Abstract
Expression of the genes in the ADE regulon of Saccharomyces cerevisiae is repressed by the presence of purine bases in the extracellular medium and derepressed when cells are grown in the absence of purines. Derepression requires the transcriptional activators Bas1 and Pho2, as well as the biosynthetic intermediates 5′-phosphoribosyl-4-succinocarboxamide-5-aminoimidazole (SAICAR) and 5′-phosphoribosyl-4-carboxamide- 5-aminoimidazole (AICAR). In this study, we investigated if nuclear localization and binding to promoter DNA by the activators are regulated by purines. Using indirect immunofluorescence, we found that Bas1 is localized to the nucleus under both repressing and derepressing conditions. Importantly, we detected Bas1 bound to promoter DNA under both conditions using chromatin immunoprecipitation assays at several ADE promoters (ADE1, ADE2, ADE4, and ADE5,7) and HIS4. We analyzed the binding of Bas1 to wild-type and mutant sequences of the ADE5,7 promoters in vivo, and found that Bas1 binds independently to each of its two binding sites. Pho2 was not required for the association of Bas1 with chromosomal DNA, but it was required for an increase in Bas1-immunoprecipitated DNA. The presence of Pho2 at promoters was dependent on Bas1 and occurred only under derepressing conditions when the ADE genes are transcribed at elevated levels. We propose a model for regulation of the ADE genes in which DNA-bound Bas1 is inactive due to masking of its activation domain and Pho2 binds poorly to promoters when cells have sufficient purine nucleotides. Upon limitation for purines, the SAICAR/AICAR regulatory signal is transmitted to the nucleus to increase Bas1 and Pho2 interaction, recruiting Pho2 to promoters and freeing the activation domains for transactivation.
In Saccharomyces cerevisiae, the adenylate biosynthesis genes (ADE genes) are transcriptionally activated in the absence of extracellular adenine (8, 9, 11, 12, 20, 37). Two transcription factors, Bas1 and Pho2 (also known as Bas2), are required for the regulated expression of the ADE genes under adenine-limiting conditions (2). However, expression of Bas1 and Pho2 is not regulated by adenine (42, 43), indicating that adenine repression occurs by down-regulating the activator functions of the Bas1 or Pho2 proteins. Apart from nine ADE genes (ADE1, ADE2, ADE4, ADE5,7, ADE6, ADE8, ADE12, ADE13, and ADE17), Bas1 and Pho2 together also transcriptionally activate the HIS1, HIS4, and HIS7 genes of the histidine biosynthesis pathway and the GLN1, SHM2, and MTD1 genes, involved in the synthesis of glutamine, glycine, and 10-formyl tetrahydrofolate, respectively (2, 9, 10).
Bas1 is a myb-related protein that binds to DNA with the consensus sequence TGACTC and flanking sequences using an amino-terminal myb motif (14, 31, 42). Bas1 binds to two sites in each of the promoters for the ADE2 and ADE5,7 genes (8, 31), and putative binding sites have been identified in the promoters for the other genes it regulates (2, 8, 9, 36). Activation and Pho2-interaction domains have also been mapped to the Bas1 protein (13, 25, 43). Direct interaction with Pho2 is critical for regulation and was proposed to unmask a latent activation domain in Bas1 (13, 43).
Pho2 binds to DNA via an amino-terminal homeodomain (4, 42). At the ADE2 and ADE5,7 promoters, Pho2 binds to a site adjacent to the proximal Bas1 binding site (8, 31). Pho2 also cooperates with two other activators, Pho4 and Swi5, to stimulate the expression of the PHO5 and HO genes (3, 6). A region between amino acids 170 and 407 is important for the ability of Pho2 to interact with and distinguish between these three coregulators (5), and an activation domain was mapped to the carboxyl-terminal 156 amino acids of Pho2 (13).
Through its interactions with its coregulators, Pho2 is necessary for the expression of over 20 genes involved in at least three different regulatory pathways (6, 8, 24). In the cases of phosphate utilization and mating type switching, the cellular regulatory signal is transmitted to the Pho4 and Swi5 proteins, respectively. When cells have sufficient intracellular phosphate, the Pho80/Pho85 kinase phosphorylates Pho4 to prevent its nuclear entry, increase its nuclear export, and inhibit its interaction with Pho2 (15). Likewise, the cyclinB/Cdk1 kinase phosphorylates Swi5 to prevent nuclear entry until chromosome segregation in M phase (21, 39). In both cases, activities of the kinases are regulated to indirectly affect the transcriptional activity of the coregulators with Pho2.
Experiments from the Daignan-Fornier laboratory showed that intermediates in the de novo purine nucleotide biosynthetic pathway, 5′-phosphoribosyl-4-succinocarboxamide-5-imidazole (SAICAR) and 5′-phosphoribosyl-4-carboxamide-5-imidazole (AICAR), are likely to be the intracellular signals needed for activation of the ADE genes (27, 28). The levels of these biosynthetic intermediates were proposed to reflect levels of the nucleotide pools indirectly through regulation of the activity of the first enzyme of the pathway, glutamine 5-phosphoribosyl-1-pyrophosphate amidotransferase, by ATP and ADP (27). Feedback inhibition decreases pathway flux and leads to low levels of SAICAR and AICAR. When feedback inhibition is relieved, flux through the pathway increases and the SAICAR/AICAR signal is generated. It is thought that the regulatory signal affects the activity of the transcription factors by promoting their interaction, although it is not known if this is direct or indirect (27, 43).
Two studies using protein fusions led to insights about the roles of Bas1 and Pho2 during adenine regulation. One study generated fusions of the DNA binding domain of LexA with Bas1 and Pho2 (43), while the other generated fusions of the VP16 activation domain with Bas1 and Pho2 (25). The LexA-Bas1 fusion protein promoted Pho2-dependent, adenine- regulated expression, and the Pho2-VP16 protein showed Bas1-dependent, adenine-regulated expression. These studies suggested that the regulatory signal does not alter DNA binding by Bas1 or transactivation by Pho2. Interestingly, both the Bas1-VP16 and LexA-Pho2 fusion proteins exhibited partner-independent, constitutive expression, suggesting that the regulatory signal stimulates the activation function of Bas1 and the DNA binding of Pho2. Since a Bas1-Pho2 fusion protein promoted constitutive gene expression, the model for adenine regulation is that the regulatory signal stimulates interaction between these factors to increase activation function of Bas1 and DNA binding by Pho2 (25, 43).
To better understand the molecular mechanism underlying the Bas1-Pho2 interaction that leads to gene activation, we addressed two questions in the present study. First, we asked if Bas1 changes its subcellular localization during adenine regulation, similar to the changes exhibited by Pho4 and Swi5, the other coregulators with Pho2. Nuclear localization of Bas1 was shown in a previous study using a Bas1-green fluorescent protein fusion (26); however, it was not shown that the fusion protein was responsive to adenine regulation. Therefore, we considered it important to determine if the subcellular localization of Bas1 changes using a protein that exhibits adenine regulation of a target gene. Second, we wanted to test directly predictions made by the two previous studies that used the LexA and VP16 fusion proteins by assessing the behavior of Bas1 and Pho2 at native genes during adenine regulation. The results of indirect immunofluorescence and chromatin immunoprecipitation experiments demonstrate that Bas1 is nuclearly localized and is bound to promoter DNA under both repressing and derepressing conditions. Derepression leads to a small increase in the binding of Bas1 that is dependent on Pho2. Unlike Bas1, Pho2 does not bind to promoters under repressing conditions or when Bas1 is absent, but derepression leads to a dramatic increase in the presence of Pho2 at the promoters.
MATERIALS AND METHODS
Strains and media. (i) Yeast.
Saccharomyces cerevisiae strains AY854 (MATα BAS1 PHO2 ura3-52), AY856 (MATa bas1-2 PHO2 ura3-52), and AY858 (MATα BAS1 pho2-2 ura3-52) were previously described (31). Strain RR376 (MATα BAS1-myc13::KanMX6 PHO2 ura3-52) containing 13 copies of the Myc epitope tagged at the C terminus of BAS1 was created from AY854 by a PCR-based method for tagging chromosomal genes by yeast transformation (18). The pFA6a-13Myc-KanMX6 plasmid was used as a template, and the module was amplified using Elongase (Invitrogen) and oligonucleotides JO-205 and JO-206 under the following conditions: 35 cycles of denaturation at 94°C for 30 s and combined annealing/elongation at 68°C for 2.5 min. Transformants were selected on yeast-peptone-dextrose plates containing 200 μg/ml G418. G418+ colonies were analyzed by genomic PCR to verify the presence of the tag. Strain RR377 (MATα BAS1-myc13::KanMX6 pho2-Δ10::hisG ura3-52) is a derivative of RR376 and was created by the replacement of PHO2 with the pho2-Δ10::hisG- URA3-hisG allele (43) and selection of transformants on synthetic complete (SC) medium lacking uracil. The pho2 deletion phenotype was confirmed by the absence of growth on YPAD medium lacking inorganic phosphate. URA3 was popped out by culturing strains on medium containing 5-fluoroorotic acid (5-FOA) to generate pho2-Δ10::hisG.
