Abstract
Candida albicans is an opportunistic fungal pathogen whose virulence is related to its ability to switch between yeast, pseudohyphal, and true-hyphal morphologies. To ask how long-distance nuclear migration occurs in C. albicans hyphae, we identified the fundamental properties of nuclear movements and microtubule dynamics using time-lapse microscopy. In hyphae, nuclei migrate to, and divide across, the presumptive site of septation, which forms 10 to 15 μm distal to the basal cell. The mother nucleus returns to the basal cell, while the daughter nucleus reiterates the process. We used time-lapse microscopy to identify the mechanisms by which C. albicans nuclei move over long distances and are coordinated with hyphal morphology. We followed nuclear migration and spindle dynamics, as well as the time and position of septum specification, defined it as the presumptum, and established a chronology of nuclear, spindle, and morphological events. Analysis of microtubule dynamics revealed that premitotic forward nuclear migration is due to the repetitive sliding of astral microtubules along the cell cortex but that postmitotic forward and reverse nuclear migrations are due primarily to spindle elongation. Free microtubules exhibit cell cycle regulation; they are present during interphase and disappear at the time of spindle assembly. Finally, a growth defect in strains expressing Tub2-green fluorescent protein revealed a connection between hyphal elongation and the nuclear cell cycle that is coordinated by hyphal length and/or volume.
Candida albicans is the leading cause of human fungal infections. As a multimorph, it grows with budding yeast, pseudohyphal, and true-hyphal morphologies. The ability to interconvert between these morphologies appears to be required for virulence (14, 34). A plausible model posits that yeast cells are important for dissemination to target organs, whereas the hyphal form facilitates tissue invasion (42). In support of this model, mutations that commit strains to one growth form or another are avirulent (28, 34). Despite the apparent importance of morphological switching to the virulence of C. albicans, little is known about the fundamental cell biological differences which coordinate cell cycle progression and morphology and how these differences regulate morphological switching.
One clear difference between the different C. albicans cell forms is their spatial coordination of nuclear dynamics and morphology (16), yet little is known about the mechanism(s) by which nuclear position is coordinated. Nuclear migration is most dramatic in hyphae: nuclei migrate into germ tubes and divide across the future site of septation 10 to 20 μm from the basal mother cell (16, 43, 48). Mother nuclei then return to the basal cell while daughter nuclei migrate toward the growing tip (16). In yeast (3, 16) and pseudohyphae (39, 43), mitosis resembles that of the budding yeast, Saccharomyces cerevisiae, where nuclei divide across the bud neck. Thus, mitosis occurs across the site of septation irrespective of cell morphology. This suggests that nuclear position varies as a function of morphology.
How nuclei migrate to the site of septation in C. albicans hyphae is unclear. However, it is likely to involve the microtubule (MT) cytoskeleton, the organization of which is strikingly similar to that of S. cerevisiae: spindle pole bodies (SPBs) embedded in the nuclear envelope (37) nucleate astral MTs (19). The antimicrotubule drugs methyl benzimidazol-2-ylcarbamate and nocodazole eliminate astral MTs (6) and inhibit nuclear migration and mitosis in hyphae (52). Interestingly, treating hyphae with benzimidazol-2-ylcarbamate also inhibits the activity of the Spitzenkörper, resulting in a pseudohyphum-like morphology (6). Thus, MTs are involved in positioning nuclei and regulating hyphal morphogenesis.
The mechanisms underlying nuclear movements in S. cerevisiae are well understood. Briefly, astral MTs mediate nuclear movements through their attachment to an SPB, the main MT-organizing center in yeast. SPBs replicate during early S phase (4) and form a short mitotic spindle that moves to the mother bud neck by a combination of processes involving SPB-bound MTs. These include the tracking of MT plus ends along polarized actin cables (plus-end tracking [23]) and the depolymerization of MT plus ends at the cortex (35). Cytoplasmic dynein then facilitates the sliding of astral MTs laterally along the cell cortex, which pulls the elongating mitotic spindle into the bud (1). In C. albicans, mutation of the dynein heavy chain causes defects in nuclear position and, in addition, defects in hyphal morphology (29), highlighting the coordination of nuclear events and morphogenesis in C. albicans.
MTs also drive nuclear movements during anaphase. In higher eukaryotes, chromosomes first move to the spindle poles by the shortening of kinetochore MTs during anaphase A (30). The spindle poles then separate, driven by MT motors that push interpolar MTs away from each other during anaphase B (20). In S. cerevisiae, anaphase A and anaphase B start at roughly the same time and anaphase B accounts for the majority of nuclear movement during anaphase (31). Thus, these four MT-based mechanisms (plus-end tracking, depolymerization, sliding, and anaphase spindle elongation) combine to faithfully segregate daughter nuclei to daughter cells.
A dynamic, cell cycle-regulated MT cytoskeleton is a hallmark of eukaryotic cells. In higher eukaryotes, when the cell enters mitosis, extensive arrays of interphase MTs break down and the components are reincorporated into the mitotic spindle (33). During this process, bulk MT polymer decreases as interphase MTs disassemble and returns to interphase levels once metaphase spindles form (reviewed in reference [25]). A similar phenomenon has not been observed in S. cerevisiae but is evident in several fungi, including the fission yeast Schizosaccharomyces pombe and the dimorphic phytopathogen Ustilago maydis, where SPB-independent interphase MTs disassemble prior to the assembly of the mitotic spindle (18, 41).
To begin to understand the cell biological mechanisms that connect cell cycle regulation and morphogenesis and to determine the mechanisms underlying nuclear migration with respect to hyphal morphology in C. albicans, we used time-lapse fluorescence microscopy to capture complete cell cycles. By following nuclear, spindle and septin dynamics, we established a clear chronology of cell cycle events during the growth of the first hyphal cell from a yeast mother cell. Short-interval time-lapse microscopy revealed that forward nuclear migration results from the repetitive sliding of astral MTs along the cell cortex, while reverse migration is due primarily to spindle elongation. A pool of SPB-independent MTs is evident during interphase and disappears coincident with spindle assembly, suggesting that the components are reincorporated into the mitotic spindle. Finally, the growth defect in strains expressing Tub2-green fluorescent protein (GFP) revealed that the nuclear cell cycle is coordinated with hyphal length and/or volume and that when the rate of hyphal-tip growth is slowed, the cell cycle delays at two events: the formation of the septin ring and the onset of anaphase.
MATERIALS AND METHODS
Strain construction.
Strains containing C-terminal fluorescent fusions were generated in strain BWP17 (50) as described previously (13) and are listed in Table 1. Transformation cassettes were generated by PCR using the appropriate gene-specific primer pair (reverse primers are designated R1, R2, or R3 for URA3, HIS1, or ARG4, respectively [Table 1]) and a plasmid template containing specific fluorescent-protein- and nutritional-marker-encoding sequences (13). Positive transformants were screened by fluorescence microscopy and confirmed by PCR. Strains were made prototrophic at the ARG4 locus by the transformation of either NotI-linearized pRS-ARG-URA-BN (9) or EcoRI-linearized pRS-ARG4ΔSpe1 (50).
TABLE 1.
