Abstract
Candida albicans is an opportunistic human pathogen that can sense environmental changes and respond by altering its cell morphology and physiology. A number of environmental factors have been shown to influence this dimorphic transition, including pH, starvation, serum, and amino acids. In this report, we investigate the function of the C. albicans CCAAT-binding factor. In Saccharomyces cerevisiae, this heterooligomeric transcriptional activator stimulates the expression of genes that encode proteins involved in respiration. To examine the function of this transcription factor in C. albicans, we cloned CaHAP5 and generated a hap5Δ/hap5Δ mutant of C. albicans. Using mobility shift studies, we identified four separate complexes from C. albicans cell extracts whose DNA-binding activities were abolished in the hap5Δ/hap5Δ mutant, suggesting that they represented sequence-specific CCAAT-binding complexes. We found that the C. albicans hap5Δ homozygote was defective in hyphal development under a variety of conditions, and the mutant displayed a carbon source-dependent “hyperfilamentation” phenotype under certain growth conditions. In addition, the mRNA levels for two enzymes involved in respiration, encoded by COX5 and CYC1, were overexpressed in the hap5Δ/hap5Δ mutant when grown in medium containing amino acids as the sole carbon and nitrogen source. Thus, the C. albicans CCAAT-binding factor appeared to function as a repressor of genes encoding mitochondrial electron transport components, in contrast to its activator function in S. cerevisiae. These data provide the first evidence that the CCAAT-binding factor can act as a transcriptional repressor and raise new and interesting questions about how carbon metabolism is regulated in this opportunistic human pathogen.
The CCAAT-binding factor is a heterooligomeric transcriptional activator that binds to promoter elements containing the pentanucleotide sequence 5′-CCAAT-3′ (7, 34, 35). The CCAAT-binding factor is unique among DNA-binding proteins in that it requires three heterologous subunits for DNA-binding activity (reviewed in reference 34). The Saccharomyces cerevisiae CCAAT-binding factor is composed of four subunits, termed Hap2p, Hap3p, Hap4p, and Hap5p (34, 37), which are involved in the transcriptional activation of numerous genes that encode proteins involved in respiration (13, 15, 29, 57), as well as other genes (11, 29, 47, 57). The Hap2/3/5p heterotrimer has been shown to be sufficient for sequence-specific DNA binding at target promoters (37), yet the complex lacks the ability to stimulate transcription and requires a fourth subunit, Hap4p, for transcriptional activation (16). Thus, the interaction of Hap4p with Hap2/3/5p modulates the ability of the CCAAT-binding factor to stimulate transcription in yeast. The expression of HAP2, HAP3, and HAP5 has been shown to be constitutive (13), whereas HAP4 is repressed in the presence of glucose (16), providing the regulatory switch between fermentation and respiration (14, 29). Null mutations in any of the HAP genes cause S. cerevisiae strains to be unable to grow in the presence of nonfermentable carbon sources (16, 24, 37, 43), underscoring their importance as regulators of respiratory genes.
Hap2p, Hap3p, and Hap5p contain essential core domains that are highly conserved in eukaryotes, from fungi to humans (reviewed in reference 34). These core regions have been shown to be sufficient for the assembly and DNA-binding activity of the heterotrimeric complex (36). Moreover, the expression of the conserved domains of Hap2p and Hap3p in their respective null mutants is sufficient for functional complementation of the respiratory defect (41, 54, 55). Previous studies have shown that Hap5p from S. cerevisiae contains a 32-amino-acid region which is lacking in the homologous proteins of higher eukaryotes but present in other fungi (36). This fungus-specific conserved domain is necessary for Hap4p to interact with the Hap2/3/5p heterotrimer; hence, this region was termed the Hap4p recruitment domain (36). The in vivo expression of a HAP5 allele containing just the Hap5p core region and the Hap4p recruitment domain (HAP5C4) is sufficient to complement an S. cerevisiae hap5Δ mutant (36).
As mentioned previously, the CCAAT-binding factor is highly conserved structurally, yet the genes regulated by this transcription factor vary with the organism being considered. For example, the S. cerevisiae CCAAT-binding factor regulates genes that encode proteins needed for respiratory metabolism (15, 57), whereas in the filamentous fungus Aspergillus nidulans, this factor is involved in the regulation of genes involved in penicillin biosynthesis and other unrelated genes (6). In higher eukaryotes, the CCAAT-binding factor is a proximal promoter factor, like Sp1 (10), that regulates the expression of a large number of unrelated genes (34, 35). In fact, it has been suggested that CCAAT-binding sites are present in ∼30% of promoters from higher eukaryotes (7, 35).
Candida albicans is the most frequently encountered fungal pathogen in humans and is responsible for a variety of mucosal and systemic infections (8, 40). With the increasing number of patients that are immunocompromised, due in part to immunosuppressive therapies and AIDS, the incidence of fungal infections has risen over the past several years (18, 56). C. albicans and S. cerevisiae diverged from a common ancestor approximately 300 million years ago (42). The two yeasts exist in different environmental niches, with C. albicans surviving primarily in warm-blooded animal hosts (40) while S. cerevisiae is found on sugary plant exudates and fresh and decaying fruits (1). The fact that these yeasts thrive in different niches raises the question of whether transcription factors such as the CCAAT-binding factor have evolved to regulate the same or different genetic pathways in these organisms. S. cerevisiae is a respirofermentative yeast that generates energy by fermentation even in the presence of oxygen, whereas C. albicans generates energy using respiratory metabolism when oxygen is available, although it is considered a facultative anaerobe (14). Thus, even if the CCAAT-binding factor regulates similar genes in both organisms, the mechanism of gene regulation may be different as a result of their differing metabolic programs.
In this article, we report the characterization of the CCAAT-binding factor in C. albicans. We have cloned the gene encoding C. albicans Hap5p, generated a hap5Δ/hap5Δ mutant, and examined its phenotypes in both liquid and solid media. In addition, electrophoretic mobility shift assays performed with cell extracts from C. albicans revealed four distinct CCAAT-binding complexes whose DNA-binding activities were abolished in a hap5Δ homozygote. The hap5Δ/hap5Δ mutant displayed defects in hypha formation as well as aberrant expression of CYC1 and COX5, which encode two proteins involved in electron transport. Our findings indicate that the CCAAT-binding factor may be involved in the regulation of genes encoding proteins involved in respiration via repression and/or activation and raise a number of questions about how this central metabolic pathway may be integrated with the dimorphic transition.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used in this study are listed in Table 1. Strains were routinely cultured in yeast extract-peptone-dextrose (YPD) medium (22) supplemented with 80 mg/liter of uridine as appropriate. For DNA transformations, synthetic complete medium (SC) lacking auxotrophic supplements or synthetic minimal medium (SD) augmented with the auxotrophic requirements was used (22). Media for evaluating hypha formation, including Lee's (30), modified Lee's (31), and Spider (31) medium, were prepared as previously described. Serum medium consisted of YPD containing 10% newborn calf serum (Sigma). Medium M199 contained Earle's salts and glutamine but lacked sodium bicarbonate (Gibco-BRL) and contained 150 mM HEPES adjusted to pH 4.5 or 7.5. Yeast nitrogen base medium (YNB) with amino acids was prepared using 0.17% yeast nitrogen base without amino acids or ammonium sulfate (Difco) and 0.1% amino acid dropout powder containing the 20 amino acids at the described concentrations (22), buffered to the indicated pH with 150 mM HEPES. Glucose and lactate were added to media at a 2% final concentration as indicated, and media were solidified with 1.5% agar as appropriate. In liquid culture, germ tube formation was assessed at 37°C following inoculation of stationary-phase cultures into prewarmed medium at a density of ∼1 × 106 cells/ml. Filamentation on agar media was assessed by serial dilution of stationary-phase cultures in phosphate-buffered saline, spreading of ∼50 to 100 cells per plate on the appropriate medium, and incubation at 37°C.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Candida albicans strains | ||
| BWP17 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG | 48 |
| DMC104 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG HAP5/hap5Δ::URA3 | This study |
| DMC108 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG hap5Δ::URA3/hap5Δ::HIS1 | This study |
| DMC117 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG-ARG4 hap5Δ::URA3/hap5Δ::HIS1 | This study |
| DMC120 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG-HIS1 arg4Δ::hisG/arg4Δ::hisG-ARG4 HAP5/hap5Δ::URA3 | This study |
| DMC126 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG-HAP5-ARG4 hap5Δ::URA3/hap5Δ::HIS1 | This study |
| DMC146 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG-HIS1 arg4Δ::hisG/arg4Δ::hisG-ARG4-URA3 | This study |
| Saccharomyces cerevisiae strains | ||
| BWG1-7a | Mataura3-52 leu2-3,112 his4-519 ade1-100 | 36 |
| DMY110 | Mataura3-52 leu2-3,112 his4-519 ade1-100 hap5Δ::hisG | 36 |
| BY4733 | Matahis3Δ200 trp1Δ63 ura3Δ0 leu2Δ0 met15Δ0 | 5 |
Oligonucleotides.
