Abstract
This is the first study to characterize the environmental conditions which contribute to the presence and proliferation of environmental mycobacteria in a major freshwater river. Over 20 different species of environmental mycobacteria were isolated, including the pathogenic M. avium and M. kansasii. Species of the rapidly growing M. fortuitum complex were the most commonly isolated mycobacteria, and one-third of all isolates were not identified at the species level, even by 16S sequencing. PCR restriction analysis of the hsp65 gene was more accurate and rapid than biochemical tests and as accurate as yet less expensive than 16S sequencing, showing great promise as a new tool for species identification of environmentally isolated mycobacteria. Total environmental mycobacteria counts positively correlated with coliform and Escherichia coli counts and negatively correlated with chemical toxicity and water temperature. Environmental mycobacteria can survive in the alkaline conditions of the river despite previous reports that especially acidic conditions favor their presence. A representative river isolate (M. fortuitum) survived better than E. coli O157:H7 at pHs below 7 and above 8 in nutrient broth. The river strain also retained viability at 8 ppm of free chlorine, while E. coli was eliminated at 2 ppm and above. Thus, in vitro studies support environmental observations that a variety of extreme conditions favor the hardy environmental mycobacteria.
Environmental mycobacteria, also referred to as nontuberculous mycobacteria, atypical mycobacteria, and mycobacteria other than tuberculosis, have been increasingly implicated in a variety in human diseases, including nosocomial infections (38), pulmonary and disseminated diseases in immunocompromised patients (40, 47), and a variety of other diseases (46). Environmental mycobacteria have also been implicated in outbreaks and pseudoutbreaks in a variety of health care settings (38). In the majority of these instances, treated water has been implicated as a possible common vehicle of transmission (3, 16).
Nontuberculous mycobacteria are ubiquitous (16) and can be found in a variety of ecosystems, including fresh and salt water (23). Mycobacteria have also been isolated from treated drinking water (17), hospital and clinic water systems (19, 20), water supplies in hemodialysis centers (6), industrial hot water systems (13), ice machines (21), swimming pools and hot tubs (27), raw milk (28), and even water-damaged buildings (2). Due to the intrinsic resistance of mycobacteria to chlorine (43) and other frequently used water treatment chemicals and surface disinfectants (22, 31), they have been able to colonize a remarkably wide range of surfaces in hospitals, factories, and residential areas (25, 30, 33). Insects (18, 36) and protozoans (9, 42) may serve as vectors for environmental mycobacteria as well. Some species of environmental mycobacteria, including M. avium subsp. paratuberculosis and M. marinum, are zoonotic pathogens that cause millions in agricultural losses each year (8, 12). Due to the importance of environmental mycobacteria, as opportunistic pathogens in both humans and animals, it is extremely important to understand the ecological conditions that contribute to their proliferation and persistence in freshwater ecosystems.
While there are many reports of environmental isolation of mycobacteria, only two examined environmental parameters which correlate with the presence of environmental mycobacteria. In Finnish woodland and peatland brook sediments, the culturable counts of total environmental mycobacteria correlated positively with total carbon content, lead content, and respiration rate of the sediment (total number of organisms) (26). Environmental mycobacteria counts correlated negatively with pH, as more acidic conditions favored the environmental mycobacteria. Species identification of the environmental mycobacteria was not reported. A second study focused on the M. avium complex (M. avium, M. intracellulare, and M. scrofulaceum) in brown- water swamps in the southeastern United States (29). M. avium complex counts correlated positively with warmer temperature, low pH, low dissolved oxygen, high soluble zinc, and concentrations of humic and fulvic acids.
The Rio Grande is located in the southwestern United States and spans a total of 3,000 km from Alamosa, Colorado, running through New Mexico and along the Texas/Mexico border, eventually emptying into the Gulf of Mexico. The river provides a major source of potable and agricultural water for the population of the Texas/Mexico border region. The cities of El Paso, Texas (population 750,000), and Ciudad Juarez, Mexico (population ∼2 million), are highly dependent on the resources of the river, which provides 60% of the potable drinking water to the area.