Strain JRY134 (MATa BAS1 PHO2-HA3::KanMX ura3-1 his3-11,-15 trp1-1 leu2-3,-112 can1-100 ash1Δ::LEU2 swi5-Myc9::KlTRP1), which expresses Pho2-HA3, was a very generous gift of Jonathan Mathias and Andrew Vershon. Strain RR409 (MATα BAS1 PHO2-HA3::KanMX6 ura3-52) contains three copies of the hemagglutinin (HA) epitope tag at the C terminus of PHO2 and was created from AY854 by integrative homologous recombination with a PCR product obtained using genomic DNA from JRY134 as a template and oligonucleotides IO-611 and IO-612, flanking the HA-tagged region of PHO2. Transformants were selected on YPAD-plus-G418 plates and were tested by PCR to confirm the presence of the tag. Expression was assessed by Western analysis using anti-HA antibodies. Strain RR412 (MATα bas1-Δ10::hisG PHO2-HA3::KanMX6 ura3-52) is a derivative of RR409 and was created by the replacement of BAS1 with the bas1-Δ10::hisG-URA3-hisG allele by integrative transformation with an EcoRI-to-SphI fragment obtained from pR166 and selection on SC medium lacking uracil. The bas1 deletion was confirmed by PCR analysis of the genomic locus and by adenine bradytrophy. The URA3 gene was popped out by culturing the strain on 5-FOA medium to generate bas1-10Δ::hisG.
Mutations at the proximal and distal Bas1-binding sites at the chromosomal ADE5,7 locus were introduced into strain RR376 by the two-step gene replacement method (32). The first step replaced positions −233 to −153 of the ADE5,7 promoter with URA3: a fragment with URA3 was produced by amplification of plasmid pRS426 using the upstream primer IO-520 and the downstream primer IO-521 that each carried 69 bases homologous to ADE5,7 and 21 bases homologous to URA3. Ura+ transformants of strain RR376 were selected on medium lacking uracil. The genotype of strain RR403 (MATα BAS1-myc13::KanMX6 ade5,7-Δ100::URA3 PHO2 ura3-52) was assessed by adenine auxotrophy and genomic PCR analysis. To introduce wild-type or mutant promoter sequences, the region from −233 to −133 of ADE5,7 was amplified from plasmids pR136 (the wild-type promoter), pR139 (substitution mutation in the distal Bas1-binding site at position −214: TGACTC to TGAATTC), and pR152 (same substitution mutation in the proximal Bas1-binding site at position −181). The wild-type promoter fragment was prepared from pR136 using primers IO-600 and IO-602; the distal binding site mutation fragment was prepared from pR136 using primers IO-601 and IO-602; and the proximal binding site mutation fragment was prepared from pR152 using primers IO-600 and IO-602. The amplified fragments were introduced into the genome by integrative transformation, as described above, selecting for loss of URA3 by growth on 5-FOA medium. Strains were characterized by assessing adenine auxotrophy, PCR from genomic DNA with EcoRI restriction, and DNA sequence analysis. Strain RR405 (MATα BAS1-myc13::KanMX6 ade5,7-d214 PHO2 ura3-52) carries a mutation in the distal binding site, and strain RR406 (MATα BAS1-myc13::KanMX6 ade5,7-p181 PHO2 ura3-52) carries a mutation in the proximal Bas1 binding site.
(ii) Media.
Strains were grown at 30°C on YPAD solid medium or SC liquid or solid medium containing 2% (wt/vol) glucose, 0.17% (wt/vol) yeast nitrogen base (Difco), and 0.5% (wt/vol) ammonium sulfate supplemented with amino acids as described previously (35). Adenine was added to a final concentration of 0.15 mM where indicated. Medium lacking inorganic phosphate was prepared by adding 10 mM MgSO4 to YPAD broth and precipitating out phosphates with 10% aqueous ammonium hydroxide; after filtration, the pH was adjusted to 5.8 with hydrochloric acid, and agar was added to 2% (33). Medium containing 5-FOA was made by supplementing SC with 0.1% 5-FOA. G418 was added to YPAD medium at 200 μg/ml.
Plasmids and oligonucleotides.
Sequences of the oligonucleotides used in this study are given in Table 1. Plasmid pR116 carries an ADE5,7-lacZ fusion on a URA3 CEN plasmid (31), and plasmid pR133 carries the heterologous UASADE5,7-lacZ reporter (31). Plasmid pR136 carries wild-type UASADE5,7, and plasmids pR139 and pR152 carry mutations in either the distal or proximal Bas1-binding site of UASADE5,7, respectively, and were described previously (31). Plasmid pR166, which contains the bas1-Δ10::hisG-URA3-hisG allele, was constructed in two steps. First, the 5′ end of the BAS1 gene on a 382-bp EcoRI-BglII fragment was inserted at the same sites in pNKY50 (1), producing an intermediate plasmid. The EcoRI-BglII fragment was generated by PCR using pCB286 (2) and oligonucleotides RO-109 and RO-110. The 3′ end of the BAS1 gene on a 141-bp BamHI-SphI fragment was inserted into the same sites in the intermediate plasmid to generate pR166. The BamHI-SphI fragment was generated using PCR and oligonucleotides RO-111 and RO-112 on pCB286.
TABLE 1.
Oligonucleotides used in this study
| Name | Comment | Sequence |
|---|---|---|
| IO-520 | ADE5,7 | 5′-CAACTGCCGTTGTTCATACCGTGACTAATAGTGGCAGTAAGCAGCTTTTTTTTTTTTTCAAAATTTTTCTCGCGCGTTTCGGTGATGACG-3′ |
| IO-521 | ADE5,7 | 5′-ACGGTTAACAGCGGTTGCACTTGCCTCTGAACATAGTTTAAATACCAAGTTCAAGCCCATCGCATAGGCCTGTGCGGTATTTCACACCGC-3′ |
| IO-600 | ADE5,7 | 5′-AACTGCCGTTGTTCATACCGTGACTAATAGTGGCAGTAAGCAGCTTTTTTTTTTTTTCAAAATTTTTCATTTTTTTTTTCAGTTGACTCG-3′ |
| IO-601 | ADE5,7 | 5′-AACTGCCGTTGTTCATACCGTGACTAATAGTGGCAGTAAGCAGCTTTTTTTTTTTTTCAAAATTTTTCATTTTTTTTTTCAGTTGAATTC-3′ |
| IO-602 | ADE5,7 | 5′-GACGGTTAACAGCGGTTGCACTTGCCTCTGAACATAGTTTAAATACCAAGTTCAAGCCCATCGCATATTCATTATTGCTGTTATTACCAG-3′ |
| RO-109 | BAS1, 5′ | 5′-CACGGAATTCTCCAGTCACAGAATAAAGCC-3′ |
| RO-110 | BAS1, 5′ | 5′-CACGAGATCTCATTCTCGATAAAATGTATTCTGCG-3′ |
| RO-111 | BAS1, 3′ | 5′-CACGGGATCCGTTGAGAAGGCAAGGCC-3′ |
| RO-112 | BAS1, 3′ | 5′-CACGGCATGCGCGAGTCATGAAACTACAACG-3′ |
| JO-205 | BAS1 | 5′-GAGCATGATATGACGTCAGGAGGTTCTACCGAGAATGGGTCAGTCCTGCCACTGAATCCT-3′ |
| JO-206 | BAS1 | 5′-GCTTATTACAAAACTAATATGTTAAACAATTGAAAGATTTGTGTTTTTTTTCGGCCTTGC-3′ |
| IO-611 | PHO2 | 5′-GGATGATACGCTCAATTTACTGGATACTACCGTCAACACG-3′ |
| IO-612 | PHO2 | 5′-GATTTATATAAGGACACATGTCTCCACCTATAACGCGAGC-3′ |
| ON275 | POL1 ORF | 5′-AGAGCTGGTAGAAATGAGTATATCTTAC-3′ |
| ON276 | POL1 ORF | 5′-TTAGAGCTTGTTGACGAATATCAC-3′ |
| ARN1-5′ | ARN1 | 5′-TGCACCCATAAAAGCAGGTG-3′ |
| ARN1-3′ | ARN1 | 5′-GAGAGCTATCGAATGTTTCC-3′ |
| RO-29 | ADE5,7 | 5′-CGTCCTCGAGCATTTTTTTTTTCAGTTGAC-3′ |
| RO-28 | ADE5,7 | 5′-CGTCCTCGAGGTTCAAGCCCATCGC-3′ |
| IO-301 | ADE1 | 5′-GCCATACTCGAAATTTCAAC-3′ |
| IO-302 | ADE1 | 5′-ACCGTCTTACCTCAAAGAAT-3′ |
| IO-305 | ADE2 | 5′-AGCCGAGAATTTTGTAACAC-3′ |
| IO-306 | ADE2 | 5′-ATGTACTTAGAAGAGAGATC-3′ |
| IO-303 | ADE4 | 5′-ACTATGAACGCTCGTAAGTA-3′ |
| IO-304 | ADE4 | 5′-GAATCTGAAAAGTACAAACC-3′ |
| IO-307 | ADE16 | 5′-CATGTTCTTTTTAGTCGCAC-3′ |
| IO-308 | ADE16 | 5′-TGGATTCTTTGTTTGGAATG-3′ |
| IO-311 | HIS4 | 5′-GCAGAATGCCCCCATCACAA-3′ |
| IO-312 | HIS4 | 5′-CCTTCTATATCGAATGACTG-3′ |
| IO-401 | ADE5,7 | 5′-TTCAGTTGACTCGCCCCGTC-3′ |
| IO-402 | ADE5,7 | 5′-TCCTGGGACGGTTAACAGCG-3′ |
| IO-403 | CYC1 | 5′-CAGTAAATTGACCTGAATAT-3′ |
| IO-404 | ADE5,7 | 5′-GTTCAAGCCCATCGCATATT-3′ |
| ACT-up | ACT1 | 5′-CTTTCTGGAGGAGCAATGATC-3′ |
| ACT-down | ACT1 | 5′-GGATTCCGGTGATGGTGTTAC-3′ |
Immunofluorescence.