The oligonucleotides used in this study are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea (5′-3′) |
|---|---|
| oDM0337 | GGCCggatccGGGTTGAAAGTCATTGCTGGTAGAGATATCAAAAGTGG |
| oDM0338 | GGCCaagcttGTGAGGAACTGAGAACTGATCTTACAAATAAGAGC |
| oDM0341 | GGCCgaattcGGTGCCACTTTATTTAAAACTAGATG |
| oDM0342 | GGCCggatccGCTTTCTTCAAATAAGTAACTAAATCG |
| oDM0361 | CAAACATTCAATTAAGACGAAGAAATACAGAAAGACCCCCCAAAACAATACTCAAACAATGTTTTCCCAGTCACGACGTT |
| oDM0362 | ATACGTAGCTTCATCATTATGACCATTTGCATTTTCCGCCCCATTTGTAATATCCTCTTCTGTGGAATTGTGAGCGGATA |
| oDM0369 | GGTGAGGCATGAGTTTCTGCTCTCTCA |
| oDM0370 | CTGTATATCGGCACCACTCAATAAGTTACAGCA |
| oDM0371 | GGTGGTGCCTTGTTTGTGGAAGAACAACAAGA |
| oDM0449 | GAGATCATTACAAAGAGCTGCCACTAAAGCC |
| oDM0450 | GGAGTTAATTCTTGCCATGGTAATTCC |
| oDM0459 | TAGCGGTTCTGACGTGCAAATCGATCGTCG |
| oDM0460 | ACTAACACCTTTTGTGGTGTCTGATGAGCG |
| UAS2UP1 top | GAATTTTGATCCACCAATCAACGCTCGCCAAATGAGC |
| UAS2UP1 bottom | GAATTGCTCATTTGGCGAGCGTTGATTGGTGGATCAA |
| UAS2 top | GAATTTTGATCCACCAACCAACGCTCGCCAAATGAGC |
| UAS2 bottom | GAATTGCTCATTTGGCGAGCGTTGGTTGGTGGATCAA |
Relevant restriction enzyme sites are shown in lowercase letters.
Plasmid construction.
Plasmid pDM569 contains a functional copy of CaHAP5 and was generated by amplification of C. albicans orf19.1973/orf19.9529 by PCR with the oligonucleotide primers oDM0337 and oDM0338, which incorporated unique BamHI and HindIII restriction sites into the 5′ and 3′ ends of the PCR product, respectively. The amplified fragment was digested with BamHI/HindIII and cloned into Yeplac181 (20) digested with the same enzymes. Plasmids pDM619 and pDM620, containing CaHAP5 in vectors pRS316 (51) and Yep352 (25), respectively, were constructed by digestion of pDM569, pRS316, and Yep352 with BamHI/HindIII, after which the CaHAP5 fragment and vectors were purified and ligated. The plasmid pDM407 contains S. cerevisiae HAP5 cloned into vector pRS316 at the XbaI and XhoI restriction sites. The hap5Δ::URA3 construct for the disruption of HAP5 in C. albicans was generated by homologous recombination in S. cerevisiae as follows. First, the plasmid pDM569 was introduced into S. cerevisiae BY4733 (5) by lithium acetate transformation (19), and Leu+ transformants were selected. A single transformant was grown to saturation in SC-Leu medium and subsequently inoculated into 10 ml of YPD medium, grown for 4 hours, and harvested for transformation with a hap5Δ::URA3 PCR product. The hap5Δ::URA3 construct was generated by PCR amplification from the template plasmid pGEM-URA3 (53) using primers oDM0361 and oDM0362, which incorporated 60 bp of homology to CaHAP5. The hap5Δ::URA3 PCR product was introduced into S. cerevisiae BY4733 containing pDM569 to allow homologous recombination between the plasmid and the hap5Δ::URA3 disruption cassette. Colonies were selected on SC-Leu-Ura and subsequently grown in SC-Leu-Ura liquid medium, and DNA was isolated by the glass bead-phenol method as previously described (26) and then introduced into Escherichia coli DH5α. Transformants containing the hap5Δ::URA3 allele were confirmed by restriction enzyme analysis, and the plasmid was designated pDM574. Plasmid pDM576, containing the hap5Δ::HIS1 disruption construct, was generated as follows. The HIS1 gene was isolated from pGEM-HIS1 (53) by digestion with SphI/NotI, and the same enzymes were used to release the URA3 gene from pDM574. The digested vector and HIS1 were ligated and transformed into E. coli DH5α. Restriction enzyme digestion was used to confirm the construct. The plasmid pDM590, containing CaHAP5 adjacent to ARG4, was generated in two steps. First, CaHAP5 was removed from the plasmid pDM569 by digestion with BamHI/HindIII and cloned into the same sites of plasmid pDM581, a derivative of pBluescript (Stratagene), to create the plasmid pDM585. To generate pDM590, the C. albicans integration vector pDM583, containing ARG4 (McNabb et al., manuscript in preparation), and pDM585 were digested with BamHI/XhoI, and the DNA fragments were purified and ligated. The plasmid pDM596 contains both ARG4 and URA3 for restoring prototrophy to C. albicans strains. It was generated by amplifying URA3 with oligonucleotide primers oDM0382 and 0DM0383, using pGEM-URA3 as the template. The primers introduce BamHI sites at both ends of the PCR product. URA3 and pDM583 were subsequently digested with BamHI, the vector was treated with shrimp alkaline phosphatase (Promega Corp.), and the DNA fragments were ligated.
Construction of C. albicans strains.
The hap5Δ/hap5Δ strain DMC108 was generated as follows. Two consecutive rounds of transformation of the parent strain BWP17 were performed using the hap5Δ::URA3 and hap5Δ::HIS1 disruption cassettes (see Fig. 3). The hap5Δ::URA3 cassette was released from plasmid pDM574 by digestion with BamHI/HindIII and introduced into BWP17 using a lithium acetate transformation kit (QBiogene, Inc.), and transformants were selected on SC-Ura medium. To verify the HAP5/hap5Δ::URA3 heterozygote, genomic DNAs were isolated from multiple transformants as described previously (26), and PCR was used to confirm the appropriate recombination. For the PCRs, oligonucleotide primers oDM0369 (anneals within the URA3 gene) and oDM0371 (anneals to CaHAP5 loci upstream of the recombination) were used. The HAP5/hap5Δ::URA3 strain, designated DMC104, was subsequently transformed with SmaI/HindIII-digested pDM576 containing the hap5Δ::HIS1 allele as described above, and transformants were selected on SC-His medium. The transformants were subsequently tested on SC-His-Ura medium to verify recombination at the HAP5 locus versus the hap5Δ::URA3 locus. Genomic DNAs were prepared from His+ Ura+ transformants, and PCR was used to verify the correct recombination, using oligonucleotide primers oDM0370 (anneals within HIS1) and oDM0371 (anneals to HAP5 loci upstream of the recombination). The final strains were confirmed by Southern blot analysis.
FIG. 3.