C. albicans strains and PCR primers used in this study
| Strain or primer | Genotype or sequence (5′-3′)a | Source |
|---|---|---|
| Strains | ||
| BWP17 | ura3Δ::imm434/ura3Δ::imm434 his1::hisG/hisI::hisG arg4::hisG/arg4::hisG | 50 |
| YMG5628 | BWP17 CDC3/CDC3-YFP:URA3 | 13 |
| YMG6449 | BWP17 NOP1/NOP1-YFP:HISI | This study |
| YJB7062 | BWP17 NOP1/NOP1-YFP:URA3 | This study |
| YJB7150 | BWP17 HHF1/HHF1-GFP::URA3 | This study |
| YJB7468 | BWP17 NOP1/NOP1-CFP::ARG4 | This study |
| YJB8179 | YJB7150 NOP1/NOP1-CFP::ARG4 | This study |
| YJB8548 | YJB7468 TUB2/TUB2-GFP::HIS1 | This study |
| YJB8860 | YJB7062 CDC3/CDC3-GFP::HIS1 | This study |
| YJB8552 | YJB7468 NUP49/NUP49-GFP::URA3 | This study |
| YJB8895 | BWP17 TUB2/TUB2-GFP::HIS1 | This study |
| YJB8897 | YMG5628 TUB2/TUB2-GFP::HIS1 | This study |
| YJB8952 | YJB8895 ARG4::URA3::arg4::hisG/arg4::hisG | This study |
| YJB8959 | YJB8860 ARG4::arg4::hisG/arg4::hisG | This study |
| YJB8961 | YJB8897 ARG4::arg4::hisG/arg4::hisG | This study |
| Primers | ||
| NOP1F | ACCTTGGAACCTTATGAAAGAGACCATTGTATTGTTGTTGGTAGATACA | 6 |
| TGAGAAGCGGAATAAAGAAAggtggtggttctaaaggtgaagaattatt | ||
| NOP1R1 | TTTTTAGTTTTCAATAATCAAATGTATTAATCCTATTGTACAAAATATTT | This study |
| TTATTTAAAATTTAGAGTATCCCtctagaaggaccacctttgattg | ||
| NOP1R2 | TTTTTAGTTTTCAATAATCAAATGTATTAATCCTATTGTACAAAATATTT | 6 |
| TTATTTAAAATTTAGAGTATCCCgaattccggaatatttatgagaaac | ||
| NOP1R3 | TTTTTAGTTTTCAATAATCAAATGTATTAATCCTATTGTACAAAATATTT | This study |
| TTATTTAAAATTTAGAGTATCCCactagtattgtagtacaaggtatc | ||
| NUP49F | TATAGTTTATTTATGGAATTGACTGAAACAATGGCTCAATTACATAATG | This study |
| AAGTGAATAGATTAACTAAAggtggtggttctaaaggtgaagaattatt | ||
| NUP49R1 | CAAAAAAAGAAAAATAAGAAAAACTATTACTATTATACATTATAATAT | This study |
| ATAATACAATTGTTGCACTTTCtctagaaggaccacctttgattg | ||
| HHF1F | GTCACTTCATTGGATGTTGTTTACGCTTTGAAGAGACAAGGTAGAACCT | This study |
| TGTATGGTTTCGGTGGTggtggtggttctaaaggtgaagaattatt | ||
| HHF1R1 | TTATACAAAAAACTTCTTAAATTAATACTATACAATAAAGAAAACGAAC | This study |
| TAAAAAGACAATTAGAAtctagaaggaccacctttgattg | ||
| TUB2F | CAAGAAGCTAGTATTGATGAAGAAGAATTAGAATATGCCGATGAAATC | This study |
| CCATTAGAAGATGCCGCCATGGAAggtggtggttctaaaggtgaagaattatt | ||
| TUB2R2 | CATTAACTAAACCAAAAAAAACCATAATTATATTAGAAGTGAACAAAA | This study |
| AAAAAATTATAATTAATTATATTGgaattccggaatatttatgagaaac | ||
| CDC3F | ACAAAAATTATTACCACAAGACCCACCAGCACAACCAGCTCCACAAAA | 13 |
| GAGTCGTAAAGGATTTTTACGTggtggtggttctaaaggtgaagaattatt | ||
| CDC3R1 | AATTAAACAAACAGATTAACAAACAAATAAACTAAATTAAGTTACATA | 13 |
| CTATTTAGCTATACCTCGGCCCtctagaaggaccacctttgattg | ||
| CDC3R2 | AATTAAACAAACAGATTAACAAACAAATAAACTAAATTAAGTTACATA | 13 |
| CTATTTAGCTATACCTCGGCCCgaattccggaatatttatgagaaac |
Uppercase indicates homology to genomic DNA for integrative purposes. Lowercase indicates homology to plasmids described in the work of Gerami-Nejad et al. (13).
Preculture and hyphal induction.
All media contained uridine at 80 μg/ml, except those used for selecting URA3 transformants. Bovine serum (Sigma, St. Louis, MO) was always used at 10% (vol/vol). To enrich cultures for unbudded singlet cells, strains were cultured overnight in YPAD (36) at 30°C. Ten to 20 μl of the culture was pelleted, resuspended in 100 to 200 μl of fresh synthetic complete medium (SDC [36]), and spread with glass beads on an SDC-serum-agar (2% agar) plate, preheated to 37°C. A round, 25-mm no. 1 coverslip (Fisher, Pittsburgh, PA) was placed over the cells, and the plate was returned to 37°C for 10 to 15 min when complete cell cycle time-lapse microscopy was performed or for longer periods to capture specific cell cycle stages.
Microscopy.
Plates were placed on the stage of a Nikon E600 epifluorescence microscope under a 60× Plan-Apo (numerical aperture, 1.4) objective preheated to 37°C with an FCS2 objective heater (Bioptechs, Butler, PA). Heat transfer through type 37 immersion oil optimized for GFP and yellow fluorescent protein (YFP) microscopy at 37°C (Cargille, Cedar Grove, NJ) was sufficient to maintain hyphal growth in and around the field of view for more than 5 h. Microscopy of cyan fluorescent protein (CFP) required type DF immersion oil (Cargille) with unheated objectives. Filter sets were from Chroma Technology Corp (Rockingham, VT). GFP images were captured with an Endow bandpass filter set (no. 41017) (11). For CFP/YFP imaging, filter set no. 86004 JP5v2, which utilizes a single dichroic mirror, was used. Images were captured with a CoolSnap HQ cooled charge-coupled-device camera (Photometrics, Tucson, AZ). Time-lapse automation, image processing, and data collection from 16-bit images were accomplished with Metamorph imaging software, v6.2 (Universal Imaging Corp., Westchester, PA). Data were exported to Microsoft Excel (Microsoft Corp., Redmond, WA), and statistical analyses were performed with the Statistical Analysis Package in Excel. Standard errors of the means (SEM) are included for values determined by calculating the arithmetic mean of the population. For figure assembly, images were converted to 8 bits and exported to Adobe Photoshop, v7.0 (Adobe Systems, Mountain View, CA).
Many acquisition details are provided in the text and figure legends. Generally, an ND4 neutral density filter was used with 200-ms exposures for Nop1-YFP or Tub2-GFP alone or in combination with Cdc3-YFP/GFP. The exposure time for Nop1-CFP was 500 ms. Two-by-two camera binning was always used. Differential interference contrast (DIC) images were taken in parallel by manually adjusting the focus prior to image capture. Where Z-stacks were acquired, four images at each time point were acquired with a step size of 1 μm. Two-dimensional images were generated using the Maximum function in Metamorph, and the Arithmetic dialogue was used to generate merged fluorescent/DIC images. Supplemental movies available at http://www.cbs.umn.edu/labs/berman/kensmovies.htm are numbered to reflect the corresponding figure numbers herein.
Analysis of nuclear movements.
The “neck” of the basal cell/germ tube (Fig. 1C) was arbitrarily defined as the zero point of a hyphal cell (position, 0 μm), where negative values represented positions within the basal cell while positive values were distal to the basal cell.
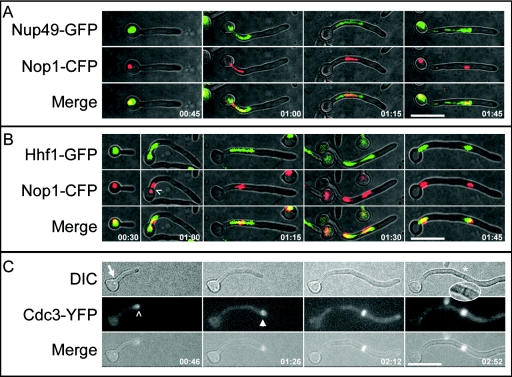
FIG. 1.
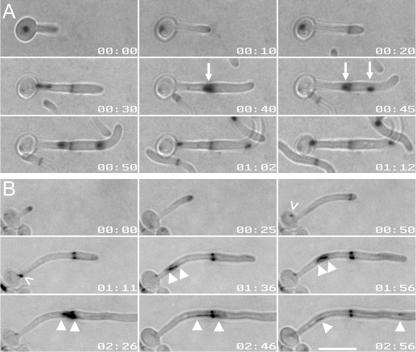
Nuclear organization and Cdc3 localization in C. albicans hyphae. (A, B) Hyphae were induced under coverslips, and images were taken at the times indicated (minutes:seconds) after hyphal induction. Hhf1-GFP and Nup49-GFP were imaged in the YFP channel to minimize CFP detection. (A) Nop1-CFP (red) occupies a smaller area than the putative nuclear membrane (Nup49-GFP) (green) during all stages of the cell cycle. (B) The separation of Nop1-CFP occurs late in mitosis (01:15), and it remains in close proximity to Hhf1-GFP throughout the cell cycle (merge). Note that when Nop1 first migrates into the germ tube, the signal stretches and sometimes separates briefly (01:00; caret). (C) Cdc3 localizes to the true septin ring at the time that the site of septation is specified. Images from a time-lapse movie of hyphae expressing Cdc3-YFP. Cdc3-YFP is detected at the hyphal tip (00:46; carat) before it localizes to the presumptum (arrowhead; 01:26). Subsequently, the septum appears as a dark band by DIC microscopy (asterisk and inset; 02:52). Bars, 10 μm. Times are expressed as (hours:minutes).
Individual hyphae were cropped from the time-lapse stacks and arranged in a montage into tif image files, where individual measurements were made frame by frame. Hyphal growth rates were calculated by linear regression analysis on plots of hyphal length versus time generated with the Track Points feature of Metamorph. We defined the time of presumptive septum (presumptum) appearance as the time when a hypha grew past the point where the septum eventually appeared in the DIC channel. This was done based on the observations that septin rings (i) develop just after the tip passes the site where the septin ring will form, (ii) are static with respect to the position of the hyphal tip (Fig. 1C) (Cheryl Gale, personal communication), and (iii) mark the future site of septation (12, 43, 48). Due to the asynchrony of the populations, we used either the mitotic frame (Nop1) or spindle disassembly frame (Tub2) to align data graphically. This approach minimized the variance of events temporally proximal to mitosis. To calculate the mean values for times between nuclear and morphogenetic events, we determined the time between any two given events within a single hypha and then found the mean for that event among all hyphae. As many hyphae grew out of the field of view and others grew out of the plane of focus, it was not possible to quantify every parameter for every hypha.
Analysis of microtubule dynamics.