Immunofluorescence microscopy was performed as described previously (29). Briefly, 5 ml of cells were grown in SC medium containing or lacking adenine for 3 to 4 h, and formaldehyde was added to 3.7%. After 30 min, cells were pelleted by centrifugation, resuspended in 2 ml of 50 mM potassium phosphate containing 4% fresh paraformaldehyde, and incubated for 12 h with shaking at room temperature. Cells were pelleted, incubated in 1 ml of 200 mM Tris-HCl, pH 8.0, 20 mM EDTA, and 1% β-mercaptoethanol, and then made into spheroplasts using 10 μg/ml oxalyticase in 1.2 M sorbitol, 50 mM Tris-HCl, pH 8.0, for 30 min. Cells were washed in 1.2 M sorbitol, permeabilized for 60 s with 1% sodium dodecyl sulfate, 1.2 M sorbitol, and washed three times in 1.2 M sorbitol. Cells (20 μl) were then adhered to polylysine-coated glass slides and labeled with anti-Myc antibody for 90 min. Bas1-Myc13 was detected using sheep antimouse immunoglobulin G (IgG) antibody conjugated to Texas Red (Molecular Probes). Mounting medium containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) was used to visualize nuclear DNA.
ChIP assays.
Yeast strains RR376 and its isogenic derivatives that express Bas1-Myc13 and strain JRY134, which expresses Pho2-HA3, were grown in SC medium with or lacking adenine to mid-log phase (optical density at 600 nm [OD600], 0.8 to 1.0). A chromatin immunoprecipitation (ChIP) assay was performed as described previously (7, 16, 38). Briefly, 50-ml cultures were treated with 1% formaldehyde for 30 min, and cross-linking was stopped by the addition of 125 mM glycine. Cells were collected by centrifugation, washed three times with 40 ml of Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 150 mM NaCl), and stored at −80°C. The cell pellet was resuspended in lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin), and cells were disrupted by intense vortexing for 2 h in the presence of glass beads. The cell extract was sonicated with four 20-s pulses to shear chromatin to an average DNA fragment size of 400 to 500 bp. DNA fragments that had been cross-linked to protein were enriched by immunoprecipitation with anti-Myc or anti-HA specific monoclonal antibody (Roche). Prior to use, 2 μl of the anti-Myc or anti-HA antibody was preincubated overnight with magnetic beads (Dynal Dynabeads M-450; coated with antimouse IgG) at 4°C. Immunoprecipitation reactions using 2 × 107 beads per 50-μl sample were performed with gentle shaking overnight at 4°C, followed by magnetic pellet formation. Pellets were washed two times with lysis buffer, two times with lysis buffer containing 360 mM NaCl, two times with wash buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and one time with 10 mM Tris-HCl, pH 8.0, 1 mM EDTA. The cross-links were reversed by heating at 65°C for 6 to 8 h, and the DNA was purified using the PCR Purification kit (QIAGEN). The extent of the enrichment was monitored by PCR amplification on different promoters using gene-specific primers. Input DNA was serially diluted and used as a template to determine the amount that fell in the linear range for DNA amplification. The PCR amplification reaction (20 μl) contained 1/50 of the precipitated DNA, 10 pmol of the primer pair (see below), 0.25 mM of each deoxynucleoside triphosphate, 0.5 μCi of [α-32P]dATP (3,000 Ci/mmol), and 0.5 units of Taq DNA polymerase (QIAGEN). For the ChIP assays, the amplification cycle was 5 min at 94°C, followed by 20 cycles of 40 s at 94°C, 40 s at 55°C, and 40 s at 72°C.
The following pairs of PCR primers (see Table 1 for the nucleotide sequences) were used to amplify promoter or open reading frame (ORF) sequences, as indicated (+1 refers to the ATG codon): POL1 ORF from positions +2377 to +2830 using primers ON275 and ON276; ARN1 promoter from positions −260 to −85 using primers ARN1-5′ and ARN1-3′; ADE5,7 promoter from positions −233 to −133 using primers RO-29 and RO-28; ADE1 promoter from positions −310 to −100 using primers IO-301 and IO-302; ADE2 promoter from positions −300 to −60 using primers IO-305 and IO-306; ADE4 promoter from positions −545 to −260 using primers IO-303 and IO-304; ADE16 promoter from positions −300 to −50 using primers IO-307 and IO-308; and HIS4 promoter from positions −310 to −170 using primers IO-311 and IO-312.
PCR products were size separated on 1.2% agarose gels. Gels were fixed with 10% trichloroacetic acid, dried, and exposed to a multipurpose phosphorimaging screen. The amount of [α-32P]dATP incorporated into the PCR fragments was determined by quantifying the band intensities as pixels on a Cyclone phosphorimager. The values were normalized to a reference PCR product of either POL1 or ARN1 that was amplified in the same reaction mixture. The severalfold enrichment of immunoprecipitated DNA under derepressing conditions relative to repressing conditions was calculated. For each experiment, ChIP assays were performed on three sets of formaldehyde-treated cells and PCRs were performed in triplicate.
β-Galactosidase assays.
Transformants to be assayed for β-galactosidase activities were inoculated in 5 ml of SC medium supplemented with 0.15 mM adenine and cultured for ∼42 h. Each saturated culture was diluted 1:50 in 25 ml of fresh medium, with and without adenine supplementation, and grown for 5 h with shaking at 30°C. Cells were harvested by centrifugation and frozen at −20°C overnight. β-Galactosidase assays were performed using whole-cell extracts (19).
RNA measurement.
Overnight cultures of strains RR376, RR405, and RR406 were inoculated into 100 ml of SC medium supplemented with adenine at an OD600 of 0.1 and were grown with shaking to an OD600 of 0.5. Half of the sample was harvested as the repressed (ade-containing [+ade]) sample. Cells from the remainder were pelleted, washed twice with synthetic dextrose medium, and resuspended in SC medium lacking adenine. After 15 min, cells were harvested by centrifugation as the derepressed (lacking adenine [−ade]) sample. RNA was isolated by using the MasterPure yeast RNA isolation kit (Epicenter) and spotted onto Gene Screen Plus membranes using a dot blot manifold (34). Radiolabeled probes for detection of ADE5,7 and ACT1 were prepared by random priming as described for ADE5,7 (30) and using a PCR fragment containing ACT1 that had been generated by amplification of genomic DNA with ACT-up and ACT-down primers. Membranes were hybridized with the probes in Quick Hyb solution (Stratagene) and washed to remove excess probe (34). Membranes were exposed to a phosphorimaging screen and were analyzed on a Cyclone phosphorimaging system. ADE5,7 RNA levels were normalized to ACT1 RNA.