Southern blot analysis of the hap5Δ/hap5Δ mutant. (A) hap5Δ::URA3 and hap5Δ::HIS1 deletion alleles were generated as outlined in Materials and Methods. The complete coding sequence of CaHAP5 was removed and replaced with the indicated selectable markers. (B) Genomic DNAs were isolated from strains BWP17 (HAP5/HAP5), DMC104 (HAP5/hap5Δ::URA3), and DMC108 (hap5Δ::URA3/hap5Δ::HIS1), digested with PstI/HindIII, and analyzed by Southern blot hybridization using the probe indicated in panel A.
To reintroduce CaHAP5 into the hap5Δ::URA3/hap5Δ::HIS1 mutant (DMC108), the plasmid pDM590 was digested with HpaI to direct integration at the arg4 locus, and the linear plasmid was introduced into the hap5Δ homozygote by lithium acetate transformation (Qbiogene, Inc.), with transformants selected on SC-Arg medium. As a control strain, the hap5Δ homozygote was transformed with pDM583 vector which had been linearized with HpaI within ARG4. To rescue the his1 and arg4 auxotrophies of the HAP5/hap5Δ::URA3 heterozygote, the strain DMC104 was sequentially transformed with pGEM-HIS1 linearized with NruI and pDM583 linearized with HpaI. The auxotrophies of BWP17 were rescued by sequential transformation with pGEM-HIS1 linearized with NruI and pDM596 digested with HpaI within ARG4. All strains used for phenotypic analyses were prototrophic, as confirmed by growth on synthetic minimal medium.
Determination of generation time.
Strains were grown to saturation in YPD medium and subsequently diluted in fresh prewarmed YPD medium at a starting density of 1 × 106 cells/ml and incubated at 30°C. Immediately after inoculation and at 2-hour intervals, samples were removed, diluted appropriately, and counted on a hemocytometer. Growth was monitored for an 8-hour period, and the doubling time was calculated from the data obtained during exponential growth. The procedure was repeated three times, and mean values with standard deviations are reported.
Yeast extract preparation and electrophoretic mobility shift assays.
Protein extracts were prepared as described previously (37), with some minor modifications. Cells were grown in YPD medium to an optical density at 600 nm (OD600) of ∼1.0 and harvested by centrifugation at 5,000 × g for 5 min. The cells were washed with a 1/25 volume of extraction buffer [200 mM Tris-HCl, pH 8.0, 400 mM (NH4)2SO4, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, 1 μg/ml leupeptin, 1 μg/ml pepstatin], and centrifugation was repeated. The cell pellet was resuspended in a 1/500 volume of extraction buffer, and the cells were disrupted by agitation at 4°C with a vortex mixer in the presence of an equal volume of glass beads (0.45-mm diameter). Following incubation for 30 min on ice, unlysed cells and glass beads were removed by centrifugation, and the proteins in the supernatant were precipitated by the addition of 100% (NH4)2SO4 in 10 mM HEPES at pH 8.0 and 5 mM EDTA to a final (NH4)2SO4 concentration of 40%. Following incubation for 30 min at 4°C with gentle agitation, the precipitated proteins were collected by centrifugation at 10,000 × g for 10 min, and the pellet was resuspended in DNA-binding buffer (20 mM HEPES-NaOH at pH 7.9, 100 mM KCl, 1 mM EDTA, 20% glycerol, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 5 mM benzamidine) and dialyzed against the same buffer before use in mobility shift assays. The UAS2UP1 probe (CCAAT) was designed as described previously (23) and is based on the sequence of UAS2UP1 from CYC1 of S. cerevisiae. The UAS2 probe (CCAAC) is identical in sequence to UAS2UP1 except for a T-to-C change in the CCAAT site (17). The DNA probe was end labeled by the Klenow fragment with [α-32P]dATP (Amersham Corp.). All binding reactions contained 20 to 30 μg of cell extract in DNA-binding buffer, 1 μg of denatured salmon sperm DNA, 1 μg of poly(dI-dC), and 0.5 to 1.0 ng of radiolabeled probe in a final reaction volume of 20 μl. Reactions were incubated at room temperature for 30 to 45 min, and the protein-DNA complexes were resolved by gel electrophoresis (4 h at 300 V) on 5% polyacrylamide gels (acrylamide:bisacrylamide ratio, 29:1) in 0.5× Tris-borate-EDTA at 4°C. For competitor reactions, the unlabeled UAS2 and UAS2UP1 probes were added at 100×, 200×, and 400× concentrations relative to the radiolabeled probe. After electrophoresis, the gels were fixed, dried, and visualized with a Molecular Dynamics PhosphorImager.
Microscopy and imaging.
Photographs of colony morphology were taken with a Nikon Biaphot microscope fitted with a high-resolution charge-coupled device camera and the AutoMontage imaging software package (Syncroscopy, Frederick, MD). Photographs of individual cells were done with a Zeiss Axioplan 2 microscope fitted with a high-resolution charge-coupled device camera and AutoMontage imaging software. Photographs of individual colonies were representative of the total population. The figures were prepared using Adobe Photoshop 7.0 and Microsoft PowerPoint 2000.
Southern blot analysis.
For preparation of C. albicans genomic DNA, strains were grown to saturation in YPD medium at 30°C. Genomic DNAs were isolated for Southern blot analysis using the glass bead-phenol protocol as previously described (26). DNAs were digested with PstI/HindIII, resolved by electrophoresis on 0.7% agarose gels, and transferred to GeneScreen Plus membranes (NEN Life Sciences Products) as described by the manufacturer's protocol. The probe for the CaHAP5 locus was a 550-bp fragment from the 5′ end of the gene obtained by the digestion of pDM569 with BamHI/EcoRI. The probe was radioactively labeled with [α-32P]dATP (Amersham) using a random-primed DNA labeling kit (Boehringer Mannheim) according to the manufacturer's instructions. Hybridizations and washes were performed at 65°C using previously described protocols (49).
Northern blot analysis.
C. albicans strains were grown to saturation in YPD medium and subsequently inoculated into the indicated medium and grown for 4 hours at 30°C. The cells were harvested by centrifugation, and total RNA was prepared by the glass bead-acid phenol method as previously described (2). Approximately 20 μg of each total RNA sample was loaded, separated by formaldehyde-1% agarose gel electrophoresis, and transferred to GeneScreen Plus membranes (Dupont-NEN Research Products) according to the manufacturer's protocol. The membranes were hybridized and washed under standard high-stringency conditions (49). The CYC1, COX5, and 26S rRNA probes for hybridization were obtained by PCR amplification from C. albicans genomic DNA using the primer pairs oDM0341/oDM342, oDM0449/oDM0450, and oDM0459/oDM0460, respectively. The probes were purified by agarose gel electrophoresis and GeneClean (Qbiogene, Inc.) and subsequently radiolabeled with [α-32P]dATP (Amersham) by use of a random primer labeling kit (U.S. Biochemicals) according to the manufacturer's protocol. The transcript levels were quantified on a Molecular Dynamics PhosphorImager.
RESULTS
Identification of CaHAP5 and functional complementation of S. cerevisiae hap5Δ.
S. cerevisiae Hap5p and its homologs in other fungi have been shown previously to have two distinct functional domains (Fig. 1A). The first domain is conserved in all eukaryotes and is comprised of 89 amino acid residues that form a histone fold structure (3, 48) necessary for interactions with Hap2p and Hap3p to form the heterotrimeric DNA-binding complex (36). The second domain consists of 32 amino acid residues (termed the Hap4p recruitment domain) immediately N-terminal to the histone fold core that are essential for the interaction of Hap4p with the Hap2/3/5p complex (36). Using these conserved regions of S. cerevisiae Hap5p, we searched the Candida albicans genome databases (http://genolist.pasteur.fr/CandidaDB and http://www.candidagenome.org/) and identified orf19.1973/orf19.9529, which showed 71% identity and 81% similarity (Fig. 1A), strongly suggesting that it was the authentic CaHAP5 gene. There was no obvious homology between the proteins outside the two conserved regions, and no other open reading frames in the C. albicans genome showed a high degree of identity (data not shown). Compared with the other Hap5p homologs known to functionally complement an S. cerevisiae hap5Δ mutant, including those encoded by Schizosaccharomyces pombe php5 (36) and Neurospora crassa aab-1 (9; D. S. McNabb, unpublished observations), the putative C. albicans Hap5p protein displayed the greatest similarity (Fig. 1A). CaHAP5 is predicted to encode a protein with 348 amino acid residues with a C-terminal 124-amino-acid segment that extends beyond the conserved domains. This region is notable only because it is absent from S. cerevisiae Hap5p and contains 25 glutamine and 22 acidic amino acid residues, raising the question of whether it may function as either an acidic or glutamine-rich transcriptional activation domain. Previous studies have demonstrated that glutamine-rich activators do not function in S. cerevisiae (44) but may function in S. pombe (46).