For analyses of MT dynamics, hyphae were induced for 90 min to enrich for short spindles. Images were captured at 3-second intervals. Rates of MT growth, shrinkage, and sliding were determined by linear regression analysis (R2 > 0.9) of MT length versus time, only for MTs that remained in focus for 15 seconds or longer. For fluorescent-speckle microscopy (FSM) and the quantification of free MTs during the cell cycle, hyphae were induced at 37°C for various periods of time to capture the relevant cell cycle stages and images were captured for ∼5 min at 1- or 3-second intervals with a 100× Plan-Apo objective (numerical aperture, 1.4).
We performed MT life history analysis (5) to quantify the frequency of MT transitions from growth to shrinkage (catastrophe) and from shrinkage to growth (rescue). A single MT life history was defined as the de novo growth of an MT from an SPB and termination by one of three events: complete MT depolymerization, MT release from the SPB (where no MT remained attached to the SPB), or MT “breakage,” which resulted in a free MT and a shorter MT remaining attached to the SPB. In the case of MT breakage, the life history terminated when the remaining SPB-bound MT had depolymerized completely; the free MT was not considered in the analysis. This analysis did not require that MTs remain in focus, only that we could discern relevant periods (growing, shrinking, and transitions).
We used FSM (49) to assess the polarity of MTs bound to and released by SPBs. We identified MTs with varied degrees of Tub2-GFP incorporation, resulting in bright and dark regions that served as reference points with respect to each other, the free MT end, and the SPB.
RESULTS
Nuclear organization in Candida albicans.
To follow the dynamics of nuclear migration in hyphae, we generated in vivo fluorescent protein fusions to the C termini (13) of putative nuclear markers (http://www-sequence.stanford.edu/group/candida), including a nuclear pore protein, Nup49 (orf19.4987), a histone H4 gene, (Hhf1; orf19.1059), and a small nucleolar ribonucleoprotein, Nop1 (orf19.3138). Nop1 localized constitutively to a bright, round structure in yeast and hyphal cells that fluoresced significantly more than the other nuclear tags (Hhf1 or Nup49), enabling the use of short exposure times that resulted in less phototoxicity. We also visualized the nucleolus with respect to the nuclear membrane with a strain coexpressing Nop1-CFP and Nup49-GFP (YJB8552). The continuity of the Nup49 signal throughout the cell cycle indicated that, as in yeast cells (37), the nuclear membrane remains intact during mitosis in hyphae. Importantly, Nop1-CFP appeared within the Nup49-GFP signal at all stages of the cell cycle (Fig. 1A).
To follow C. albicans nucleolar movement with respect to the chromosomes, we coexpressed Nop1-CFP and Hhf1-GFP (histone H4) in strain YJB8179. The nucleolus remained closely associated with Hhf1-GFP and divided with the last of the chromosomal DNA (Fig. 1B). This resembles nucleolar behavior in S. cerevisiae, where nucleolar antigens (including Nop1 [10]) are closely associated with chromosomal DNA (17, 51), even though nucleolar separation is delayed with respect to the separation of the rest of the genome (8, 44, 46, 47). Thus, Nop1 is a useful nuclear marker that resides within the nuclear membrane and is closely associated with genomic DNA.
Cdc3-YFP reveals the site of septation.
Previous studies showed that the true septin ring specifies the site of septation (12, 43, 48) but did not establish the timing of septin ring appearance with respect to the position of the hyphal tip. Time-lapse microscopy of germ tubes expressing Cdc3-YFP (YJB5628) revealed that, like Cdc10 (48), Cdc3 appears in growing hyphal tips (Fig. 1C). More importantly, in all hyphae examined (n = 13), Cdc3 localized to the future site of septation coincident with the time that the hyphal tip passed this position in the germ tube. This gave the impression that Cdc3 was left behind the extending tip as it passed this position, rather than that the ring appeared within the tube at a later time. Consistent with previous reports (43, 48), the septin ring had divided into two rings when the septum appeared in the DIC channel (Fig. 1C and inset). We conclude that the true septin ring appears when the eventual site of septation is specified, and this occurs at the time that tip growth passes this site. Similar to what occurs with S. cerevisiae, where septins bearing epitope tags (GFP or hemagglutinin) at their C termini are not fully functional (M. Longtine, personal communication), fluorescently tagged C. albicans septins affect cell cycle dynamics (K.R.F., unpublished observations). Thus, most of our studies were performed in strains that do not carry fluorescent-protein-tagged septins and we retrospectively designate the position where the true septin ring appears (at the time when the hyphal tip passes that position) by noting the eventual position of the septum by DIC microscopy. We term this presumptive site of septation the presumptum (presumptive septum).
The dynamics of nuclear migration in hyphae.
We next quantified the real-time dynamics and properties of nuclear migration (detected with Nop1-YFP in strain YJB6449) with respect to the basal cell and the presumptum. We defined the hyphal neck position (y) as 0.0 μm, such that positions within the basal cell were negative values and positions within the germ tube were positive values. Figure 2A shows representative Nop1 behavior through the first hyphal cell cycle that started from an unbudded cell and ended with the appearance of the septum in the DIC channel (Fig. 2.mov at the above-named website). When Nop1 first entered the germ tube, it frequently stalled and stretched at the neck, sometimes resolving into separate Nop1 signals (e.g., Fig. 1B, 01:00 [where 01:00 indicates 1 min, 0 s). These nucleoli, which likely reflect transient separation of the two rRNA gene-containing chromosomes, reassociated after they had passed the neck, and subsequent mitoses were unambiguous (Fig. 2A, 01:23, and 2.mov).
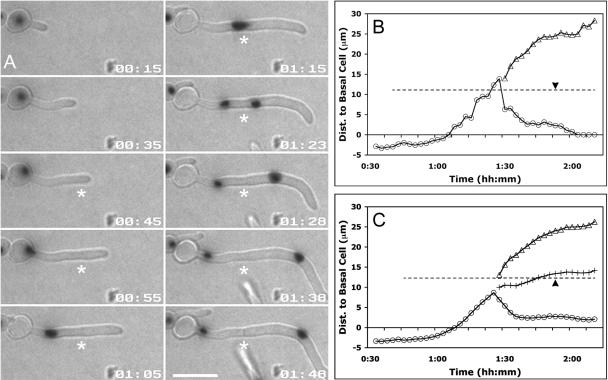
FIG. 2.
Dynamics of Nop1 nuclear migration relative to the presumptum. Hyphae expressing Nop1-YFP were analyzed at 2.5-min intervals by capturing a single DIC image and a Z-stack of YFP images at each time point. (A) Selected frames from Fig. 2.mov show a representative example of Nop1 dynamics in a typical hypha. Nop1 remains in the basal cell until after the presumptum forms (asterisk; 00:45). Subsequently, Nop1 migrates to the presumptum (position indicated by asterisks) and divides (01:23). Both mNop1 and dNop1 move away from the presumptum (01:23 to 01:48) and mNop1 remains within the germ tube when septation occurs (asterisk; 01:48). Time is given as hours:minutes (hh:mm) from the time of hyphal emergence. Bar, 10 μm. (B, C) The hyphal neck is defined as 0 μm. Negative values represent positions within the basal cell, while positive values represent positions within the germ tube. The dashed line represents the position of the presumptum, where the left end represents the time that the hyphal tip extends beyond the eventual site of septation. The black arrowheads indicate the time that septation occurs. mNop1, circles; dNop1, triangles; midpoint of mNop1 and dNop1, plus signs. See the text for details. (B) Graphical representation of Nop1 position in Fig. 2.mov. (C) Mean Nop1 positions were plotted against time by aligning the data at the time of mitotic Nop1 separation. Values are the averages for 29 cells. Dist., distance.
Inspection of premitotic nuclear movement in individual hyphae revealed that forward nuclear migration to the presumptum was nonlinear but that postmitotic movements were less erratic (Fig. 2.mov and 2B). Postmitotically, the mother Nop1 signal (mNop1) moved toward the basal cell while the daughter Nop1 signal (dNop1) moved forward in the hypha. Interestingly, the movement of both nuclei stalled before the septum became visible (Fig. 2B, 01:45), implicating a common mechanism of movement.
To ask about the general nature of nuclear movements, we averaged the Nop1 data using the time of mitotic Nop1 separation as the common reference point (Fig. 2C) (n = 29). We found four rules of nuclear movement in these hyphae. First, nuclei never moved into germ tubes until after presumptum formation. Second, on average, Nop1 first divided when it was within 2.4 ± 0.4 μm of the presumptum. Third, mitotic separation of the nuclei was biased toward the mother cell: the midpoint between mNop1 and dNop1 (Fig. 2C) did not perfectly coincide with the presumptum (Fig. 2C). Surprisingly, in 22/24 instances, mNop1 movement toward the basal cell stalled before septation, and nuclei remained within the germ tube, 3.5 ± 0.3 μm away from the basal cell. Only after septation did mNop1 migrate slowly into the basal cell (Fig. 2.mov and 2C). Together, these results show that premitotic forward nuclear migration is nonlinear and initiates only after the site of septation is specified. These results also show that nuclear movement away from the presumptum is rapid before septation and slow after septation and that mother nuclei tend to reenter the basal cell slowly during this postseptation phase.
Nuclear migration is due to spindle dynamics.