RESULTS
Bas1 is localized to the nucleus under repressing and derepressing conditions.
Of the three coregulators known to interact with Pho2, both Pho4 and Swi5 are regulated by phosphorylation, which alters their subcellular localization (22, 23). We asked whether Bas1 is regulated similarly, such that it is localized to the nucleus only when purines are limiting. We modified the chromosomal copy of BAS1 by introducing 13 copies of the Myc epitope at the carboxyl terminus. Expression of tagged Bas1-Myc13 protein was confirmed by immunoblot analysis using anti-Myc antibodies. We detected a single band of ∼110 kDa, corresponding to the predicted molecular mass (data not shown).
It was important to determine if the Myc-tagged Bas1 protein retained its normal functions in regulation and transactivation. To address this issue, we assessed whether an ADE5, 7-lacZ reporter showed wild-type expression and adenine regulation in a BAS1-myc13 strain. Cells that expressed no Bas1 protein, native Bas1, or Myc-tagged Bas1 protein were transformed with the lacZ reporter and were grown in synthetic complete medium containing or lacking adenine. β-Galactosidase assays were performed on extracts prepared from these cells. As shown in Table 2, cells lacking the Bas1 protein expressed a basal level of β-galactosidase that was not derepressible. However, the Myc-tagged Bas1 protein was able to derepress expression of β-galactosidase to the same extent as native Bas1 protein. This result indicates that the Myc epitope tags do not interfere with transactivation or regulation by Bas1.
TABLE 2.
Activator-dependent lacZ reporter expression in epitope-tagged strains
| Strain/plasmid | Relevant genotype | β-Galactoside sp act (U)a
|
Fold repressionb | |
|---|---|---|---|---|
| +ade | −ade | |||
| AY854/pR116 | BAS1 PHO2 | 8.0 | 29.0 | 3.6 |
| AY856/pR116 | bas1-2 PHO2 | 8.0 | 7.2 | 0.9 |
| RR376/pR116 | BAS1-myc13 PHO2 | 9.0 | 29.3 | 3.3 |
| AY854/pR133 | BAS1 PHO2 | 0.7 | 3.6 | 5.1 |
| AY858/pR133 | BAS1 pho2-2 | 0.2 | 0.3 | 1.5 |
| JRY134/pR133 | BAS1 PHO2-HA3 | 0.5 | 6.6 | 13.2 |
The specific activity of β-galactosidase is expressed as nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per mg of protein and was measured in triplicate.
Fold repression is calculated as the β-galactosidase activity from cells grown in medium lacking adenine (−ade) divided by the activity from cells grown in medium containing adenine (+ade).
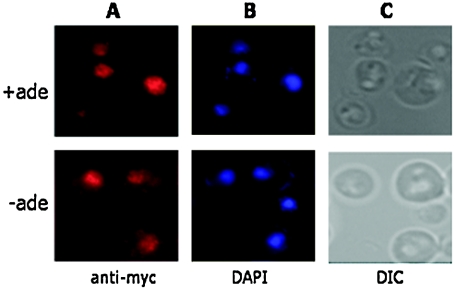
To determine if the localization of the Bas1 protein changes when adenine is present in the growth medium, we performed indirect immunofluorescence microscopy using antibody against the Myc epitopes in a strain expressing Bas1-Myc13. We detected colocalization of Bas1-Myc13 with the DAPI-stained nucleus in a tagged strain (Fig. 1) and saw no fluorescence in the untagged strain (data not shown). Importantly, Bas1-Myc13 is found predominantly in the nucleus under both repressing and derepressing conditions, and it is not excluded from the nucleus under repressing conditions (Fig. 1A). Therefore, unlike the other transcriptional coregulators that function with Pho2, Bas1 does not shuttle in and out of the nucleus in response to extracellular signals.
FIG. 1.
Bas1 localizes to the nucleus under repressing and derepressing conditions. Cells expressing Myc-tagged Bas1 were grown in SC medium containing adenine (+ade, top row) or lacking adenine (−ade, bottom row) and prepared for microscopy. (A) Bas1-Myc13 was localized by indirect immunofluorescence using anti-Myc antibody and Texas Red-conjugated sheep antimouse IgG. (B) Nuclear DNA was visualized using DAPI fluorescence in the same cells. (C) The corresponding differential interference contrast (DIC) image.
Bas1 binds to ADE genes under repressing and derepressing conditions.
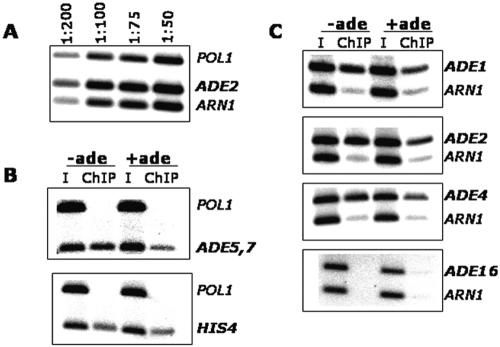
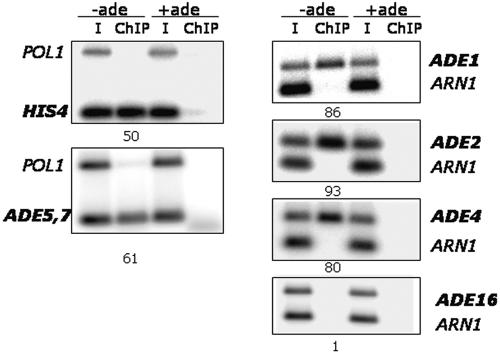
Our results above demonstrate the nuclear localization of Bas1; however, this finding does not indicate if Bas1 is bound to DNA under both repressing and derepressing conditions or if it binds to promoters only when cells lack adenine. To address the question of DNA binding in vivo, we performed ChIP assays using the Myc-tagged Bas1 protein. Cells were grown under both repressing (+ade) and derepressing (−ade) conditions and were treated with formaldehyde. After harvesting and lysing of cells and sonication of the DNA, Bas1-Myc DNA complexes were immunoprecipitated with anti-Myc antibodies. As described in greater detail below, we analyzed eight genes to assess the binding of Bas1 in the ADE regulon. We studied HIS4 and five ADE genes, ADE5,7, ADE1, ADE2, ADE4, and ADE16; each of these genes is regulated by Bas1 except ADE16, which has constitutive expression (40). As controls, we used primers corresponding to the ORF of POL1 and to the promoter of ARN1. Binding of Bas1-Myc13 to each of these promoters under repressing and derepressing conditions was assessed by quantitative PCR using promoter-specific primers. PCR assays were first performed on serial dilutions (50- to 200-fold diluted) of the input DNA using control and ADE-specific primers to ensure that the PCR amplification was in the linear range; an example of this analysis for ADE2 is shown in Fig. 2A.
FIG. 2.
Bas1 binds promoter sites in vivo under repressing and derepressing conditions. Cells expressing Myc-tagged Bas1 were grown in SC medium containing adenine (+ade) or lacking adenine (−ade) and prepared for chromatin immunoprecipitation assays using anti-Myc antibodies. (A) PCR amplification of serially diluted input DNA using primers specific to the POL1 open reading frame, the ARN1 promoter, and the ADE2 gene promoter. (B) PCR amplification of immunoprecipitated DNA using primers specific to POL1, ADE5,7, and HIS4. (C) PCR amplification of immunoprecipitated DNA using primers specific to ADE1, ADE2, ADE4, ADE16, and ARN1. The intensity of PCR bands was quantified on a phosphorimager, and calculations were performed as described in Table 3. I, input DNA.
The results of the ChIP assays on the ADE5,7 and HIS4 promoters are shown in Fig. 2B. We found that Myc-tagged Bas1 binds to each promoter under both +ade and −ade growth conditions. The same result was obtained by ChIP assays at three other ADE loci examined, ADE1, ADE2, and ADE4 (Fig. 2C). However, we did not detect ChIP DNA at ADE16, which exhibits Bas1-independent expression. These data demonstrate that Bas1 is not only localized in the nucleus but is also bound to promoter DNA, whether or not the gene is activated.