FIG. 1.
C. albicans HAP5 complements an S. cerevisiae hap5Δ mutant. (A) Amino acid sequence alignment of the conserved domains of Hap5p homologs. Above the alignment is a schematic representation of the Hap4p recruitment and histone fold domains (48) of the homologs. α1, -2, -3, and -C designate the alpha helices of the histone fold motif, and L1, L2, and LC represent the loop/beta-strand segments. The GenBank accession numbers for sequences are as follows: C. albicans Hap5p, EAK96721.1; S. cerevisiae Hap5p, NP015003.1; Schizosaccharomyces pombe Php5p, NP596412.1; Neurospora crassa Aab1p, EAA28004.1; Arabidopsis thaliana NF-YC, NP176013.1; and Mus musculus NF-YC, AAH53723.1. (B) Complementation of the respiratory deficiency of S. cerevisiae hap5Δ with CaHAP5. Strains BWG1-7a (HAP5) and DMY110 (hap5Δ) were transformed with the indicated genes carried on either an ARS/CEN (A/C) or 2μm vector. The strains were grown at 30°C on YPD or yeast extract-peptone-lactate, as indicated, for 3 days and then photographed.
To demonstrate that orf19.1973/orf19.9529 was the authentic CaHAP5 gene, we tested whether the gene could functionally complement the respiratory deficiency of an S. cerevisiae hap5Δ mutant. For these studies, S. cerevisiae strain DMY110 (hap5Δ) was transformed with an ARS/CEN vector containing either ScHAP5 or CaHAP5 or a 2μm vector containing CaHAP5. As controls, S. cerevisiae BWG1-7a (HAP5) and DMY110 (hap5Δ) were transformed with an ARS/CEN vector only. Transformants were selected on SC-Ura medium and subsequently replica plated on YPD or yeast extract-peptone-lactate and incubated at 30°C. As previously reported (37), the hap5Δ strain failed to grow on rich medium containing lactate as the carbon source, but the CaHAP5 gene restored normal growth when carried on a low- or high-copy-number plasmid (Fig. 1B). These data demonstrated that the C. albicans Hap5p protein is competent to assemble with Hap2p and Hap3p to bind DNA. Furthermore, the complementation also implies that C. albicans Hap5p can recruit Hap4p to the complex and stimulate transcription in vivo, suggesting that C. albicans may encode a Hap4p homolog. In fact, using the conserved domain within Hap4p homologs (4), we and others (52) have identified three open reading frames that encode putative Hap4p proteins in C. albicans, and the open reading frames have been designated HAP41 (orf19.740/orf19.8359), HAP42 (orf19.1481/orf19.9056), and HAP43 (orf19.681/orf19.8298) (McNabb, unpublished observations). While it remains to be established whether these open reading frames encode functional Hap4p subunits, the data presented in this report strongly support the existence of multiple Hap4p proteins in C. albicans.
Identification of multiple CCAAT-binding complexes in C. albicans.
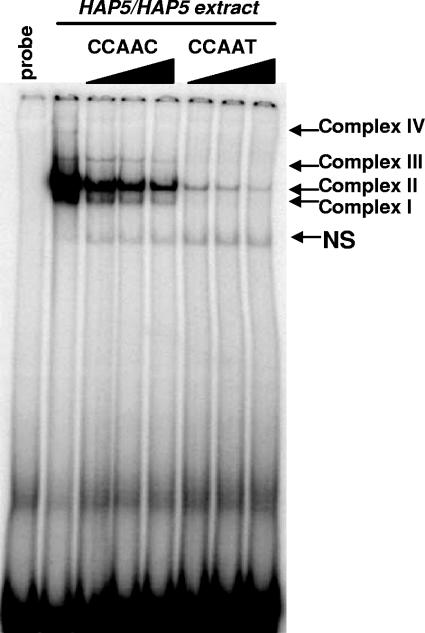
With the identification of C. albicans HAP5, we wanted to determine whether CCAAT-binding activity could be detected in cell extracts prepared from C. albicans. Thus, C. albicans DMC146 was grown in YPD medium at 30°C to an OD600 of ∼1, and the cells were harvested and cell extracts prepared as described in Materials and Methods. To assess CCAAT-binding activity, we used a 37-bp double-stranded DNA oligonucleotide derived from the sequence of UAS2UP1 (CCAAT) from the S. cerevisiae CYC1 gene (17, 23). With this probe, mobility shift assays using DMC146 extracts revealed four distinct CCAAT-binding complexes that were designated complexes I through IV (Fig. 2). Complex II was the most abundant, with complex I migrating slightly faster and being less abundant. Complexes III and IV were much less obvious but were clearly present. To evaluate the specificities of these DNA-binding complexes, competition experiments were performed using two different unlabeled DNA probes either identical to the radiolabeled CCAAT probe or containing a T-to-C mutation (CCAAC) that reduces the DNA-binding affinity (17). The CCAAC sequence competed with the radioactive probe for the binding of all four complexes (Fig. 2), but the competition with the CCAAT probe was more efficient due to its higher binding affinity. These data strongly suggested that complexes I through IV are sequence-specific CCAAT-binding complexes. A faster-migrating DNA-protein complex (termed NS) was considered nonspecific since it was not competed by either competitor sequence. In summary, the mobility shift studies identified multiple CCAAT-binding complexes in C. albicans. At this stage, we cannot rule out the existence of additional CCAAT-binding complexes since the probe was not derived from a native C. albicans promoter and only a single growth condition was used for extract preparation.
FIG. 2.
Identification of four CCAAT-binding complexes in C. albicans cell extracts. Mobility shift assays were performed with DNA-binding reaction mixtures containing a radiolabeled CCAAT-box oligonucleotide probe incubated with cell extracts prepared from strain DMC146 grown in YPD medium (HAP5/HAP5 extract) at 30°C. The four complexes that bound specifically to the CCAAT-box probe are indicated on the right (complexes I through IV). A nonspecific DNA-binding complex (NS) was also observed. The specificity of the CCAAT-binding complexes was shown using competition experiments, with an unlabeled CCAAC or CCAAT oligonucleotide probe added to the reactions at a 100-, 200-, or 400-fold excess, as indicated. The first lane contains the radiolabeled CCAAT-box probe alone (probe).
Deletion of HAP5 abolishes CCAAT-binding activity in C. albicans.
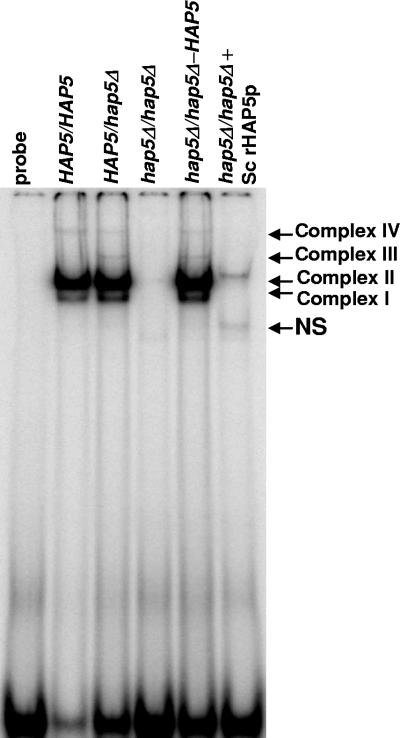
To determine whether the CCAAT-binding complexes detected by gel shift assays were Hap5p dependent, HAP5/hap5Δ and hap5Δ/hap5Δ mutants of Candida albicans were generated as outlined in Materials and Methods. The null mutations were initially confirmed by PCR, and the genomic structure of each locus was subsequently verified by Southern blot analysis (Fig. 3). A CaHAP5 integration construct was targeted to the arg4 locus of the hap5Δ homozygote to reintroduce the wild-type gene. Once the mutants and rescued strains were generated, the remaining auxotrophies in each strain were marker rescued to prototrophy.