Seven sites were sampled once per month over a 1-year period along a region of the Rio Grande spanning 95 km. Water quality data, including temperature, pH, dissolved oxygen, conductivity, total dissolved solids, total chloride, total sulfate, fecal coliforms, and total coliforms, were measured for each site (Table 3) at the time of collection by the International Boundary and Water Commission (IBWC), under the Texas Clean Rivers Program. We obtained aliquots of the water samples for isolation of environmental mycobacteria. Recovered mycobacteria were enumerated, and a selected representative proportion of recovered isolates were identified to species level using a combination of PCR restriction analysis (PRA), biochemical assays, and 16S rRNA analysis. This study is the first attempt to characterize the environmental conditions that correlate with the presence of environmental mycobacteria in a major freshwater river.
TABLE 3.
Summary of environmental parametersa
| Site | TC | FC | CTX | HP | Fl | Tmp | O2 | pH | Con | DS | Cl | Sl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.3e4 ± 5.3e4 | 2.2e4 ± 5.7e4 | 33 ± 18 | 0.40 ± 0.38 | NA | 18.1 ± 8.1 | 8.4 ± 1.3 | 8.2 ± 0.3 | 1.2e3 ± 3.8e2 | NA | NA | NA |
| 2 | 4.2e4 ± 6.4e4 | 1.1e4 ± 1.3e4 | 40 ± 18 | 0.20 ± 0.26 | 580 ± 500 | 17.0 ± 6.9 | 8.2 ± 1.5 | 8.2 ± 0.2 | 1.6e3 ± 6.0e2 | 980 ± 450 | 170 ± 100 | 290 ± 140 |
| 3 | 3.9e4 ± 5.2e4 | 3.2e4 ± 6.4e4 | 41 ± 18 | 0.41 ± 0.69 | NA | 16.2 ± 7.3 | 9.7 ± 2.1 | 8.3 ± 0.2 | 1.7e3 ± 7.4e2 | 1,030 ± 540 | 180 ± 110 | 310 ± 160 |
| 5 | 4.1e4 ± 7.2e4 | 3.3e4 ± 6.3e4 | 43 ± 15 | 0.56 ± 0.68 | 28 ± 37 | 16.8 ± 6.9 | 10.9 ± 2.7 | 8.6 ± 0.4 | 1.7e3 ± 7.1e2 | 1,000 ± 490 | 180 ± 110 | 300 ± 150 |
| 6 | 2.7e4 ± 5.6e4 | 2.0e4 ± 3.5e4 | 37 ± 19 | 0.44 ± 0.64 | 2.9 ± 4.6 | 17.8 ± 7.9 | 10.0 ± 3.4 | 8.4 ± 0.4 | 1.6e3 ± 6.5e2 | 810 ± 410 | 150 ± 100 | 250 ± 120 |
| 7 | 9.5e4 ± 1.4e5 | 2.7e4 ± 3.0e4 | 33 ± 18 | 0.47 ± 0.54 | 32.5 ± 45.0 | 16.2 ± 7.0 | 8.6 ± 3.8 | 8.1 ± 0.4 | 2.3e3 ± 1.2e3 | 1,600 ± 710 | 390 ± 250 | 420 ± 180 |
Data presented as mean ± standard deviation, with typically 13 observations (1/month). NA, not available. Abbreviations are as in Table 2.
MATERIALS AND METHODS
Sample collection and analysis.
Water samples were collected by the Texas Commission on Environmental Quality (TCEQ) from seven surface water sites along a 95-km area of the Rio Grande. Site 1 is at the Anapra Bridge (latitude 31°78.0′N, longitude 106°55.0′W). Site 2 is the Courchesne Bridge (latitude 31°80.3′N, longitude 106°54.0′W). Site 3 is 2.4 km upstream of the Haskell wastewater treatment plant (latitude 31°76.0′N, longitude 106°47.0′W). Site 4 is the effluent from the Haskell plant. Site 5 is 1.3 km downstream of the plant (latitude 31°75.3′N, longitude 106°41.9′W). Site 6 is the Riverside Canal (latitude 31°65.8′N, longitude 106°32.9W). Site 7 is the Alamo Control Structure (latitude 31°31.7′N, longitude 105°93.6′W). Information on sites in the Rio Grande Basin utilized by the TCEQ is available at www.ibwc.state.gov/CRP/Welcome.htm.
Samples were collected in sterile 1-liter polypropylene containers and stored at 4°C. All samples were processed within 1 week of collection and typically on the same day. All chemical tests were performed according to TCEQ standardized procedures using a Hydrolab Surveyor II (14, 44). The chemical toxicity of the water was determined by a test determining cytotoxicity to Rhizobium spp. (4). Helicobacter pylori levels were determined using an antigen enzyme-linked immunosorbent assay (Diagnostic Automation, Calabasas, CA). Total and fecal coliforms were determined by plating river water and 10-fold dilutions in triplicate onto mFC and m-endo agar plates, respectively. Colonies were enumerated after 24 h incubation at 37°C.