We also observed a significant difference in the amount of the PCR product obtained under the derepressing condition relative to the repressing condition. We quantified the results from all the promoters studied and found a 1.7- to 2.7-fold increase in binding of Myc-tagged Bas1 at the four regulated ADE genes and at HIS4 when cells are limited for adenine (Table 3). Calculations were performed separately relative to each control, i.e., POL1 and ARN1, to guard against an unexpected difference due to the location of primer binding sites in promoters relative to an ORF, but we did not see significant differences relative to the controls. These results indicate that there is an approximately 2.2-fold increase in the amount of immunoprecipitated DNA under derepressing conditions, thus indicating an increase in the binding of Bas1-Myc13 to the promoter under the derepressing conditions when the genes are transcribed.
TABLE 3.
Quantification of the relative amounts of chromatin-immunoprecipitated DNA under repressing and derepressing conditions
| Locus | Amt of ChIP DNA relative to that fora:
|
|||||
|---|---|---|---|---|---|---|
|
POL1
|
ARN1
|
|||||
| +ade | −ade | Ratiob | +ade | −ade | Ratio | |
| ADE5,7 | 2.8 | 6.1 | 2.2 | 1.5 | 3.1 | 2.0 |
| ADE1 | 2.6 | 7.1 | 2.7 | 1.2 | 2.9 | 2.4 |
| ADE2 | 3.1 | 7.9 | 2.5 | 1.0 | 2.2 | 2.2 |
| ADE4 | 3.0 | 7.0 | 2.3 | 1.3 | 2.9 | 2.2 |
| HIS4 | 2.6 | 4.5 | 1.7 | NDc | ND | ND |
The value in each +ade or −ade column is a normalized ratio and was calculated as [ChIP (ADE)/input (ADE)] divided by [ChIP (control)/input (control)]. These values are an average of triplicate measurements with a standard deviation of less than 20%.
The derepression ratio is the signal detected in −ade medium divided by that in +ade.
ND, not determined.
Binding site selection by Bas1 does not change during derepression.
The apparent ∼2-fold increase in the binding of Bas1 under derepressing conditions could represent a change in the selection of binding sites under the different growth conditions. Multiple binding sites for Bas1 have been identified in the adenine-regulated gene promoters, and the sites generally occur in pairs (8, 10, 31, 37, 42). When these binding sites have been investigated in detail, as was done for the promoters of ADE2, ADE5,7, and MTD1 (8, 10, 31), one of the two sites is critical for both expression and regulation whereas the other is important for augmenting expression. One simple model to explain the increase in the ability to ChIP Bas1 to promoters is that Bas1 might bind to the noncritical binding site under both repressing and derepressing conditions and only bind to the critical site under derepressing conditions. This model is consistent with the observation that Bas1-VP16 promotes constitutive (Pho2- and adenine-independent) expression (25) if Bas1 binds constitutively to the noncritical binding site.
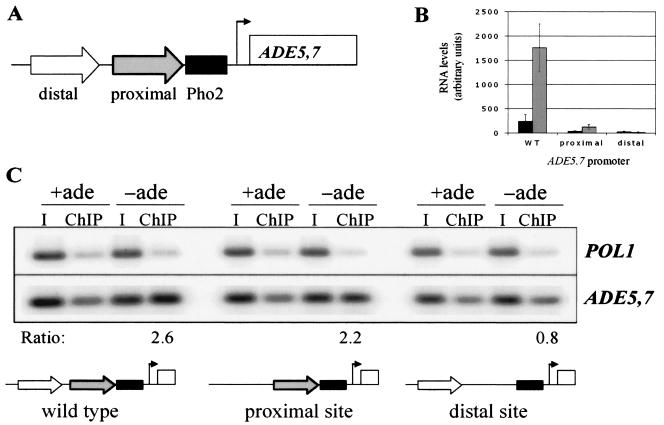
To determine if Bas1 exhibited a difference in site preference in vivo, we used substitution mutations constructed previously to analyze the cis-acting elements and trans-acting factors at ADE5,7 (31). The ADE5,7 promoter has two Bas1-binding sites, schematically depicted in Fig. 3A, that have different effects on reporter expression. Expression and adenine regulation occur when either the promoter is wild type or there is a substitution in the distal site, although expression and the extent of derepression are decreased in the promoter with the mutation (Table 4). However, the proximal Bas1-binding site is critical for both expression and regulation as the expression from the lacZ reporter is basal and nonderepressible when it is mutated (Table 4).
FIG. 3.
Bas1-Myc13 binds to ADE5,7 promoters carrying a single binding site in vivo. (A) Schematic representation of the ADE5,7 promoter and binding sites for Bas1 and Pho2. The Bas1 binding sites are represented by arrows: the distal site (white arrow) is located at −217, and the proximal site (gray arrow) is located at position −184. The Pho2 binding site (black bar) is located at −169. (B) Northern analysis was performed on strains with the wild-type or mutant ADE5,7 promoters at the native locus. Strain RR376 carries the wild-type promoter (WT), strain RR405 carries the proximal Bas1 binding site, and strain RR406 carries the distal Bas1 binding site. RNA was prepared from strains that had been grown in SC medium containing adenine (+ade) and lacking adenine (−ade) and blotted to membranes. Radiolabeled probes recognizing the ADE5,7 and ACT1 genes were hybridized, and the amount of label bound was quantified using a phosphorimager. The levels were normalized to ACT1 and are graphically represented in arbitrary units. Repressed levels, black bars; derepressed levels, gray bars. (C) ChIP assays were performed on the same three isogenic strains as described in panel B that had been grown in SC medium containing (+ade) and lacking (−ade) adenine. PCR was performed with ADE5,7- and POL1-specific primers. ChIP values and the ratio were calculated as described in Table 3 and are listed above the schematic of the promoter. The ratios were calculated as 2.6 for wild type, 2.2 for the proximal site alone, and 0.8 for the distal site alone. I, input DNA.
TABLE 4.
Chromatin immunoprecipitation assays performed on plasmid and chromosomal DNA templates
| Plasmidd | Chromosome ChIPa
|
Plasmid ChIPb
|
β-Galactosidasec
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| −ade | +ade | Ratioe | −ade | +ade | Ratioe | −ade | +ade | Ratioe | |
| None | 3.0 | 1.6 | 1.9 | ||||||
| Wild type | 1.2 | 0.5 | 2.2 | 0.7 | 0.3 | 2.3 | 290 | 15 | 19 |
| Proximal site | 1.2 | 0.6 | 2.0 | 0.7 | 0.4 | 1.8 | 64 | 11 | 5.8 |
| Distal site | 0.9 | 0.5 | 1.9 | 0.3 | 0.5 | 0.6 | 5.4 | 6.1 | 0.9 |
ChIP assays were performed using the primer pair (IO-401 and IO-402) specific to the chromosomal locus of ADE5,7 and relative to POL1, as in Table 3.
ChIP assays were performed using the primer pair (IO-403 and IO-404) specific to the plasmid-borne locus of ADE5,7 and relative to POL1, as in Table 3.
β-Galactosidase activities reported previously (31) are reprinted here for ease of comparison.
Plasmids used were pR136, carrying the wild-type ADE5,7 promoter, pR139, carrying the proximal site (distal site mutation), and pR152, carrying the distal site (proximal site mutation) (31).
Ratio is calculated as the −ade value divided by the +ade value.
We reasoned that if the increase in the ability to ChIP Bas1 to promoters under the derepressing conditions (Fig. 2 and Table 3) was due to a change in the selection of Bas1 binding sites, we would detect the twofold increase only when both sites were present. In the promoter carrying only the distal (noncritical) binding site, we would see Bas1 binding without an increase in the ability to ChIP Bas1, whereas in the promoter carrying only the proximal (critical) binding site, we would detect the binding of Bas1 only under derepressing conditions. We performed ChIP assays with Bas1-Myc13 at the wild-type and mutated promoters (31), first when they were located on plasmids and then from the native chromosomal locus after integration.