Microtubules are likely to be responsible for the dynamics of C. albicans nuclei. Based on studies of S. cerevisiae, forward movement of Nop1 could be the result of MT plus-end tracking and/or plus-end depolymerization or it could result from MT sliding along the cortex. The postmitotic movement of Nop1 signals could be the result of similar processes or could be the result of chromosome dynamics during spindle elongation. To identify relevant periods of the cell cycle where MT and/or spindle dynamics may mediate C. albicans nuclear migration and to compare and contrast spindle dynamics with Nop1 dynamics, we performed time-lapse microscopy on hyphae expressing GFP fused to the β-subunit of tubulin (Tub2; strain YJB8952), which labels SPBs, mitotic spindles, and MTs (19).
During early hyphal growth, an unduplicated SPB with or without astral MTs was sometimes evident (Fig. 3A, 01:01). During early hyphal emergence, however, individual MTs and unduplicated SPBs did not image well and were not always discernible because of abundant short MTs that were not associated with the SPB (see Fig. 9). Nonetheless, shortly after presumptum formation, SPBs suddenly became prominent (Fig. 3A, 01:31), which implies that SPB duplication occurs after the site of septation is specified. After entering the germ tube, the duplicated SPBs adopted a short spindle configuration and migrated toward the presumptum in a series of nonlinear steps (Fig. 3B). In this example, the short spindle migrated past the presumptum, into the daughter compartment, and back to the mother side prior to the onset of spindle elongation. Similarly, as spindles elongated, they were sometimes displaced forward and backward without significant changes in spindle length (Fig. 3B, arrows).
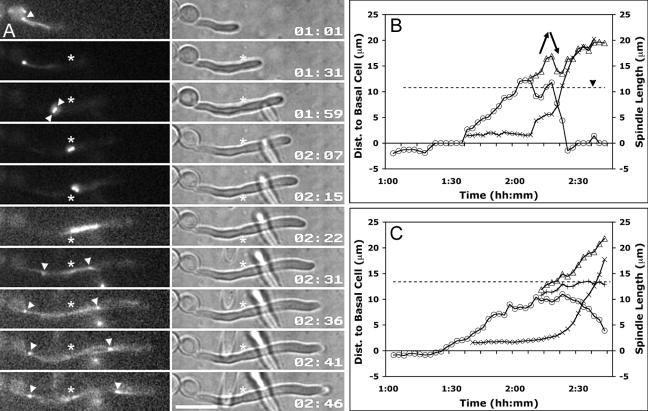
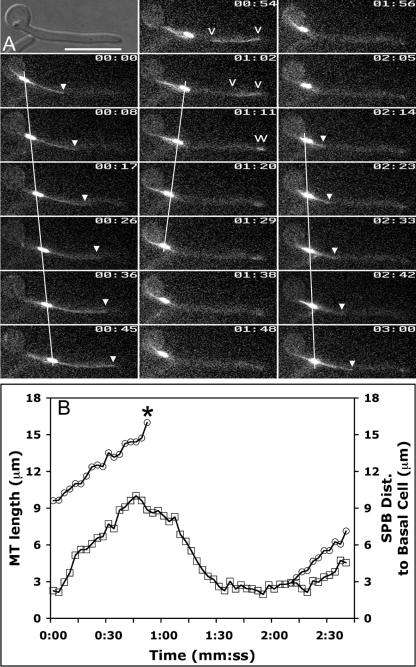
FIG. 3.
Spindle dynamics relative to the presumptum. Hyphae were induced and analyzed at 2.5-min intervals as for Fig. 2. (A) Selected frames from Fig. 3.mov show a representative example of spindle dynamics in a typical hypha. The SPB becomes prominent after the presumptum forms (01:31) and forms a short spindle (01:59) as it migrates forward in the germ tube. As the short spindle approaches the presumptum, spindle elongation ensues (02:22). The septum appears shortly after spindle disassembly (02:46). Asterisks indicate presumptum/septum position, and white arrowheads indicate SPBs. Time is measured in hours:minutes (hh:mm). Bar, 10 μm. (B, C) Chart values, dashed lines, and the black arrowhead are as defined in the legend for Fig. 2. mSPB, circles; dSPB, triangles; spindle midpoint, plus signs; spindle length, multiplication signs. See the text for details. (B) Graphical representation of spindle position and spindle length from Fig. 3.mov. During mitosis, elongating mitotic spindles remain subject to external forces that move them asymmetrically about the presumptum (arrows). (C) Mean SPB positions were plotted against time by aligning the data at the time of spindle disassembly. Septation occurred 3.7 ± 1.2 min after spindle disassembly (not shown).Values are the averages for 15 cells.
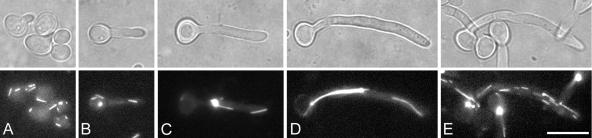
FIG. 9.
Free MTs are regulated during the cell cycle. Hyphae were induced under coverslips at 37°C for various times to count free MT appearance over time at different stages of the cell cycle. Using the 100× objective at ∼22°C, a single DIC image was taken at the start of imaging and then single-plane GFP images were taken at 1- or 3-second intervals, as indicated below. (A) Free-MT singlet cells and short hyphae with unduplicated SPBs (1-second intervals). (B) Hyphae with duplicated SPBs prior to movement into the germ tube (1-second intervals). (C) Hyphae with short migratory spindles (3-second intervals). (D) Mitotic hyphae (3-second intervals). (E) Hyphae after spindle disassembly in G1 (3-second intervals). Bar, 10 μm.
To compare spindle dynamics with nuclear dynamics, we averaged the data from 15 cells and aligned the data at the time of spindle disassembly, the final reference point common to all spindles (Fig. 3C). We found that the four observations made with Nop1-YFP were almost identical for the studies with Tub2-GFP. First, the SPB did not enter the germ tube until after presumptum formation. Second, short spindles began elongating when they were within 1.7 ± 0.7 μm (n = 15) of the presumptum. Third, separation of the mitotic spindle was biased toward the basal cell. The midpoint of the mitotic spindle (Fig. 3C) was closer to the basal cell during the early stages of anaphase. Later in mitosis the bias was less evident. Finally, the mother SPB (mSPB) approaches but does not enter the basal cell prior to septation. Thus, the data are consistent with the hypothesis that SPB-bound microtubules mediate the forward spindle migration into the germ tube and that mitotic spindle elongation accounts for most of the movement that returns the mother nucleus toward the basal cell prior to septation. However, the reentry of the mother nucleus into the basal cell usually occurs postmitotically and is due to a mechanism that mediates slower migration rates.
Cell cycle events occur at constant hyphal lengths.
Strains carrying fluorescent proteins fused to Tub2 have been reported to grow more slowly than wild-type strains (19). Consistent with this, the rate of hyphal-tip growth was lower in the Tub2-GFP strain (YJB8952; 0.23 ± 0.02 μm/minute) than in the Nop1-YFP strain (YJB6449; 0.37 ± 0.01 μm/minute). This resulted in a significant increase (P value < 0.001) in mean cell cycle length (time from hyphal emergence to septation) in hyphae expressing Tub2-GFP (156.8 ± 7.8 min, n = 15) compared to that of hyphae expressing Nop1-YFP (108.1 ± 2.3 min, n = 27). We hypothesized that the GFP moiety on β-tubulin interfered with nuclear cell cycle events, universally protracting all cell cycle events and resulting in decreased production and/or delivery of hyphal growth stimuli to the hyphal tip.
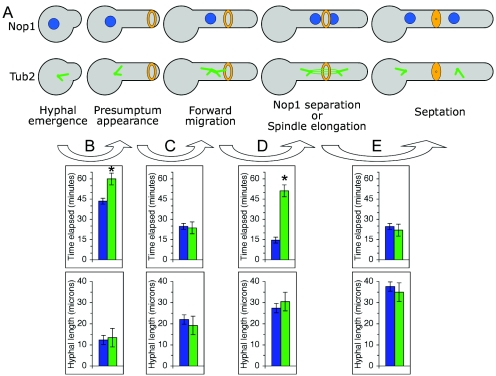
To test this hypothesis, we calculated the time elapsed between a number of cell cycle events for all hyphae in the Nop1-YFP and Tub2-GFP experiments and averaged the resulting values (Fig. 4). Surprisingly, cell cycle events were not universally protracted. Rather, two specific cell cycle stages were delayed in the Tub2-GFP strain relative to the Nop1-YFP strain. First, the time elapsed between hyphal emergence (germ tube evagination) and the time of presumptum appearance (hyphal-tip growth past the point of eventual septation) was 43.5 ± 2.2 min in the Nop1-YFP strain and 60.0 ± 4.4 min in the Tub2-GFP strain (P value < 0.001) (Fig. 4B). Subsequently, however, the times elapsed between presumptum appearance and the onset of forward nuclear/spindle migration (movement of Nop1-YFP or Tub2-GFP through the hyphal neck) were not statistically different (Fig. 4B) (24.8 ± 1.3 min and 23.7 ± 2.7 min, respectively; P value > 0.1). Second, the times between the initiation of forward nuclear/spindle migration and the initiation of mitosis were 14.6 ± 1.3 min in the Nop1-YFP strain and 51.2 ± 5.0 min in the Tub2-GFP strain (P value < 0.001) (Fig. 4D). Finally, no delay was detected in the time elapsed between anaphase (Nop1 separation or onset of spindle elongation) and septation (Fig. 4D) (24.7 ± 1.1 min and 22.0 ± 1.5 min, respectively; P value > 0.1). Importantly, all of these events, including the delayed events, occurred when hyphal lengths of the two strains were statistically similar (P value > 0.1) (Fig. 4B to E). Similar to previous observations of yeast and pseudohyphae (3, 38, 39), these results suggest that hyphal cell cycle events occur only after germ tubes have grown to critical lengths and/or volumes. We conclude that when the hyphal growth rate is delayed by Tub2-GFP, cell size determinants delay the specification of the site of septation and spindle elongation until these threshold sizes are achieved.