To examine the effect of the hap5Δ null mutations on CCAAT-binding activity, the heterozygous mutant (HAP5/hap5Δ), homozygous mutant (hap5Δ/hap5Δ), and HAP5-rescued (hap5Δ/hap5Δ ARG4-HAP5) strains were grown in YPD at 30°C to an OD600 of ∼1, and cell extracts were prepared and used for mobility shift assays. The heterozygous strain displayed CCAAT-binding complexes I through IV, while the hap5Δ homozygote showed no detectable CCAAT-binding activity (Fig. 4). When HAP5 was reintroduced, the four CCAAT-binding complexes were restored, demonstrating that Hap5p is essential for the DNA-binding activities of complexes I through IV. In this experiment, complex III was not seen in the wild-type extract, a result that was likely due to the lower concentration of the CCAAT-box probe in the reaction, as indicated by the amount of free probe. The same wild-type extract preparation was used for the mobility shifts shown in Fig. 2, where complex III is present. To determine whether CCAAT-binding activity could be reconstituted by the addition of exogenous Hap5p, recombinant S. cerevisiae Gst-Hap5p (37) was added to the hap5Δ/hap5Δ extract. A single DNA-bound complex was observed which was likely equivalent to complex II based on the relative size of Gst-ScHap5p to C. albicans Hap5p. In summary, these data provide additional evidence that we have cloned the authentic HAP5 gene from C. albicans and that the encoded protein is involved in the assembly of at least four sequence-specific CCAAT-binding complexes.
FIG. 4.
CCAAT-binding complexes I through IV are Hap5p dependent. Mobility shift assays were performed with DNA-binding reaction mixtures containing a radiolabeled CCAAT-box probe incubated with crude extracts prepared from strains DMC146 (HAP5/HAP5), DMC120 (HAP5/hap5Δ), DMC117 (hap5Δ/hap5Δ), and DMC126 (hap5Δ/hap5Δ-HAP5), as indicated. For extract preparation, the strains were grown in YPD medium at 30°C. Purified recombinant S. cerevisiae GST-Hap5p (Sc rHAP5p) was included in the reaction as indicated. The CCAAT-binding complexes I through IV and the nonspecific DNA-binding complex (NS) are indicated to the right of the panel. As a control, the binding reaction in the first lane contains the unbound probe.
The C. albicans hap5Δ/hap5Δ mutant has a slow growth phenotype.
To further characterize the hap5Δ homozygote, we examined the generation time of the strain in rich medium with glucose as the carbon source (YPD). Stationary-phase cultures of the wild-type (HAP5/HAP5), heterozygous mutant (HAP5/hap5Δ), homozygous mutant (hap5Δ/hap5Δ), and HAP5-rescued (hap5Δ/hap5Δ ARG4-HAP5) strains of C. albicans were inoculated into YPD medium at an initial density of 1 × 106 cells/ml and grown at 30°C. Growth was monitored by cell counts with a hemocytometer every 2 hours after inoculation, and doubling times were calculated from these data. The growth experiment was carried out three times independently, and the data reported are means ± standard deviations. The HAP5/HAP5 parent strain and the HAP5/hap5Δ heterozygote had generation times of 55 ± 6 and 53 ± 3 minutes, respectively, while the hap5Δ homozygote had a longer doubling time (97 ± 6 minutes). The long generation time of the mutant was restored to normal (59 ± 2 min) by reintroducing HAP5, implying that the C. albicans CCAAT-binding factor regulates a genetic pathway(s) that is important for optimal growth.
The C. albicans hap5Δ/hap5Δ mutant has defects in the morphogenetic transition.
To determine whether the CCAAT-binding factor functions in the yeast-to-hypha transition, appropriate strains were grown in several media known to induce hyphal development. On solid medium, the hap5Δ homozygote displayed defective hyphal growth on all of the media examined. The most prominent defects in filamentation occurred with YPD containing 10% calf serum and with Spider medium (Fig. 5). The hap5Δ homozygote developed hyphae on modified Lee's medium, although the colonies were flat and lacked a wrinkled appearance, while the HAP5 strains formed wrinkled colonies that protruded upward and developed extensive filaments (Fig. 5). Hyphal growth on Lee's medium was significantly delayed, and filamentation was observed only after 9 days of incubation at 37°C (data not shown), which may reflect the slow growth of the hap5Δ/hap5Δ mutant. The hap5Δ homozygote displayed a “hyperfilamentation” phenotype on M199 medium at pH 7.5 (Fig. 5), showing significant hyphal growth after 2 days at 37°C. In contrast, the HAP5-containing strains did not form hyphal extensions until day 4. Interestingly, the hap5Δ/hap5Δ mutant formed a much smaller colony prior to filamentation, suggesting that as the colony develops the cells become more sensitive to the changing environment and respond by switching to filamentous growth. When the same strains were grown on M199 medium at pH 4.5, the hap5Δ/hap5Δ mutant grew slightly slower than the wild-type strain, but otherwise was unremarkable (data not shown), suggesting that the hyperfilamentation seen on M199 medium required an alkaline pH environment.
FIG. 5.
Defects in filamentation caused by loss of the CCAAT-binding factor. Prototrophic strains DMC146 (HAP5/HAP5), DMC117 (hap5Δ/hap5Δ), and DMC126 (hap5Δ/hap5Δ ARG4::HAP5) were grown on the indicated media for 4 days at 37°C, except for Spider medium, which was incubated for 6 days at 37°C.
To examine whether the defective hyphal growth on solid media occurred in liquid culture, the wild-type (HAP5/HAP5), heterozygous mutant (HAP5/hap5Δ), homozygous mutant (hap5Δ/hap5Δ), and HAP5-rescued (hap5Δ/hap5Δ ARG4-HAP5) strains of C. albicans were inoculated into Lee's medium, modified Lee's medium, serum, and M199 (pH 7.5) liquid medium, and germ tube formation was examined at 2-hour intervals for a total of 8 hours. For most of these media, the hap5Δ/hap5Δ mutation had no effect on hyphal development, and the kinetics of germ tube formation was normal (data not shown). The only exception was Spider medium (Fig. 6). The hap5Δ/hap5Δ mutant did not form germ tubes in Spider medium at 37°C, and normal germ tube growth was restored by the reintroduction of HAP5 (Fig. 6). To determine whether this represented a difference in the kinetics of germ tube formation related to the low growth rate, the incubation of the hap5Δ/hap5Δ cultures in Spider medium was extended to 10 h at 37°C, yet no germ tubes were observed (data not shown). In summary, the hap5Δ/hap5Δ mutant displayed delays or defects in the yeast-to-hypha transition on all of the hypha-inducing solid media that were examined; however, Spider medium was the only liquid medium that showed defective germ tube formation.
FIG. 6.
Loss of the CCAAT-binding factor results in defective germ tube formation in liquid Spider medium. Prototrophic strains DMC146 (HAP5/HAP5), DMC117 (hap5Δ/hap5Δ), and DMC126 (hap5Δ/hap5Δ ARG4::HAP5) were grown in liquid Spider medium at 37°C, and samples were removed every 2 hours to evaluate germ tube formation. Photomicrographs are shown for the 0-h and 6-h time points, as indicated at the left of the figure.
The hyperfilamentation phenotype of the hap5Δ/hap5Δ mutant occurs in the absence of glucose.