In the first approach, we performed ChIP assays on strains bearing plasmids carrying the wild-type and mutant promoters. The Myc-tagged Bas1 strain was transformed with one of the plasmids, and transformants were grown under repressing (+ade) and derepressing (−ade) conditions. ChIP assays were repeated as previously, with one change. We were concerned that the plasmid DNA may not shear during sonication, so we substituted restriction digestion with AluI for sonication to fragment the DNA. PCR amplification was performed using locus-specific primers to assess Bas1 binding at the plasmid-borne ADE5,7 promoter as well as at the native chromosomal ADE5,7 promoter as a control. The plasmid- and chromosome-specific primer pairs amplified products of 230 bp and 150 bp, respectively, so that the resulting fragments could be easily distinguished by size on the same gel.
The results of this ChIP analysis are summarized in Table 4. As expected, we observed a twofold increase in the ability to ChIP the control chromosomal ADE5,7 locus in all three transformed strains, independent of which plasmid was present. Using primers specific to the plasmid, we detected Bas1 binding to both mutant promoters and to the wild-type promoter under repressing conditions (Table 4). Under derepressing conditions (−ade), we saw a ∼2-fold increase in the immunoprecipitated DNA with the wild-type promoter, as predicted, and also with the mutant promoter that carried only the proximal Bas1-binding site. However, we did not detect an increase in the immunoprecipitated DNA from the promoter that carried only the distal site. Thus, the increase in the binding of Bas1 was detected in both cases when the critical proximal site was present. Since the levels of expression as measured by β-galactosidase reporter assays are very different between the wild-type promoter and the mutant promoter with only the proximal Bas1 binding site (Table 4), the increase in the binding of Bas1 correlates with derepressed transcription but not with the strength of transcription.
To extend these plasmid-based results, we repeated the ChIP assay using the same promoter mutations after they had been introduced onto the chromosome at the native ADE5,7 locus. The point mutations in the Bas1 binding sites were introduced into the genome using a two-step method (32) in which we first replaced the ADE5,7 promoter with the URA3 gene (selecting for Ura+ transformants) and then replaced the URA3 gene with either wild-type or mutant promoter fragments (selecting for 5-FOA resistance). Strains bearing the ADE5,7 gene with the wild-type promoter (RR376) or a promoter with only the proximal or distal Bas1-binding site (RR405 and RR406, respectively) were grown in medium containing or lacking adenine. ADE5,7 expression from the native and mutant promoters was assessed by Northern assays. Strain RR376 exhibited regulated expression of ADE5,7 (Fig. 3B). Strain RR405 (which has the proximal site only) exhibited reduced expression and decreased ability to derepress. In strain RR406 (which has the distal site only), transcripts were barely detectable and no regulation was seen. The results of this RNA analysis (Fig. 3B) are consistent with the results of the plasmid-borne β-galactosidase reporter (Table 4).
These same three strains were used to detect the binding of Bas1 to the wild-type and mutant promoters by ChIP analysis (Fig. 3C). We found that Bas1 is bound to the promoter in all of the strains under repressing (+ade) conditions. We observed an ∼2-fold increase in immunoprecipitating DNA in the strain with the wild-type promoter (2.6-fold) and in the mutant strain with only the proximal site (2.2-fold), but we detected no increase in the strain that carried the distal site alone (0.8-fold). These chromosomal data are essentially the same that we obtained with Bas1 on the plasmid-borne promoters. Thus, the increase in the ability to ChIP Bas1 to DNA required an intact proximal site but not the presence of two Bas1-binding sites. In contrast to the model suggested above, the increase in the ChIP assay is not due to a change in the selection of binding sites by Bas1, but rather, it is associated with the proximal site that is adjacent to the Pho2-binding site (31).
Bas1 binds to the ADE promoters independently of Pho2.
ADE gene expression requires Pho2 (8, 42). We wanted to determine if Bas1 binding to promoters in vivo is independent of Pho2, as was indicated by the binding of the Bas1-VP16 fusion protein (25). Second, we wanted to determine if the twofold increase in immunoprecipitated DNA (Fig. 2 and 4; Table 3) is dependent upon Pho2. We reasoned that the derepression signal (the pathway intermediates SAICAR and AICAR) is generated in cells during growth in medium lacking adenine whether or not Pho2 is present. If the increase in ChIP requires only the reception of the regulatory signal by Bas1, then it would be independent of Pho2. However, if the increase in ChIP occurs because of a change in Bas1, i.e., its conformation or affinity for DNA due to Pho2 interaction (43), or because of a change in the chromatin environment due to transcription, then the increase in ChIP would be dependent on Pho2.
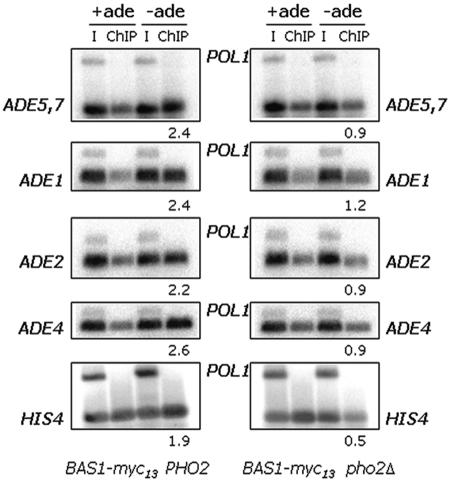
FIG. 4.
Bas1 binds to the ADE regulon promoter in a strain lacking Pho2. ChIP analysis of Bas1-Myc13 binding to the ADE1, ADE2, ADE4, ADE5,7, and HIS4 promoters in strains that express the Pho2 protein (left) and in strains that lack Pho2 (right). Gene-specific primers were used to amplify the ADE, HIS4, and POL1 promoters. ChIP values and the ratio were calculated as described in Table 3. The ratio of Bas1 bound under derepressing to repression conditions is listed under each −ade lane. I, input DNA.
We deleted the PHO2 gene in a strain expressing the Myc-tagged Bas1 protein and then performed ChIP assays using the anti-Myc antibodies and PCR primers directed to the ADE1, ADE2, ADE4, ADE5,7, and HIS4 loci. The results of this experiment are shown in Fig. 4. For the strain that expresses Pho2, we detect Bas1 bound to the promoter under repressing conditions, and we see an increase in the binding of Bas1 of 1.9- to 2.6-fold (Fig. 4, left side). For the strain lacking Pho2 (Fig. 4, right side), we detect binding of Bas1 under both conditions, but we failed to observe the increase in immunoprecipitated DNA in the ChIP assay and instead see no change in Bas1 at the ADE loci (the ratios range from 0.9 to 1.2) or a reduction in the binding of Bas1 to HIS4 (ratio is 0.5). This result demonstrates that the increase in the association of Bas1 with promoters is dependent on Pho2.
Pho2 binds at the ADE genes only under derepressing conditions.
We have shown that Bas1 is nuclear and bound to DNA under repressing conditions and that we can detect an increase in the ability to ChIP Bas1 under derepressing conditions; this increase is dependent on the proximal binding site at ADE5,7, the presence of Pho2, and the growth conditions of medium lacking adenine. We want to determine if Pho2 binds to the ADE promoters under both conditions or only under the derepressing condition using ChIP assays to investigate its binding. Pho2 was tagged with three copies of the HA epitope at the C terminus; we confirmed that the Pho2-HA3 protein was expressed by Western analysis using an anti-HA antibody (data not shown) and that the tag did not disrupt function by analyzing β-galactosidase activity from a UASADE-lacZ reporter construct (Table 2). We grew a strain that expressed the Pho2-HA3 protein in medium containing or lacking adenine and then performed ChIP analyses using anti-HA antibodies. Primers were directed against the ADE1, ADE2, ADE4, ADE5,7, ADE16, and HIS4 loci as well as the ARN1 and POL1 controls. The results of this analysis are shown in Fig. 5. We see virtually no binding of Pho2 under repressing conditions at any of the loci and can readily detect Pho2 binding under derepressing conditions at HIS4 and the ADE loci (ADE1, ADE2, ADE4, and ADE5,7). The ratios of derepression binding to repression binding range from 50- to 93-fold. As expected, the constitutively expressed ADE16 gene exhibits no binding of Pho2. These results clearly show that unlike Bas1, Pho2 binds poorly to DNA under repressing conditions and is present at the promoters only when the genes are expressed.
FIG. 5.
Pho2 binds to ADE gene promoters in vivo only under derepressing conditions. Strain JRY134 (BAS1 PHO2-HA3::KanMX6) was grown on SC medium containing adenine or lacking adenine and prepared for ChIP assays using anti-HA antibodies. Primers specific to the indicated loci were used as described in the legend to Fig. 2. ChIP values and the ratios were calculated as described in Table 3. The ratio of Bas1 bound under derepressing to repressing conditions is listed under each −ade lane. I, input DNA; ChIP, chromatin immunoprecipitation.
