-terminal region of the Aspergillus transcription factor mediating nitrogen metabolite repression. Mol. Microbiol. 17:877-888. [DOI] [PubMed] [Google Scholar]
- 29.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 30.Margelis, S., C. D'Souza, A. J. Small, M. J. Hynes, T. H. Adams, and M. A. Davis. 2001. Role of glutamine synthetase in nitrogen metabolite repression in Aspergillus nidulans. J. Bacteriol. 183:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozov, I. Y., M. G. Martinez, M. G. Jones, and M. X. Caddick. 2000. A defined sequence within the 3′ UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol. Microbiol. 37:1248-1257. [DOI] [PubMed] [Google Scholar]
- 33.Muro-Pastor, M. I., R. Gonzalez, J. Strauss, F. Narendja, and C. Scazzocchio. 1999. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 18:1584-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oakley, C. E., C. F. Weil, P. L. Kretz, and B. R. Oakley. 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53:293-298. [DOI] [PubMed] [Google Scholar]
- 36.Platt, A., T. Langdon, H. N. Arst, Jr., D. Kirk, D. Tollervey, J. M. Sanchez, and M. X. Caddick. 1996. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 15:2791-2801. [PMC free article] [PubMed] [Google Scholar]
- 37.Polkinghorne, M. A., and M. J. Hynes. 1982. l-Histidine utilization in Aspergillus nidulans. J. Bacteriol. 149:931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravagnani, A., L. Gorfinkiel, T. Langdon, G. Diallinas, E. Adjadj, S. Demais, D. Gorton, H. N. Arst, Jr., and C. Scazzocchio. 1997. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16:3974-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, A. J., R. B. Todd, M. C. Zanker, S. Delimitrou, M. J. Hynes, and M. A. Davis. 2001. Functional analysis of TamA, a coactivator of nitrogen-regulated gene expression in Aspergillus nidulans. Mol. Genet. Genomics 265:636-646. [DOI] [PubMed] [Google Scholar]
- 40.Tate, J. J., R. Rai, and T. G. Cooper. 2005.. Methionine sulfoximine-treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J. Biol. Chem. 280:27195-27204. [DOI] [PMC free article] [PubMed]
- 41.Xiao, X., Y. H. Fu, and G. A. Marzluf. 1995. The negative-acting NMR regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein NIT2. Biochemistry 34:8861-8868. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, X. D., and G. A. Marzluf. 1993. Amino-acid substitutions in the zinc finger of NIT2, the nitrogen regulatory protein of Neurospora crassa, alter promoter element recognition. Curr. Genet. 24:212-218. [DOI] [PubMed] [Google Scholar]