In an effort to understand the phenotypes of the hap5Δ homozygote, we considered the known regulatory functions of the CCAAT-binding factor in the highly divergent yeasts S. cerevisiae and S. pombe. In both organisms, the CCAAT-binding factor has been shown to regulate the expression of genes needed for respiratory metabolism (6, 15, 32, 36, 57). For C. albicans, it was previously suggested that defects in respiration resulted in cells switching from yeast to hyphal growth (28), and the phenotypes of the C. albicans hap5Δ homozygote on hypha-inducing media that contained little to no glucose, namely M199 (pH 7.5), Spider, and modified Lee's media, were consistent with a defect in carbon source utilization that resulted in aberrant filamentation. Since C. albicans uses primarily respiratory metabolism when grown in an oxygenated environment (14), the slow growth of the hap5Δ/hap5Δ mutant in YPD medium could also be explained by the inefficient oxidation of glucose to yield energy. In support of this argument, we have empirically noted that the total biomass yield for the hap5Δ/hap5Δ mutant grown in YPD medium was reduced relative to that for the HAP5 strains (data not shown). Considering the phenotypes of the C. albicans hap5Δ homozygote on various media in conjunction with the known functions of the CCAAT-binding factor in budding and fission yeasts, we decided to investigate the relationship between the carbon source, the hyperfilamentation phenotype, and respiration.
To evaluate the association between the carbon source and the anomalous filamentation phenotype, a synthetic solid medium was used that contained yeast nitrogen base with amino acids as the nitrogen source and either glucose (YNB-glucose + AA), lactate (YNB-lactate + AA), or no additional carbon source (YNB + AA). Since the hyperfilamentation on M199 medium appeared to be pH dependent, the synthetic media were prepared at both pH 5.6 and pH 6.8 for comparison. Thus, saturated cultures of the wild-type (HAP5/HAP5), homozygous mutant (hap5Δ/hap5Δ), and HAP5-rescued (hap5Δ/hap5Δ ARG4-HAP5) strains of C. albicans were plated on these media and incubated at 37°C (Fig. 7). The strains containing HAP5 grew normally on YNB + AA (pH 5.6), albeit slower than in the presence of glucose, and the colony morphology was consistent with the yeast growth form (Fig. 7A). In contrast, the hap5Δ/hap5Δ mutant developed a small yeast colony and switched to filamentous growth, resembling the phenotype on M199 medium (pH 7.5) (Fig. 5). This was unexpected given the acidic pH of the medium. However, when the strains were grown at pH 6.8 (Fig. 7B), the colony morphology was exclusively filamentous. In addition, the HAP5-containing strains also displayed hyphae at the more alkaline pH (Fig. 7B). To further evaluate the hap5Δ/hap5Δ colony morphology at pH 6.8, the HAP5/HAP5 and hap5Δ/hap5Δ strains were grown on YNB + AA (pH 6.8) at 37°C, and the colony morphology was evaluated at 24-h intervals (Fig. 8). The HAP5/HAP5 strain formed a typical yeast colony that progressed to hyphal development after 3 days of incubation. In contrast, the hap5Δ/hap5Δ mutant formed hyphae within the first 24 h, and the macroscopic colony was exclusively filamentous.
FIG. 7.
The hyperfilamentation of a C. albicans hap5Δ homozygote is carbon source dependent. (A) Strains DMC146 (HAP5/HAP5), DMC117 (hap5Δ/hap5Δ), and DMC126 (hap5Δ/hap5Δ ARG4::HAP5) were grown at 37°C for 5 days on YNB + AA medium buffered to pH 5.6 with 150 mM HEPES. Glucose or lactate (2%) was added to the medium as indicated. (B) Strains shown in panel A grown in YNB + AA medium buffered to pH 6.8 with 150 mM HEPES. Photographs in each row were done at the same magnification for comparisons of colony sizes.
FIG. 8.
Kinetics of hyperfilamentation for the hap5Δ/hap5Δ mutant. Strains DMC146 (HAP5/HAP5) and DMC117 (hap5Δ/hap5Δ) were grown at 37°C on YNB + AA medium buffered to pH 6.8 with 150 mM HEPES. Photomicrographs were taken at 24-h intervals to monitor colony morphology. The photographs in each row were done at the same magnification for comparisons of the size and growth of the strains. The sizes and morphology of strains DMC120 (HAP5/hap5Δ) and DMC126 (hap5Δ/hap5Δ ARG4::HAP5) were identical to those of DMC146 (data not shown).
If the hyperfilamentation of the mutant were due to the poor carbon source or the inadequate assimilation of amino acids, then the addition of glucose to the medium should rescue the defect, whereas the addition of lactate (a poor carbon source) would fail to restore normal growth. Thus, the hap5Δ/hap5Δ mutant was grown on YNB-glucose + AA and YNB-lactate + AA at pH 5.6 or 6.8 for 5 days at 37°C (Fig. 7A and B). The colony morphology of the mutant was similar to that for the HAP5 strains in the presence of glucose, regardless of the pH. In contrast, the addition of lactate to the medium did not rescue the hyperfilamentation of the hap5Δ/hap5Δ mutant at either pH, although the defect was not as severe as that with the medium containing only amino acids. Importantly, the hyperfilamentation defect of the hap5Δ/hap5Δ mutant was rescued by reintroduction of the HAP5 allele, demonstrating that the phenotype was due solely to the loss of Hap5p. These data argue that the hap5Δ homozygote is defective in the use of alternative carbon sources that require respiration and suggest that the regulation of genes involved in respiratory metabolism may be impaired by a loss of CCAAT-binding activity. The fact that the more alkaline pH seemed to exacerbate the phenotype is likely indirect, since the hyperfilamentation was apparent at either pH 5.6 or pH 6.8.
The CCAAT-binding factor is involved in the regulation of CYC1 and COX5.
To establish a connection between the hyperfilamentation phenotype and respiratory metabolism, we examined the promoters of several C. albicans genes that are necessary for respiration and also known to be regulated by the CCAAT-binding factor in S. cerevisiae (13, 17, 32, 57) and S. pombe (36). We found that the promoters for COX5 and CYC1, encoding cytochrome oxidase subunit V and cytochrome c, respectively, contained canonical CCAAT boxes at positions −409 and −251, respectively, relative to the AUG translational start codon. To evaluate whether the transcription of these genes was altered in a hap5Δ/hap5Δ mutant, the appropriate strains were grown in YNB + AA, pH 5.6, at 30°C in the presence or absence of glucose, total RNAs were isolated, and the COX5 and CYC1 mRNA levels were examined by Northern blot analysis (Fig. 9A). In the presence of glucose, CYC1 mRNA levels were the same in the HAP5/HAP5 strain and the hap5Δ/hap5Δ mutant. A twofold decrease in COX5 mRNA levels was observed, which suggests that the CCAAT-binding factor may be important for the optimal expression of this gene. These data argue that the CCAAT-binding factor is not essential for regulating the level of CYC1 transcription in the presence of glucose. The fact that neither of these genes is repressed by glucose in the wild-type strain is consistent with the use of respiration by this organism and differs from the glucose repression observed with these genes in the respirofermentative yeast S. cerevisiae (14). In the medium lacking glucose, the levels of COX5 and CYC1 mRNAs decreased two- to threefold and four- to fivefold, respectively, in the HAP5 strains. In contrast, the COX5 mRNA level was unchanged and the CYC1 mRNA level was twofold more abundant in the hap5Δ/hap5Δ mutant. Thus, the hap5Δ/hap5Δ mutant expressed an eightfold higher level of CYC1 mRNA than did the HAP5-containing strains grown in YNB + AA. These data suggested that the CCAAT-binding factor may be involved in the repression of CYC1 and COX5 gene expression in the absence of glucose. Alternatively, a morphological difference between the hap5Δ/hap5Δ mutant and the HAP5 strains could account for the altered gene expression. To address this possibility, cells were taken at the time of RNA isolation and examined microscopically (Fig. 9B). For all strains, the cells appeared to have a pseudohyphal morphology, regardless of the genotype or growth conditions, indicating that the effect of the CCAAT-binding factor on CYC1 and COX5 transcription was unrelated to morphology. It was unclear why the strains were pseudohyphal, but this may reflect the use of amino acids as the nitrogen source rather than ammonium salts under the liquid culture conditions used for these studies.
FIG. 9.