FIG. 4.
Cell cycle events are coordinated by germ tube length. (A) Schematic representation of the relevant cell cycle events in hyphae expressing Nop1-YFP (top row) or Tub2-GFP (bottom row). Nuclei, dark blue; SPBs and MTs, green; presumptum, narrow orange ring; septum, thick orange ring. SPBs are arbitrarily shown with two astral MTs. (B to D) A comparison of time elapsed and hyphal length between the Nop1-YFP strain (blue bars) and the Tub2-GFP strain (green bars). The top row of charts shows the mean time elapsed between the cell cycle events indicated by the respective arrow. The bottom row of charts shows the mean hyphal length at the latter of the two cell cycle events. (B) Time elapsed between hyphal emergence and presumptum appearance. (C) Presumptum appearance and the initiation of forward migration. (D) Forward migration and Nop1 separation or the onset of spindle elongation. (E) Spindle elongation and septation. Error bars are ± SEM. Asterisks represent significant differences (P value < 0.001) between the Nop1 strain and the Tub2-GFP strain.
Nuclear dynamics with respect to the (true) septin ring.
Cdc3 encodes a septin protein that localizes only to the true septin ring that marks the future site of septation (presumptum) (12) and not to the basal septin band that is seen with some septins at the hyphal neck (43, 48). We used strains coexpressing fluorescent fusions to Cdc3 and either Nop1 (YJB8959) or Tub2 (YJB8961) to ask when the true septin ring appears with respect to SPB duplication and the initiation of forward migration. DIC and GFP images were acquired at 5-minute intervals, and the Cdc3 and Nop1 or Cdc3 and Tub2 signals were easily distinguished (Fig. 5A and B). In 24/24 hyphae, Cdc3 appeared at the presumptum prior to Nop1 movement into the tube. Subsequently, nuclei migrated toward the ring of Cdc3 and divided across it (Fig. 5A and 5A.mov).
FIG. 5.
Nuclear dynamics with respect to the true septin ring. (A) Hyphae expressing Nop1-YFP and Cdc3-GFP were imaged at 5-minute intervals. Nop1 moved into the germ tube after Cdc3-GFP localized to the presumptum. Arrows indicate Nop1 just before (00:40) and after (00:45) mitosis. (B) Hyphae expressing Tub2-GFP and Cdc3-YFP were imaged at 5-minute intervals. SPB duplication (carats) was detected after Cdc3 revealed the presumptum. Arrowheads indicate spindle poles. Time is indicated in hours:minutes. Bar, 10 μm.
Unduplicated SPBs are difficult to detect, while duplicated SPBs become prominent as their fluorescence increases, presumably due to the incorporation of GFP-labeled tubulin into the new SPB. We used this property to determine when SPBs duplicated with respect to the appearance of Cdc3 at the presumptum. In 12/12 hyphae, the fluorescence of the SPB began increasing shortly after Cdc3 appeared at the presumptum (Fig. 5B, 00:50 to 01:11, and 5B.mov), suggesting that SPB duplication occurs after the appearance of the septin ring, although we cannot rule out the possibility that the appearance of Cdc3 and the initiation of SPB duplication occur at the same time. In either case, the two events are likely to be linked, and we conclude that the site of septation in hyphae is specified either at the same time as or shortly before SPB duplication and prior to forward nuclear migration.
Forward nuclear migration is the result of repetitive MT sliding events.
In S. cerevisiae, spindle alignment is facilitated by the combination of three iterative mechanisms (reviewed in reference [22]): the tracking of MT plus ends along the cortex (23, 27), the depolymerization of MT plus ends at the cortex (35), and the sliding of MTs laterally associated with the cortex (1). The nonlinear nature of forward spindle migration suggests that C. albicans short spindles move forward into the germ tube by iterative displacements. Short-interval (3-second) time-lapse microscopy of strain YJB8952, which carries Tub2-GFP, allowed us to distinguish between different types of MT dynamics associated with spindle movements. Selected frames from a movie (Fig. 6 _1.mov at the above-named website) demonstrating the dynamics and quantification of the dynamics are shown in Fig. 6A and B, respectively. A second example of spindle displacements resulting from repetitive MT sliding events is also shown in Fig. 6_2.mov.
FIG. 6.
Hyphal spindles align by repetitive MT sliding events. Hyphae were induced under coverslips for ∼90 min to enrich for hyphae with short spindles. Using a preheated (37°C) 60× objective, a single DIC image was taken at the start of imaging and single-plane GFP images were taken at 3-second intervals. (A) Selected frames from Fig. 6.mov. The distal end of a dSPB-bound MT (arrowheads; 00:00 to 00:45) moved toward the hyphal tip faster than the short spindle that anchored it (slanted line; 00:00 to 00:45). At 00:54, the MT detached from the SPB, slid toward the hyphal tip, and depolymerized (carats). Upon MT detachment, the short spindle moved back toward the basal cell (slanted line; 01:02 to 01:29), presumably due to the MT that grew from the mSPB (00:36 to 01:02). Time is indicated in minutes:seconds (mm:ss). Bar, 10 μm. (B) Quantification of MT length and spindle displacement in panel A. The spindle moved toward the hyphal tip (squares) as MT length increased (circles). When the SPB-bound MT detached from the SPB (asterisk), the dSPB moved back toward the basal cell, presumably due to the mSPB-bound MT to that was mostly out of focus in the basal cell. Forward movement resumed when this MT depolymerized (01:56 to 02:05 in panel A), and a new MT grew from dSPB. Dist., distance.
The major type of MT dynamics seen in germ tubes during forward migration of the spindle was MT sliding. At 00:00, a short spindle (SS) (19) was positioned at the hyphal neck, with a single MT or MT bundle (referred to as MT) emanating from each pole. The MT bound to the daughter SPB (dSPB) associated laterally along the contour of the cortex and was oriented toward the hyphal tip, while the mSPB-bound MT remained oriented away from the germ tube within the basal cell. The mSPB-bound MT began to depolymerize (cf. images at 00:00 and 00:08) while the SS moved forward into the germ tube (Fig. 6A, left column, and 6_1.mov, 00:00 to 00:45). Interestingly, the dSPB-bound MT became longer during the time that the SS moved toward the hyphal tip (Fig. 6A). Thus, spindle displacements occur while MTs grow, and therefore forward movements cannot be due to MT depolymerization. Furthermore, it is unlikely that tracking of MT plus ends along the cortex is a major contributor to forward migration as this process generally occurs without significant changes in MT length (23, 27).
Detachment of this MT from the SPB (00:54) was accompanied by the loss of forward movement (Fig. 6A, middle column). Furthermore, the SS moved back toward the basal cell, most likely due to the mSPB-bound MT, which appeared at 00:36 and persisted until 1:38, when it began to depolymerize, finally disappearing at 02:05. Forward movement resumed when a new dSPB-bound MT appeared and grew (Fig. 6A, right column, 02:14). This repetitive behavior of MT growth and simultaneous displacement of the short spindle was observed in all (n = 13) hyphae that had short, migratory spindles. Although our analysis does not exclude minor contributions from either plus-end tracking or MT depolymerization, we conclude that forward spindle migration is predominantly the result of repetitive MT growth and sliding events.
After chromosome separation, anaphase B (spindle elongation) drives most nuclear movements.
A remaining question regarding the return of mSPB toward the basal cell is whether the movement is due largely to anaphase A (the shortening of kinetochore MTs which moves chromosomes to the spindle poles) or to anaphase B (the movement of the chromosomes as a function of SPB position during spindle elongation). If anaphase A predominates, then spindle length would be long when the Nop1 signal divides. Furthermore, the separation between a Nop1 signal and its spindle pole would be relatively large. Alternatively, if anaphase B predominates, then spindle length would be short when Nop1 divides and the Nop1 signal would be relatively close to the SPB. We followed the dynamics of SPBs and nucleoli in a strain coexpressing Tub2-GFP and Nop1-CFP (YJB8548) and acquired DIC, CFP, and YFP images of hyphae with migratory short spindles at 30-second intervals.