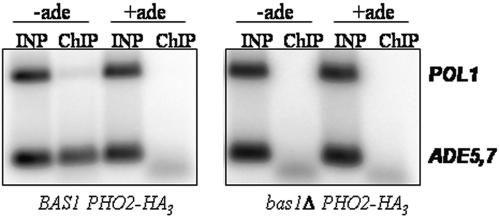
This last result suggests that the binding of Pho2 is dependent on Bas1, although other explanations may be possible. To test if Pho2 binding is dependent on Bas1, we compared the ability to ChIP Pho2-HA in a strain with a deletion of the bas1 gene versus an isogenic strain that is BAS1. We found that Pho2 does not bind to the ADE5,7 promoter when Bas1 is absent (Fig. 6). These results demonstrate that Bas1 is necessary for Pho2 to bind to the ADE gene promoters when cells are depleted of purines.
FIG. 6.
Pho2 is not recruited to ADE5,7 in a strain lacking Bas1. Strains RR409 (BAS1 PHO2-HA3) and RR412 (bas1-Δ10 PHO2-HA3) were grown in SC medium containing and lacking adenine and prepared for ChIP assays using anti-HA antibodies. Primers specific to ADE5,7 and POL1 were used in the PCRs.
DISCUSSION
This work reports on our investigations into the mechanism for regulation of the ADE regulon by the transcription factors Bas1 and Pho2. The ADE regulon consists of the genes encoding the adenylate biosynthetic pathway enzymes, several genes of the histidine biosynthetic pathway, HIS4, HIS1, and HIS7, and genes involved in one-carbon metabolism, SHM2, MTD1, and GLN1 (8-10). Using indirect immunofluorescence microscopy and chromatin immunoprecipitation assays, we found that Bas1 is localized to the nucleus and is bound to the promoters of HIS4 and several ADE genes, ADE1, ADE2, ADE4, and ADE5,7, but not to an unregulated ADE locus, ADE16. Nuclear localization and promoter binding is independent of the presence of its coregulator Pho2 as well as the presence of adenine in the growth medium and thus is presumably independent of the intracellular signal SAICAR/AICAR (27, 28). However, Pho2 binding to these promoters occurs only when the cells are grown in the absence of adenine and when its coregulator Bas1 is present.
We showed that the Bas1-Myc13 protein is localized to the nucleus during both repressing and derepressing conditions. It was important to show that the tagged protein conferred adenine regulation on an ADE-lacZ reporter, demonstrating that the Bas1-Myc13 fusion was capable of receiving and responding to the regulatory signal. Our results showing the nuclear localization of Myc-tagged Bas1 are consistent with a report showing the nuclear localization of a green fluorescent protein-Bas1 fusion protein (26). Importantly, the finding of nuclear localization does not indicate a priori that Bas1 is bound to DNA. For example, Hap1 is nuclear but exhibits a low affinity for DNA in the absence of heme because it is bound in a high-molecular-weight complex (17). Thus, in this study, we extend the observation of nuclear localization to demonstrate in vivo binding to promoter DNA that is independent of the presence of adenine in the growth medium. Our ChIP assays directly demonstrate constitutive promoter binding, a result suggested by the behavior of a Bas1-VP16 fusion protein (25). The finding that Bas1 but not Pho2 is associated with promoters independently of the growth medium is consistent with the observation that overexpression of BAS1 from a high-copy-number plasmid did not increase expression of an ADE5,7-HIS3 reporter whereas overexpression of PHO2 did increase reporter expression (43).
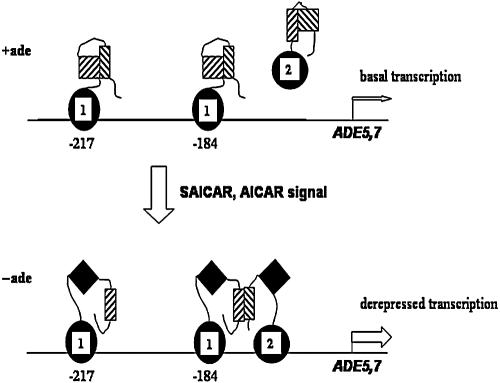
A model for regulation of genes in the ADE regulon is shown in Fig. 7. When cells are grown in adenine-containing medium, Bas1 is bound to promoter DNA in a conformation that is unable to stimulate transcription because its activation domain is masked. Pho2 binds poorly to DNA under these conditions, and genes of the ADE regulon are expressed at the basal level. When cells are grown under conditions lacking adenine, the SAICAR/AICAR regulatory signal is generated, which increases interaction between Bas1 and Pho2. Because of this interaction with Pho2, a region of Bas1 that inhibits the activation domain (BIRD) interacts with Pho2, freeing the activation domain for transactivation (25, 43). Unmasking of the activation domain requires interaction between the BIRD region of Bas1 with a region of Pho2, most likely in the region of Pho2 from 343 to 390 that is important for coregulator interaction (13, 25, 43).
FIG. 7.
Model for regulation of the ADE regulon by Bas1 and Pho2. The binding of Bas1 and Pho2 at the ADE5,7 promoter is depicted under repressing and derepressing conditions. The DNA binding domains of Bas1 and Pho2 are shown as a solid black oval and circle, respectively, marked with a “1” for Bas1 and a “2” for Pho2. The activation domains for both proteins are shown as hatched squares under repressing conditions (+ade) and as solid diamonds under derepressing conditions (−ade). The mutual interaction domains are shown as hatched rectangles. The basal and derepressed levels of transcription under the two conditions are indicated by narrow and broad arrows, respectively.
According to this model, the regulatory signal must be transmitted into the nucleus. The regulatory signal was proposed to be the biosynthetic intermediates SAICAR and AICAR (27, 28), although it is possible that the signal is a metabolic derivative of one of these molecules. SAICAR/AICAR may enter the nucleus to directly promote Bas1-Pho2 interaction, although in vitro experiments were unable to establish a direct effect by either molecule (27). Alternatively, the regulatory signal may interact with another protein to indirectly affect activity of the transcriptional activators.
We investigated whether one of the isozymes encoded by the genes ADE16 and ADE17 could have a secondary role in regulation, analogous to the repressive function of Gal80 that masks the activation domain of Gal4. The duplicated genes ADE16 and ADE17 encode the bifunctional enzyme AICAR transformylase-IMP cyclohydrolase, catalyzing the last two steps in the de novo biosynthetic pathway (41). Both gene products have similar catalytic activity, but their expression is regulated differently. ADE17 is strongly derepressed in the absence of adenine, and ADE16 is expressed at a low constitutive level (40). We hypothesized that the ADE16-encoded enzyme has an additional regulatory role, because this enzyme binds the putative signaling molecule AICAR and is expressed constitutively, while the strong derepression of ADE17 (40) is important to remove the regulatory signal. However, expression of an ADE5,7-lacZ reporter in cells carrying a single mutation of either ADE16 or ADE17 did not exhibit abnormal regulation (Stoler and Rolfes, unpublished data). Thus, it remains to be determined how the regulatory signal affects activity of the activators.
It is noteworthy that Bas1 is nuclear and bound to promoters. Pho2 interacts with three coregulators, Bas1, Pho4, and Swi5, that each respond to three very different cellular signals. Pho4 responds to the depletion of inorganic phosphate to stimulate expression of secreted phosphatases (24). The activity of Pho4 is regulated by phosphorylation, which inhibits nuclear import, promotes nuclear export, and inhibits Pho2 interaction (15). Swi5 responds to cell cycle cues to initiate the expression of the HO endonuclease (7). The activity of Swi5 is regulated by protein instability and phosphorylation, which prevents nuclear entry (21, 39). Thus, two of the three known coregulators with Pho2 change their subcellular localization through changes in phosphorylation in response to their regulatory signal. Bas1 is unique among the Pho2 partners in remaining nuclear localized. The mechanism for ADE gene regulation by Bas1 and Pho2 (Fig. 7) is thus significantly different from the mechanisms used by the other Pho2 partners.