C. albicans CCAAT-binding factor represses the expression of COX5 and CYC1. (A) Northern blot analysis of total RNAs isolated from C. albicans DMC146 (HAP5/HAP5), DMC120 (HAP5/hap5Δ), DMC117 (hap5Δ/hap5Δ), and DMC126 (hap5Δ/hap5Δ ARG4::HAP5) grown at 30°C in YNB-glucose + AA (pH 5.6) or YNB + AA (pH 5.6), as indicated. The membrane was hybridized with radiolabeled probes specific for COX5 and CYC1 mRNAs, as indicated. 26S rRNA was used to normalize RNA loading, and the ethidium bromide-stained rRNA is shown at the bottom of panel A. (B) Morphology of cells at the time of RNA isolation. Cells with the indicated genotypes were taken at the time of RNA isolation, fixed with 10% formaldehyde, and photographed. The growth medium for the strains is indicated to the left of each row.
In summary, these data indicate that the C. albicans CCAAT-binding factor is important for the regulation of genes involved in respiration. Interestingly, it functions primarily as a transcriptional repressor in C. albicans, in contrast to its role as an activator in S. cerevisiae. However, our results with COX5 suggest that the CCAAT-binding factor may also be important in transcriptional activation on glucose-containing medium. Thus, this transcription factor may have a dual function in C. albicans, depending on the growth environment. If the CCAAT sites we have identified in the CYC1 and COX5 promoters are functional binding sites, these results would imply that the CCAAT-binding factor represses respiratory gene transcription in the presence of poor quality carbon sources such as amino acids. The requirement for other transcription factors in the regulation of CYC1 and COX5 transcription will require further investigation of these two promoters.
DISCUSSION
In this work, we described the identification of the C. albicans homolog of the HAP5 gene encoding a subunit of the CCAAT-binding factor, and by generating a hap5Δ/hap5Δ mutant, evaluated the function of this transcription factor in C. albicans. We identified four distinct CCAAT-binding complexes by mobility shift assays and have shown that the deletion of CaHAP5 abolishes the DNA-binding activity of the four complexes, suggesting that they are sequence-specific CCAAT-binding factors. Moreover, we have demonstrated that the hap5Δ homozygote is defective in the yeast-to-hypha transition on various hypha-inducing media, and the hap5Δ/hap5Δ mutant displayed a hyperfilamentation phenotype on yeast nitrogen base medium containing a poor carbon source. Hyperfilamentation was suppressed by the addition of glucose, but not lactate, to the medium, suggesting that the mutant may have defects in the expression of genes related to respiratory metabolism. Northern blot analysis of two genes, COX5 and CYC1, encoding components of electron transport revealed that the CCAAT-binding factor was important for the repression of COX5 and CYC1 in amino acid-based medium lacking an additional carbon source and may be important in the activation of COX5 in glucose-containing medium. To our knowledge, this is the first evidence showing that the CCAAT-binding factor can function in transcriptional repression. We are cautious in stating that the CCAAT-binding factor acts directly to affect CYC1 and COX5 expression, since we have not demonstrated a direct interaction of the DNA-binding complex with the COX5 or CYC1 promoter, but this would be the most logical explanation for our results. Nevertheless, our future studies will focus on identifying cis-acting elements in the COX5 and CYC1 promoters important for their expression in the presence of different carbon sources. Unfortunately, our current understanding of metabolic gene regulation in respiratory yeasts is limited, making it difficult to place our results in the context of a regulatory pathway. It is also problematic to use S. cerevisiae as a model for understanding our data since the metabolic controls for respiratory versus respirofermentative yeasts may differ dramatically (14). Nevertheless, this study provides the framework for our future endeavors to evaluate the regulatory function of the CCAAT-binding factor in C. albicans.
Why did the higher pH environment seem to enhance the hyperfilamentation phenotype? At this stage, we cannot answer this question, although we did examine the mRNA levels of two known pH-responsive genes, PHR1 and PHR2 (38, 50), in a hap5Δ/hap5Δ mutant grown in M199 medium at pH 4.5 versus pH 7.5. The PHR1 mRNA levels showed normal induction in response to the alkaline pH compared to those in a HAP5 strain, while PHR2 mRNA levels were higher under acidic conditions (data not shown). Thus, the CCAAT-binding factor does not appear to be involved in the regulation of these pH-responsive genes, which have been previously shown to be regulated by Rim101p (12, 45). Another explanation that we tend to favor relates to the fact that the amino acid permeases may function less efficiently at pH 6.8 than at pH 5.6. For example, previous studies have shown that the Km of the Can1p permease is <2 μM at pH 5.0 and rises eightfold, to 16 μM, at pH 6.5 (39). The less efficient import of amino acids at pH 6.8, coupled with inefficient respiration, may have resulted in the cells sensing nitrogen and/or carbon starvation more rapidly and switching to hyphal growth.
As previously mentioned, we have identified three genetic loci that encode Hap4p homologs, and these were designated CaHap41p, CaHap42p, and CaHap43p. The functional relevance of these alternative Hap4p proteins remains to be established; to this end, we have generated null mutants of each CaHAP4 gene to evaluate their phenotypes, effects on CCAAT-binding activity in vitro, and gene expression. By analogy with the size and relative abundance of each S. cerevisiae CCAAT-binding complex (16, 37), it is reasonable to suggest that complexes I and II may contain Hap2/3/5p heterotrimers bound to DNA, while complexes III and IV represent Hap2/3/4/5p heterotetramers. Mobility shift studies with strains having deletions of the appropriate HAP genes will allow us to evaluate this supposition. The fact that we observed two complexes (III and IV) that may contain Hap4p-like proteins is consistent with the multiple HAP4 loci in C. albicans. In considering complexes I and II, we have identified two genetic loci that encode Hap3p homologs, namely, orf19.517/orf19.8148 and orf194647/orf19.12116 (McNabb, unpublished observations). We have designated orf19.517/orf19.8148 CaHAP31 and orf194647/orf19.12116 CaHAP32. The major difference between the encoded proteins lies within the first alpha helix of the Hap3p histone fold motif (3, 48). This alpha helix, designated α1, is absent from CaHap32p but present in CaHap31p (data not shown); however, the role of this evolutionarily conserved alpha helix in DNA binding and gene regulation remains unclear (48). We hypothesize that Hap31p and Hap32p could form distinct heterotrimeric complexes by associating with Hap2p and Hap5p. This would provide a logical explanation for complexes I and II seen in our mobility shift assays. One could further hypothesize that CaHap31p associates with one of the CaHap4p proteins while CaHap32p has specificity for a different CaHap4p protein. In support of this idea, mutational analysis with S. cerevisiae Hap3p suggests that ScHap4p interacts with ScHap3p in the heterotetramer (54). Additionally, Lan et al. (27) have recently shown that the C. albicans open reading frames corresponding to HAP43 and HAP32 are coordinately induced in response to a low-iron growth environment, while HAP31 is repressed. Thus, the different CCAAT-binding complexes could function in regulating transcription, by activation or repression, through binding CCAAT sites in the promoters of target genes under differing environmental conditions, analogous to what we observed with COX5 (Fig. 9). We are in the process of generating mutants of HAP31 and HAP32 in order to test this hypothesis.
The presence of a gene family encoding each of the subunits of the CCAAT-binding factor would not be unique to C. albicans. Arabidopsis thaliana has been shown to contain six different NF-YAs, nine NF-YBs, and eight NF-YCs (21), which are homologs of Hap2p, Hap3p, and Hap5p, respectively. The different genes show both tissue and developmental specificities, suggesting that distinct combinations of subunits assemble to form the DNA-binding complexes that control gene expression during different stages of plant growth and development (21). Thus, it will be of interest to determine whether HAP31 and HAP32 or the three HAP4 genes show differential expression in the yeast versus hyphal forms of C. albicans and how the different subunits affect target gene expression.