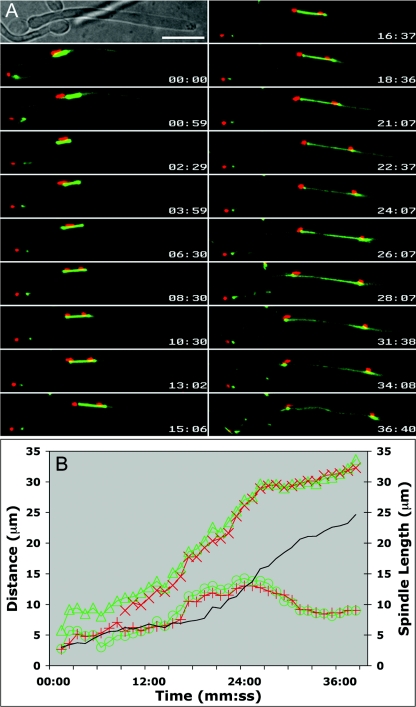
A representative example of Nop1 behavior with respect to the spindle is shown in Fig. 7. In all of the hyphae examined (n = 12), the Nop1 signal separated when spindles were short (3.1 ± 0.31 μm). When short spindles were displaced within hyphae, Nop1 exhibited similar movements with a slight delay (Fig. 7.mov), consistent with the idea that Nop1 position is a function of SPB position. Nop1 signals maintain close, yet variable, proximity to the short spindle prior to mitotic Nop1 separation and to the respective SPBs after mitosis (Fig. 7B). We conclude that the movement of mNop1 toward the basal cell (reverse migration), as well as the forward movement of dNop1 after Nop1 separation until spindle disassembly, is due primarily to forces that push the poles apart during anaphase B. Additionally, repetitive forces acting upon astral MTs contribute to the overall position of the elongating spindle (Fig. 7.mov), although that contribution is much more evident during premitotic forward migration than during postmitotic reverse migration.
FIG. 7.
Reverse nuclear movement is due primarily to anaphase B. Hyphae expressing Tub2-GFP and Nop1-CFP were grown under coverslips at 37°C for 90 min, and images were captured at 30-second intervals. (A) Selected images from Fig. 7.mov. Nop1 signals (red) remain in close proximity to SPBs (green) and trail SPBs during spindle displacements (i.e., 00:59). Time is indicated in minutes:seconds (mm:ss). Bar, 10 μm. (B) Plot of Nop1 position and SPB position shown in panel A. Separation of either Nop1 signal from its SPB is detectable early during spindle elongation but represents only a small fraction of total nuclear movement. mNop1, red plus signs; mSPB, green circles; dNop1, red multiplication signs; dSPB, green triangles. Spindle length is shown in black for reference.
Polarity and stability of MT ends.
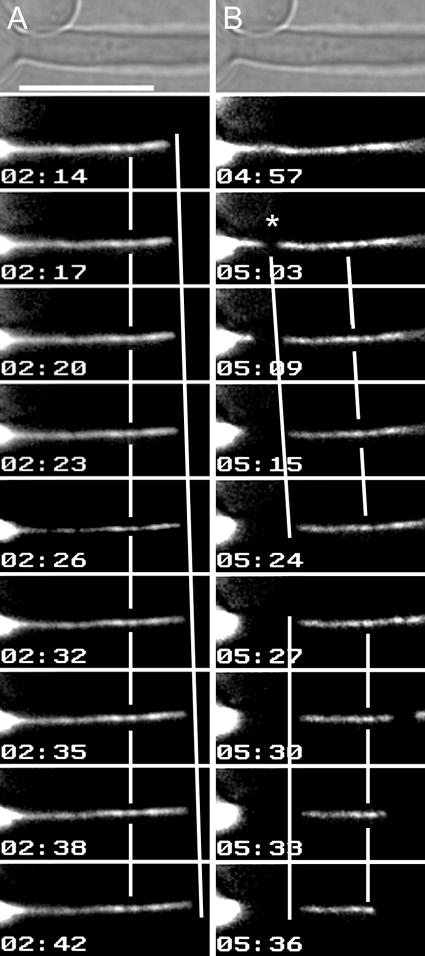
The organization of the C. albicans MT cytoskeleton suggests that SPB-anchored MTs grow and shrink preferentially at the ends distal to the SPB, while MT ends proximal to the SPB are stably associated with the SPB. To test this hypothesis, we performed fluorescent-speckle microscopy (49) by analyzing MTs that had differential levels of incorporation of Tub2-GFP (speckles) and measuring the movement of these speckles with respect to other MT landmarks, such as SPBs or MT distal ends, to determine where subunit addition and/or removal were occurring (49). A speckled MT is shown in Fig. 8, which is an enlarged region from movie Fig. 8.mov that also includes the distal end of the hypha. As the MT grew from 02:14 to 02:42, the speckles remained a constant distance from the SPB and from each other, indicating that tubulin subunits were not being added within the MT or at the SPB. Rather, the distal end moved further away from the speckles, indicating that MT growth occurs at the distal ends of SPB-bound MTs.
FIG. 8.
SPB-bound MTs grow and shrink at their distal ends. Hyphae were induced under coverslips for ∼90 min to enrich for hyphae with short spindles. Using the 100× objective at ∼22°C, a single DIC image was taken at the start of imaging and then single-plane GFP images were taken at 3-second intervals. (A) MT growth occurs preferentially at SPB-distal ends. Lines are provided for visual reference with respect to speckles, SPBs, and MT ends. The speckles remained stationary (broken vertical line), while the distal end of the MT moved away from them and the SPB (continuous slanted line). (B) Broken MTs have stable proximal ends and move toward the hyphal tip without treadmilling. An MT broke near the SPB at 05:03 (asterisk) and moved toward the hyphal tip. The SPB-proximal end moved forward (continuous slanted line, 05:03 to 05:24), as did the SPB-distal end (broken slanted line). Speckles within the MT did not move with respect to either MT end, and this length of the MT remained constant (the lines are parallel). When the MT broke again at 05:30, both new MT ends shrank away from the break site (for the right-hand fragment, see Fig. 8.mov). The SPB-proximal end of the left fragment did not move after breakage (continuous vertical line, 05:27 to 05:36), while the distal end depolymerized from the break site (broken vertical line). Time is indicated in minutes:seconds. Bar, 10 μm.
Using FSM to analyze the fates of broken MTs also revealed important features regarding the relative stability of MT ends in C. albicans hyphae. In Fig. 8B (and Fig. 8.mov), the MT broke (05:03). The remaining SPB-bound MT fragment shrank rapidly (05:03 to 05:24), while the free MT fragment moved toward the hyphal tip (05:03 to 05:24). Importantly, there was no change in either the interspeckle distances or the distance between a given speckle and the SPB-proximal end, indicating that the new SPB-proximal end of the free MT was stable after breakage. The subsequent movement of the free MT toward the hyphal tip further indicated that free MTs are displaced by translocation toward their proximal ends, as opposed to treadmilling (the simultaneous removal of subunits from the proximal end and subunit addition at the distal end). SPB proximal-end stability upon breakage is not absolute, however. When the free MT broke again (Fig. 8B, 05:27), it generated left and right fragments, both of which shrank from the break site. Importantly, however, the loss of speckles at the distal end of the left fragment occurred exclusively from the break site. We conclude that SPB-bound MTs grow and shrink preferentially from their distal ends and that broken or released MTs generally have stable ends and are transported, largely intact, toward the hyphal tips.
C. albicans microtubule dynamics differ from those of S. cerevisiae and U. maydis.
The dynamics of the C. albicans MT cytoskeleton associated with the short spindle stage in hyphae at 37°C are summarized in Table 2. We compared our results with data published for the S. cerevisiae MT cytoskeleton at 37°C (1) and with data published for the yeast-like cells of U. maydis (40). MT dynamics were similar among these fungi in that C. albicans MTs were in a perpetual state of flux termed dynamic instability, frequently switching between growing and shrinking states. However, in contrast to S. cerevisiae, where MT growth and shrinkage occurred at approximately the same rates (1, 45), but like U. maydis, C. albicans MTs grew considerably more slowly (6.4 ± 0.3 μm/minute; n = 31) than they shrank (26.6 ± 1.9 μm/minute; n = 27), spending 4.2-fold more time in growth than in shrinkage.
TABLE 2.
G2/M microtubule dynamics
| Organism | Rate (μm/min) ± SEM (no. of life histories)
|
No. of events/mind
|
|||||
|---|---|---|---|---|---|---|---|
| Growth rate | Shrinkage rate | SPB-bound MT slide rate | Free MT slide rate | Free MT shrinkage ratea | Catastrophe frequencyb | Rescue frequencyc | |
| C. albicans | 6.4 ± 0.3 (31) | 26.6 ± 1.9 (27) | 9.2 ± 0.7 (17) | 20.5 ± 1.1 (17) | 27.2 ± 0.1 (13) | 0.34 | 0.02 |
| S. cerevisiaee | 12.6 ± 1.2 | 12.0 ± 0.6 | 1.18 ± 0.1 | 5.1 ± 0.7f | NA | 2.52 | 2.04 |
| U. maydisg | 10.2 ± 1.0 | 24.9 ± 3.7 | 9.3 ± 2.4 | 41.2 ± 5.6 | NA | 1.94 | 1.82 |
Free MTs did not have a growth rate associated with them because of difficulties determining their origins.
Frequency of transition from growth to shrinkage.
Frequency of transition from shrinkage to growth.
Nine cells; 31 life histories; 58 minutes.
S. cerevisiae values are from the work of Adames and Cooper (1).
When Yeb1 is overexpressed.