One of the interesting observations we made is that under derepressing conditions the binding of Pho2 increases by nearly two orders of magnitude but the binding of Bas1 increases about twofold. The most common explanation for an increase in ChIP is a change in occupancy of the binding sites by the DNA binding protein. For Bas1 and Pho2, this could occur if the binding sites become available or if the affinity for the sites is increased in derepressing conditions. It seems clear that the affinity of Pho2 for DNA is greatly increased under derepressing conditions. Our data show that the two Bas1-binding sites at ADE5,7 are occupied with Bas1 under both growth conditions, indicating there is no change in the selection of binding sites. The ∼2-fold increase in binding that occurs at the proximal binding site is dependent on Pho2. Pho2 cooperates with Pho4 and Swi5 for binding at the PHO5 and HO promoters, respectively (3, 6), and Pho2 interacts with Bas1 (13). However, in vitro experiments failed to observe cooperativity between Bas1 and Pho2 at HIS4 (42) or at ADE1 and ADE17 (26). The most likely interpretation of the twofold increase in DNA binding by Bas1 in the ChIP assay is an increase in occupancy, perhaps reflecting cooperative binding between Bas1 and Pho2 in vivo. Alternatively, the difference in binding may reflect a change in the ability of Bas1-Myc13 to be cross-linked to DNA by formaldehyde under derepressing conditions. Greater cross-linking would increase the amount of DNA detected by quantitative PCR after immunoprecipitation (16). Thus, the increase in the ChIP assay may reflect a conformational change in Bas1 that occurs due to interaction with Pho2. A conformational change in Bas1 is consistent with studies that show that the interaction of Bas1 with Pho2 exposes a masked activation domain (25, 43). Additional experiments will be necessary to determine if the ChIP assay is detecting conformational changes or cooperative DNA binding.
Acknowledgments
This work was supported by grants MCB-9734170, MCB-0316539, and MCB-0344371 from the National Science Foundation to R.J.R.
Thank you to Jonathan Mathias and Andrew Vershon, Waksman Institute, for supplying strain JRY134, which carried the PHO2- HA3 gene.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. [DOI] [PubMed] [Google Scholar]
- 3.Barbaric, S., M. Münsterkötter, J. Svaren, and W. Hörz. 1996. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 24:4479-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berben, G., M. Legrain, and F. Hilger. 1988. Studies on the structure, expression and function of the yeast regulatory gene PHO2. Gene 66:307-312. [DOI] [PubMed] [Google Scholar]
- 5.Bhoite, L. T., J. Allen, E. Garcia, L. R. Thomas, I. D. Gregory, W. P. Voth, K. Whelihan, R. J. Rolfes, and D. J. Stillman. 2002. Mutations in the Pho2 (Bas2) transcription factor that differentially affect activation with its partner proteins Bas1, Pho4, and Swi5. J. Biol. Chem. 277:37612-37618. [DOI] [PubMed] [Google Scholar]
- 6.Brazas, R. M., and D. J. Stillman. 1993. The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc. Natl. Acad. Sci. USA 90:11237-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosma, M., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 8.Daignan-Fornier, B., and G. R. Fink. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89:6746-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, V., H. Boucherie, C. Monribot, and B. Daignan-Fornier. 1998. Role of the Myb-like protein Bas1p in Saccharomyces cerevisiae: a proteome analysis. Mol. Microbiol. 30:557-566. [DOI] [PubMed] [Google Scholar]
- 10.Denis, V., and B. Daignan-Fornier. 1998. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 259:246-255. [DOI] [PubMed] [Google Scholar]
- 11.Gedvilaite, A., and K. Sasnauskas. 1994. Control of the expression of the ADE2 gene of the yeast Saccharomyces cerevisiae. Curr. Genet. 25:475-479. [DOI] [PubMed] [Google Scholar]
- 12.Giani, S., M. Manoni, and D. Breviario. 1991. Cloning and transcriptional analysis of the ADE6 gene of Saccharomyces cerevisiae. Gene 107:149-154. [DOI] [PubMed] [Google Scholar]
- 13.Hannum, C., O. I. Kulaeva, H. Sun, J. L. Urbanowski, A. Wendus, D. J. Stillman, and R. J. Rolfes. 2002. Functional mapping of Bas2—identification of activation and Bas1-interaction domains. J. Biol. Chem. 277:34003-34009. [DOI] [PubMed] [Google Scholar]
- 14.Høvring, P. I., A. Bostad, E. Ording, A. H. Myrset, and O. S. Gabrielsen. 1994. DNA-binding domain and recognition sequence of the yeast BAS1 protein, a divergent member of the Myb family of transcription factors. J. Biol. Chem. 269:17663-17669. [PubMed] [Google Scholar]
- 15.Komeili, A., and E. O'Shea. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977-980. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, M., and C. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H. C., T. Hon, and L. Zhang. 2002. The molecular chaperone Hsp90 mediates heme activation of the yeast transcriptional activator Hap1. J. Biol. Chem. 277:7430-7437. [DOI] [PubMed] [Google Scholar]
- 18.Longtine, M., D. McEnzie III, N. Demarini, A. Shah, A. Wach, P. Brachat, P. Phillippsen, and J. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 19.Lucchini, G., A. G. Hinnebusch, C. Chen, and G. R. Fink. 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mäntsälä, P., and H. Zalkin. 1984. Nucleotide sequence of Saccharomyces cerevisiae ADE4 encoding glutamine phosphoribosylpyrophosphate amidotransferase. J. Biol. Chem. 259:8478-8484. [PubMed] [Google Scholar]
- 21.Moll, T., G. Tebb, U. Surana, H. Robitsch, and K. Nasmyth. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66: 743-758. [DOI] [PubMed] [Google Scholar]
- 22.Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell 62:631-647. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 24.Oshima, Y. 1982. Regulatory circuits for gene expression: the metabolism of galactose and phosphate, p. 159-180. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), Molecular biology of the yeast Saccharomyces cerevisiae: metabolism and gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Pinson, B., T. L. Kongsrud, E. Ording, L. Johansen, B. Daignan-Fornier, and O. S. Gabrielsen. 2000. Signalling through regulated transcription factor interaction: mapping of a regulatory interaction domain in the Myb-related Bas1p. Nucleic Acids Res. 28:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinson, B., I. Sagot, F. Borne, O. Gabrielsen, and B. Daignan-Fornier. 1998. Mutations in the yeast Myb-like protein Bas1p resulting in discrimination between promoters in vivo but not in vitro. Nucleic Acids Res. 26:3977-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rébora, K., C. Desmoucelles, F. Borne, B. Pinson, and B. Daignan-Fornier. 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol. Cell. Biol. 21:7901-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rébora, K., B. Laloo, and B. Daignan-Fornier. 2005. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson, J., D. Klionsky, L. Banta, and S. Emr. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolfes, R. J., F. Zhang, and A. G. Hinnebusch. 1997. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine-regulation of ADE5,7. J. Biol. Chem. 272:13343-13354. [DOI] [PubMed] [Google Scholar]
- 32.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 33.Rubin, G. M. 1973. The nucleotide sequence of Saccharomyces cerevisiae 5.8S ribosomal ribonucleic acid. J. Biol. Chem. 248:3860-3875. [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods of yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Springer, C., M. Künzler, T. Balmelli, and G. H. Braus. 1996. Amino acid and adenine cross-pathway regulation act through the same 5′-TGACTC-3′ motif in the yeast HIS7 promoter. J. Biol. Chem. 271:29637-29643. [DOI] [PubMed] [Google Scholar]
- 37.Stotz, A., P. P. Muller, and P. Linder. 1993. Regulation of the ADE2 gene from Saccharomyces cerevisiae. Curr. Genet. 24:472-480. [DOI] [PubMed] [Google Scholar]
- 38.Swanson, M. J., H. Qiu, L. Sumibcay, A. Kruegger, S.-J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23: 2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tebb, G., T. Moll, C. Dowzer, and K. Nasmyth. 1993. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 7:517-528. [DOI] [PubMed] [Google Scholar]
- 40.Tibbetts, A. S., and D. R. Appling. 2000. Characterization of two 5-aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase isozymes from Saccharomyces cerevisiae. J. Biol. Chem. 275:20920-20927. [DOI] [PubMed] [Google Scholar]
- 41.Tibbetts, A. S., and D. R. Appling. 1997. Saccharomyces cerevisiae expresses two genes encoding isozymes of 5-aminoimidazole-4-carboxamide ribonucleotide transformylase. Arch. Biochem. Biophys. 340:195-200. [DOI] [PubMed] [Google Scholar]
- 42.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246:931-935. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, F., M. Kirouac, N. Zhu, A. G. Hinnebusch, and R. J. Rolfes. 1997. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene expression. Mol. Cell. Biol. 17:3272-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]