At this stage, we cannot explain why respiratory gene transcription is repressed when wild-type C. albicans is grown with amino acids as the carbon source, since these conditions would require the Kreb's cycle, oxidative phosphorylation, and gluconeogenesis to be highly active. In fact, we expected the transcription of both CYC1 and COX5 to be induced. However, if one considers that the lack of glucose or another readily usable carbon source will cause the growth rate of cells to be significantly reduced, then the transcription of some genes may be repressed in response to the lack of nutrients. If the expression of the respiratory machinery were to continue at a high level in a nutrient-poor environment, as with the hap5Δ homozygote, then alterations in the ADP:ATP and/or NAD+:NADH ratio, as well as in the level of many other allosteric and feedback effectors, could cause cells to sense starvation and undergo hyphal development, as observed with the hap5Δ/hap5Δ mutant. Since the CCAAT-binding factor in C. albicans represses the transcription of genes encoding electron transport proteins, it may also regulate the genes encoding enzymes of the Kreb's cycle, analogous to the case in S. cerevisiae (32). If this were true and those enzymes were similarly overexpressed in the hap5Δ/hap5Δ mutant grown in the presence of amino acids as the sole carbon and nitrogen source, then the resulting flux of intermediates through the Kreb's cycle could bypass or partially bypass the glyoxylate cycle, an important step in carbon assimilation. As previously noted by Lorenz and Fink (33), the two decarboxylation steps of the Kreb's cycle do not permit the assimilation of C2 compounds. The glyoxylate cycle bypasses those steps, allowing C2 compounds to enter gluconeogenesis. The medium used in this study contained a mixture of 20 amino acids, of which 10 are oxidatively degraded to acetyl-coenzyme A (a C2 compound) for entry into the Kreb's cycle. Thus, the glyoxylate cycle must play a central role in carbon assimilation when cells are grown in this medium. If the glyoxylate cycle were bypassed, then cells might sense starvation and hence hyphal development. In contrast, if glucose were added to the medium, then the starvation would be corrected (Fig. 7).
Interestingly, Land et al. (28) demonstrated that perturbations in the flow of electrons through respiratory metabolism resulted in cells switching from yeast to hyphal growth. Given the phenotype of the wild type versus the hap5Δ/hap5Δ mutant on solid YNB + AA medium, the control of respiratory gene transcription must be important for maintaining the yeast versus hyphal morphology. While we suspect that the effects of the hap5Δ/hap5Δ mutation on the yeast-to-hypha transition are an indirect effect of changes in cell physiology, this represents an important avenue of investigation toward understanding how metabolic changes contribute to the dimorphic transition and ultimately to pathogenesis. Moreover, understanding the regulatory mechanism of the CCAAT-binding factor in C. albicans may offer some clues into how central metabolism is regulated in respiratory yeasts and how genes that encode proteins involved in carbon metabolism are regulated in yeasts versus hyphae.
Acknowledgments
We thank Aaron Mitchell and Fred Winston for generously supplying strains and plasmids. We also thank Douglas Rhoads and Inés Pinto for advice and discussions during this work and Marsha Rhoads and Giselle Almeida for technical assistance. Sequence data from the C. albicans genome were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida. Annotations of C. albicans genes were obtained from the Candida databases at http://genolist.pasteur.fr/CandidaDB/and http://www.candidagenome.org/.
Photographs were taken in the Light Microscopy Imaging Facility, which is supported by funds from the Arkansas Biosciences Institute. This research was supported by grants from the Arkansas Biosciences Institute and by NIH grant R01AI51470 to D.S.M.
REFERENCES
- 1.Alexopoulous, C. J., C. W. Mims, and M. Blackwell. 1996. Introductory mycology, 4th ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 3.Baxevanis, A. D., G. Arents, E. N. Moudrianakis, and D. Landsman. 1995. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 23:2685-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgarel, D., C. C. Nguyen, and M. Bolotin-Fukuhara. 1999. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 31:1205-1215. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and J. J. Hynes. 1999. HAP-like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 7.Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. [DOI] [PubMed] [Google Scholar]
- 8.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, D.C.
- 9.Chen, H., J. W. Crabb, and J. A. Kinsey. 1998. The Neurospora aab-1 gene encodes a CCAAT binding protein homologous to yeast HAP5. Genetics 148:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 11.Dang, V. D., C. Bohn, M. Bolotin-Fukuhara, and B. Daignan-Fornier. 1996. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J. Bacteriol. 178:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 14.Flores, C. L., C. Rodriguez, T. Petit, and C. Gancedo. 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 24:507-529. [DOI] [PubMed] [Google Scholar]
- 15.Forsburg, S. L., and L. Guarente. 1989. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 5:153-180. [DOI] [PubMed] [Google Scholar]
- 16.Forsburg, S. L., and L. Guarente. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3:1166-1178. [DOI] [PubMed] [Google Scholar]
- 17.Forsburg, S. L., and L. Guarente. 1988. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol. Cell. Biol. 8:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, J. L. 1993. Fungal infection rates are increasing. ASM News 10:515-518. [Google Scholar]
- 19.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 21.Giuliana, G., C. Tonelli, and R. Mantovani. 2001. Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264:173-185. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 23.Hahn, S., and L. Guarente. 1988. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science 240:317-321. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, S., J. Pinkham, R. Wei, R. Miller, and L. Guarente. 1988. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol. Cell. Biol. 8:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 27.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53:1451-1469. [DOI] [PubMed] [Google Scholar]
- 28.Land, G. A., W. C. McDonald, R. L. Stjernholm, and L. Friedman. 1975. Factors affecting filamentation in Candida albicans: changes in respiratory activity of C. albicans during filamentation. Infect. Immun. 12:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lascaris, R., H. J. Bussemaker, A. Boorsma, M. Piper, H. van der Spek, L. Grivell, and J. Blom. 2003. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 4:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Z., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz, M. C., and G. R. Fink. 2002. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 35.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNabb, D. S., K. A. Tseng, and L. Guarente. 1997. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 17:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNabb, D. S., Y. Xing, and L. Guarente. 1995. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9:47-58. [DOI] [PubMed] [Google Scholar]
- 38.Muhlschlegel, F. A., and W. A. Fonzi. 1997. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17:5960-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee, P. K., and R. Prasad. 1998. Purified arginine permease of Candida albicans is functionally active in a reconstituted system. Yeast 14:335-345. [DOI] [PubMed] [Google Scholar]
- 40.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 41.Olesen, J. T., and L. Guarente. 1990. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 4:1714-1729. [DOI] [PubMed] [Google Scholar]
- 42.Pesole, G., M. Lotti, L. Alberghina, and C. Saccone. 1995. Evolutionary origin of nonuniversal CUGSer codon in some Candida species as inferred from a molecular phylogeny. Genetics 141:903-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinkham, J. L., J. T. Olesen, and L. P. Guarente. 1987. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol. Cell. Biol. 7:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponticelli, A. S., T. S. Pardee, and K. Struhl. 1995. The glutamine-rich activation domains of human Sp1 do not stimulate transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remacle, J. E., G. Albrecht, R. Brys, G. H. Braus, and D. Huylebroeck. 1997. Three classes of mammalian transcription activation domain stimulate transcription in Schizosaccharomyces pombe. EMBO J. 16:5722-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riego, L., A. Avendano, A. DeLuna, E. Rodriguez, and A. Gonzalez. 2002. GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 293:79-85. [DOI] [PubMed] [Google Scholar]
- 48.Romier, C., F. Cocchiarella, R. Mantovani, and D. Moras. 2003. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278:1336-1345. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. G. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sybirna, K., B. Guiard, Y. F. Li, W. G. Bao, M. Bolotin-Fukuhara, and A. Delahodde. 2005. A new Hansenula polymorpha HAP4 homologue which contains only the N-terminal conserved domain of the protein is fully functional in Saccharomyces cerevisiae. Curr. Genet. 47:172-181. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing, Y., J. D. Fikes, and L. Guarente. 1993. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 12:4647-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xing, Y., S. Zhang, J. T. Olesen, A. Rich, and L. Guarente. 1994. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 91:3009-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zinner, S. H., and W. M. Scheld. 1993. Brief review of fungal infections. Eur. J. Clin. Microbiol. Infect. Dis. 12(Suppl. 1):S146-S149. [Google Scholar]
- 57.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]