U. maydis values are from the work of Steinberg et al. (40). Analyses are presumed to have been performed at <37°C.
While MT growth and shrinkage rates were comparable among these organisms, C. albicans transition frequencies were strikingly lower (Table 2). Transitions from growth to shrinkage (catastrophe) and from shrinkage to growth (rescue) occurred at 0.34 events minute−1 and 0.02 events minute−1, respectively, in C. albicans and at ≥2 events minute−1 and at ∼2 events minute−1, respectively, in the other two fungi. Thus, the rates of C. albicans MT growth and shrinkage are comparable to those of S. cerevisiae and U. maydis, while the transition frequencies are very different.
The presence of free MTs is regulated during the cell cycle.
During our analysis of MT dynamics, it became apparent that some cells had an abundance of free MTs but that others had relatively few. We asked if the behavior of free MTs was cell cycle dependent by quantifying the number of free MTs observed per minute throughout the cell cycle. Free MTs appeared at a rate of 2.8 ± 0.2 MTs minute−1 prior to hyphal emergence and in short hyphae with unduplicated SPBs (Fig. 9A and 9A.mov) (n = 14 cells; 56 min). Free MTs appeared at the comparable rate of 3.4 ± 0.3 MTs minute−1 in hyphae with duplicated SPBs that had not yet entered the germ tube (Fig. 9B and 9B.mov) (n = 13 cells; 54 min). However, once short spindles entered the germ tubes, the number of free MTs min−1 dropped dramatically to 1.0 ± 0.2 MTs minute−1 (Fig. 9C and 9C.mov) (n = 13 cells; 61 min) and remained low at 0.9 ± 0.1 MTs minute−1 through mitosis (Fig. 9D and 9D.mov) (n = 8 cells, 35 min). The frequency of free MT appearance increased rapidly to 3.3 ± 0.7 MTs minute−1, during and after spindle disassembly (Fig. 9E and 9E.mov) (n = 12 cells; 43 min). We observed similar behavior in yeast cells at 30°C (Fig.9_Supp.mov at the above-named website). These results indicate that, like U. maydis (21), but unlike S. cerevisiae, C. albicans regulates a cell cycle-dependent pool of free MTs.
A high proportion of free MTs in the short spindle and mitotic phases clearly resulted from breakage or detachment of SPB-bound MTs (Fig. 9C.mov and 9D.mov). In contrast, we could not conclusively determine the origin of free MTs during interphase because our analysis was performed using single focal planes. As a result, we could not be certain of the origin of the majority of free MTs during interphase. However, in the region between the SPBs in the postmitotic hypha in Fig. 9E.mov, free MTs appeared to arise independently of the SPBs (see also the appearance of a free MT in the basal cell of Fig. 9C.mov). Free MTs also grew and shrank at arbitrary positions (Fig. 9 movies). Using free MTs that maintained a constant length during displacement, we determined the rate of free MT movement and found that, as in U. maydis and S. cerevisiae, free MTs moved faster than SPB-bound MTs (20.5 ± 1.1 μm/minute [n = 17] and 9.2 ± 0.7 μm/minute [n = 17], respectively) (Table 2). This suggests that either the two mechanisms are different or, more likely, that the nucleus provides significant resistance to the motive force that displaces short spindles. Interestingly, free MTs shrank at roughly the same rate as SPB-bound MTs (26.6 ± 1.9 μm/minute [n = 27] and 27.2 ± 0.1 μm/minute [n = 13], respectively), suggesting that, like SPB-bound MTs, the shrinkage of free MTs occurred preferentially at the distal end. If simultaneous shrinkage from both ends occurred, it would be expected to result in a greater shrinkage rate of free MTs over SPB-bound MTs. Thus, shrinkage of both free and SPB-bound MTs is likely to be stimulated by the same factors and occurs predominantly at distal MT ends.
DISCUSSION
This study elucidates mechanisms underlying the movement of nuclei in C. albicans hyphal cells. By using time-lapse microscopy to follow nuclear, spindle, and microtubule dynamics through the first hyphal cell cycle, we discerned the movements of these structures relative to germ tube growth and the sites of the septin rings as well as the degree to which different microtubule dynamics contribute to nuclear movement. Furthermore, we quantitatively correlated critical transitions of the nuclear and morphogenetic cell cycles.
Nuclear and spindle dynamics are tightly coordinated with morphology.
Nuclear (Nop1) and spindle (Tub2) dynamics paralleled each other throughout our studies (Fig. 2 and 3). For example, the premitotic migration of Nop1 (Fig. 2) and short spindles (Fig. 3) was nonlinear. Furthermore, Nop1 division occurred and spindle elongation began near the presumptum. Finally, mNop1 resided near the neck of the basal cell and germ tube at the time of septation, while mSPBs were similarly positioned at the completion of spindle disassembly. Interestingly, neither mNop1 signals nor mSPBs had yet entered the basal cell when septation occurred. Together, these results suggested that Nop1 position is a strict function of SPB position and, more importantly, that nuclear position is regulated as a function of morphogenesis.
The appearance of the septin ring in cell cycle progression events was also quite evident. First, septin ring deposition appeared to occur immediately after tip growth passed the future site of septation (presumptum). This is consistent with the dependence of septin ring assembly upon polarisome components in S. cerevisiae (24). Secondly, spindle elongation and Nop1 separation occurred as the nucleus and short spindle approached the presumptum, suggesting that, as in S. cerevisiae (26), the septin ring may capture MTs to facilitate spindle alignment across the site of septation.
Hyphal morphogenesis is tied to the cell cycle.
By quantifying the times elapsed between nuclear and morphogenetic events, we identified two cell cycle transitions that suggest coordination between hyphal morphogenesis and the nuclear cell cycle. First, strains expressing Tub2-GFP grew more slowly than the parental strains (19). In our strains, this reduced the rate of hyphal-tip growth, consistent with the idea that the MT cytoskeleton is important for hyphal growth (2, 6). Importantly, this did not affect the timing of all nuclear events (Fig. 4). Rather, two specific events, presumptum appearance (G1/S transition) and spindle elongation (metaphase/anaphase transition), were specifically delayed. These results are consistent with the idea that the delayed hyphal-tip growth rate in hyphae expressing Tub2-GFP stimulates checkpoints at the G1/S and metaphase/anaphase transitions which inhibit the nuclear cell cycle until appropriate hyphal sizes are attained.
Premitotic forward nuclear migration occurs primarily by the sliding of SPB-bound MTs along the cortex.
Our results suggest that hyphal spindle alignment occurs primarily by the repetitive sliding of SPB-bound MTs along the cortex. First, MTs interact with the cortex along their lengths, as judged by the bending of the MT along the contours of the hyphal wall. Second, SPB displacements occur in the same direction as MT growth (Fig. 6). This observation rules out the possibility that MT depolymerization is a significant contributor to spindle alignment and makes MT plus-end tracking unlikely in part because tracking occurs without significant changes in MT length. Furthermore, MT sliding in S. cerevisiae requires cytoplasmic dynein (1), which likely occupies multiple sites between an MT and the cortex. Plus-end interactions involve a single site, the MT plus end (23), and thus would not be expected to result in the observed conformation of MTs along the hyphal wall.
Support for the sliding model of spindle alignment also comes from our analysis of MT organization in C. albicans. FSM revealed that MTs preferentially grew and shrank at their distal ends but that proximal MT ends were stable even after MT breakage (Fig. 8). These properties are consistent with MT organization in other systems (reviewed in reference 7) and support the idea that C. albicans MT minus ends are SPB proximal and MT plus ends are SPB distal. This also implies that a minus-end-directed MT motor protein facilitates hyphal spindle alignment. Consistent with this idea, C. albicans dynein heavy-chain mutants fail to segregate nuclei to daughter hyphal cells (29; K.R.F. and J.B., unpublished data).
Postmitotic reverse nuclear migration and forward migration result from anaphase B.
Spindle elongation was insufficient to position mSPBs within basal cells, suggesting that reverse nuclear migration occurs primarily as a function of the elongating spindle. Furthermore, Nop1 division occurred when spindles were relatively short compared to the overall length of mitotic spindles, suggesting that anaphase A (movement of chromosomes to the poles) occurs early with respect to the onset of anaphase B (movement of chromosomes due to spindle elongation). As a result, anaphase A makes little contribution to total nuclear movement in hyphae and reverse nuclear migration (as well as forward nuclear migration after chromosome separation) results from spindle elongation.
Free MTs are cell cycle regulated.
Most fungi exhibit spindle-independent MT organization. For example, in the fission yeast S. pombe, two microtubule-organizing centers at the cell equator organize SPB-independent MT arrays during interphase (18). In U. maydis, polar microtubule-organizing centers nucleate SPB-independent MTs during interphase (41). In both of these fungi, the interphase organization of MTs disappears during mitosis. We found a similar phenomenon in C. albicans yeast and hyphal cells: free MTs were abundant during interphase, much less prevalent during G2 and mitosis, and rapidly reappeared during spindle disassembly.
Microtubules are required for the maintenance of the Spitzenkörper in C. albicans (6). We considered the possibility that free MTs might support hyphal growth but considered this unlikely because hyphal growth is linear throughout the cell cycle (15). Similarly, in Aspergilllus nidulans, cytoplasmic MTs are cell cycle regulated, yet hyphal growth rates do not decrease (21, 32). Thus, we propose that free MTs constitute a cytoplasmic pool of tubulin that facilitates the assembly of the long mitotic spindles observed in C. albicans hyphae.
Model of nuclear migration.
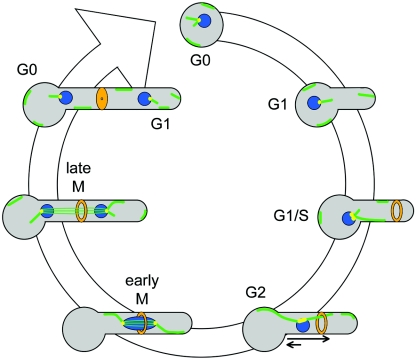
We propose the following model for nuclear and MT dynamics in C. albicans hyphae (Fig. 10). During G0/G1, tubulin saturates the cytosol of unbudded cells and short hyphae. Once a hypha reaches a critical size, it transitions into S phase: the septin ring appears, SPBs duplicate, and the MT sliding machinery is activated. As the hypha proceeds into G2, tubulin is incorporated into the developing spindle and the number of free MTs is reduced. Also in G2, repetitive sliding events from SPB-bound MTs facilitate nuclear migration to the septin ring. In early M phase, the short spindle approaches the ring and the hypha transitions into anaphase. This transition also is dependent upon the hypha reaching a critical size. During anaphase A, kinetochore MTs shorten and pull the chromosomes the short distance to the spindle poles. Subsequently, during anaphase B, interpolar MTs push the spindle poles apart, preferentially pushing the mother nucleus toward the hyphal neck. As the spindle disassembles, the concentration of cytosolic tubulin reaches saturation and free MTs reappear. Following septation, a slower, unknown mechanism moves the G0 mother nucleus back into the basal cell. Thus, in C. albicans hyphae, nuclei and MTs exhibit behaviors that, in some ways, resemble those of S. cerevisiae (appearance of the septin ring prior to nuclear movement and nuclear movement to and division across the septin ring) and in other ways are more like those of other organisms (cell cycle-regulated MT cytoskeleton).
FIG. 10.
Model of nuclear and microtubule dynamics during a hyphal cell cycle in C. albicans. See the text for details. Cellular structures are as defined for Fig. 4, except that SPBs are yellow. Arrows (G2) reflect premitotic nuclear migration.
Acknowledgments
We thank Maryam Gerami-Nejad for strain construction and Cheryl Gale and Maryam Gerami-Nejad for providing plasmids and data prior to publication. We thank Mark Longtine for communication of unpublished data. We are grateful to Mark McClellan for his technical expertise. We thank Pete Sudbery, Tom Hays, Duncan Clarke, and Cheryl Gale for critical reading of the manuscript.
This work was supported by NIH grant AI/DE 14666 to J.B. K.R.F. was supported, in part, by NIH Biotechnology Training Grant GM08347.
REFERENCES
- 1.Adames, N. R., and J. A. Cooper. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akashi, T., T. Kanbe, and K. Tanaka. 1994. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology 140:271-280. [DOI] [PubMed] [Google Scholar]
- 3.Bedell, G. W., A. Werth, and D. R. Soll. 1980. The regulation of nuclear migration and division during synchronous bud formation in released stationary phase cultures of the yeast Candida albicans. Exp. Cell Res. 127:103-113. [DOI] [PubMed] [Google Scholar]
- 4.Byers, B., and L. Goetsch. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carminati, J. L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crampin, H., K. Finley, M. Gerami-Nejad, H. Court, C. Gale, J. Berman, and P. Sudbery. 2005. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 118:2935-2947. [DOI] [PubMed] [Google Scholar]
- 7.Dammermann, A., A. Desai, and K. Oegema. 2003. The minus end in sight. Curr. Biol. 13:R614-R624. [DOI] [PubMed] [Google Scholar]
- 8.D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117:455-469. [DOI] [PubMed] [Google Scholar]
- 9.Davis, D., J. E. Edwards, Jr., A. P. Mitchell, and A. S. Ibrahim. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvorkin, N., M. W. Clark, and B. A. Hamkalo. 1991. Ultrastructural localization of nucleic acid sequences in Saccharomyces cerevisiae nucleoli. Chromosoma 100:519-523. [DOI] [PubMed] [Google Scholar]
- 11.Endow, S. A., and D. J. Komma. 1997. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 137:1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell. 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 14.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 15.Gow, N. A., and G. W. Gooday. 1982. Vacuolation, branch production and linear growth of germ tubes in Candida albicans. J. Gen. Microbiol. 128:2195-2198. [DOI] [PubMed] [Google Scholar]
- 16.Gow, N. A., G. Henderson, and G. W. Gooday. 1986. Cytological interrelationships between the cell cycle and duplication cycle of Candida albicans. Microbios 47:97-105. [PubMed] [Google Scholar]
- 17.Granot, D., and M. Snyder. 1991. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskel. 20:47-54. [DOI] [PubMed] [Google Scholar]
- 18.Hagan, I. M., and J. S. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343-357. [DOI] [PubMed] [Google Scholar]
- 19.Hazan, I., M. Sepulveda-Becerra, and H. Liu. 2002. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell. 13:134-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan, C. J., H. Wein, L. Wordeman, J. M. Scholey, K. E. Sawin, and W. Z. Cande. 1993. Inhibition of anaphase spindle elongation in vitro by a peptide antibody that recognizes kinesin motor domain. Proc. Natl. Acad. Sci. USA 90:6611-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horio, T., and B. R. Oakley. 2005. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell. 16:918-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huisman, S. M., and M. Segal. 2005. Cortical capture of microtubules and spindle polarity in budding yeast—where's the catch? J. Cell Sci. 118:463-471. [DOI] [PubMed] [Google Scholar]
- 23.Hwang, E., J. Kusch, Y. Barral, and T. C. Huffaker. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadota, J., T. Yamamoto, S. Yoshiuchi, E. Bi, and K. Tanaka. 2004. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:5329-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kline-Smith, S. L., and C. E. Walczak. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 15:317-327. [DOI] [PubMed] [Google Scholar]
- 26.Kusch, J., A. Meyer, M. P. Snyder, and Y. Barral. 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16:1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liakopoulos, D., J. Kusch, S. Grava, J. Vogel, and Y. Barral. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112:561-574. [DOI] [PubMed] [Google Scholar]
- 28.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 29.Martin, R., A. Walther, and J. Wendland. 2004. Deletion of the Dynein heavy-chain gene DYN1 leads to aberrant nuclear positioning and defective hyphal development in Candida albicans. Eukaryot. Cell 3:1574-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchison, T. J., and E. D. Salmon. 1992. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119:569-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, C. G., P. S. Maddox, E. D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riquelme, M., R. Fischer, and S. Bartnicki-Garcia. 2003. Apical growth and mitosis are independent processes in Aspergillus nidulans. Protoplasma 222:211-215. [DOI] [PubMed] [Google Scholar]
- 33.Rusan, N. M., C. J. Fagerstrom, A. M. Yvon, and P. Wadsworth. 2001. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell. 12:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, S. L., E. Yeh, P. Maddox, E. D. Salmon, and K. Bloom. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139:985-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 37.Soll, D. R. 1985. Candida albicans, p. 167-195. In P. J. Szaniszlo (ed.), Fungal dimorphism: with emphasis on fungi pathogenic to humans. Plenum Publishing Co., New York, N.Y.
- 38.Soll, D. R., G. Bedell, J. Thiel, and M. Brummel. 1981. The dependency of nuclear division on volume in the dimorphic yeast Candida albicans. Exp. Cell Res. 133:55-62. [DOI] [PubMed] [Google Scholar]
- 39.Soll, D. R., M. Stasi, and G. Bedell. 1978. The regulation of nuclear migration and division during pseudo-mycelium outgrowth in the dimorphic yeast Candida albicans. Exp. Cell Res. 116:207-215. [DOI] [PubMed] [Google Scholar]
- 40.Steinberg, G., R. Wedlich-Soldner, M. Brill, and I. Schulz. 2001. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114:609-622. [DOI] [PubMed] [Google Scholar]
- 41.Straube, A., M. Brill, B. R. Oakley, T. Horio, and G. Steinberg. 2003. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell. 14:642-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudbery, P., N. Gow, and J. Berman. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324, 2004. [DOI] [PubMed]
- 43.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan, M., T. Higuchi, V. L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117:471-482. [DOI] [PubMed] [Google Scholar]
- 45.Tirnauer, J. S., E. O'Toole, L. Berrueta, B. E. Bierer, and D. Pellman. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres-Rosell, J., F. Machin, A. Jarmuz, and L. Aragon. 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 3:496-502. [PubMed] [Google Scholar]
- 47.Wang, B. D., V. Yong-Gonzalez, and A. V. Strunnikov. 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 3:960-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warenda, A. J., and J. B. Konopka. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732Đ2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterman-Storer, C. M., A. Desai, J. C. Bulinski, and E. D. Salmon. 1998. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8:1227-1230. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, C. H., E. J. Lambie, J. Hardin, J. Craft, and M. Snyder. 1989. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98:123-128. [DOI] [PubMed] [Google Scholar]
- 52.Yokoyama, K., H. Kaji, K. Nishimura, and M. Miyaji. 1990. The role of microfilaments and microtubules in apical growth and dimorphism of Candida albicans. J. Gen. Microbiol. 136:1067-1075. [DOI] [PubMed] [Google Scholar]