Isolation and enumeration of environmental mycobacteria.
Water samples were decontaminated using a modification of the method described by Neumann et al. (34). Ten ml of water was transferred into a sterile 50-ml tube and combined with an equal volume of brain heart infusion broth (Gibco, Carlsbad, CA). The samples were then incubated at 37°C for 5 h to allow fungal and bacterial spores to germinate. After incubation, samples were transferred to sterile 100-ml flasks and each was mixed with the following solutions: 20 ml malachite green (0.2%), 5 ml cycloheximide (0.4 mg ml−1), and 20 ml of 1 M NaOH. After a 30-min room temperature exposure period, samples were titrated with 1 M HCl until dark green color reappeared (∼pH 3). Samples were then vacuum filtered through 0.44-μm nylon membranes (Millipore, Billerica, MA) which were placed onto selective 7H11 medium plates containing 10 μg ml−1 malachite green, 36 μg ml−1 cycloheximide, 50 μg ml−1 ampicillin, and 25 U ml−1 nystatin, and incubated at 37°C for 8 weeks. A typical clinical method from Cumitech 16A was examined during the first month of the study as well (7). Briefly, 50 ml of water was centrifuged at 4,000 × g for 5 min, then the supernatant was removed and the pellet was resuspended in 400 μl of deionized water. Decontamination was for 10 min by 0.6% NaOH at room temperature, followed by neutralization by 12 μl of glacial acetic acid. The entire mixture was plated onto one of the selective plates described above.
Colonies growing on primary culture plates were preliminarily identified as mycobacteria based on positive Kinyoun acid-fast staining (7) and colonial morphology. Most contamination was fungal in nature. Colonies were enumerated by visual counting and representative colonies were subcultured onto standard 7H11 medium plates and LJ medium slants (BBL, Franklin Lakes, NJ) for further analyses.
Biochemical assays.
Representative acid-fast-positive isolates were subjected to a reduced set of biochemical assays including Tween 80 hydrolysis, urease production, 5% NaCl tolerance, chromogenicity, and growth rate (7, 45). Biochemical results served as comparisons to the PRA data. A full set of biochemical assays (approximately 15) would have been prohibitively time consuming and expensive.
DNA extraction.
DNA was extracted from isolates by suspending a loopful of bacteria in 300 μl of TE buffer (10 mM Tris, 1 mM EDTA, pH 8) in 1.5-ml microcentrifuge tubes followed by two freezing (−20°C) and thawing (80°C) cycles. The tubes were then spun at 13,000 rpm using a Sorvall MC 12V centrifuge to pellet cell debris. Two hundred μl of the supernatant was then transferred to sterile 0.5-ml microcentrifuge tubes and stored at −20°C until use.
PRA analysis.
PCR mixtures were prepared by combining 5 μl of genomic lysate, 19 μl of 10% glycerol in sterile water, and 0.5 μl each of hsp65-specific primers tb11 (ACCAACGATGGTGTGTCCAT) and tb12 (CTTGTCGAA CCGCATACCCT) in Ready-to-Go PCR tubes (Amersham Biosciences, Piscataway, NJ) to yield a final volume of 25 μl. The PCRs were run in a Dyad thermocycler (MJ, Hercules, CA) at the following specifications: one cycle at 95°C for 5 min, 45 cycles of 94°C, 60°C, and 72°C, each for 1 min, and then a final extension at 72°C for 4 min. A product of 439 bp was generated by all mycobacterial species. Restriction digestion of PCR products and electrophoretic analysis were performed as previously described (10). The PRAsite database (app.chuv.ch/prasite/index.html) was used for comparative identification (10).
Ribosomal gene sequencing.
Identification by comparison of 500 bp of the 16S rRNA gene (obtained by the MicroSeq 500 method, Applied Biosystems) from river isolates to a sequence database was performed by Accugenix (Newark, DE). Greater than 1% mismatch between the sample and the closest database entry was considered unable to match to a specific species.
Coincubation experiments.
E. coli O157:H7 strain Sakai was obtained from the American Type Culture Collection (ATCC BAA-460). M. fortuitum strain 4G was isolated in October from site 4, and all identification procedures (biochemicals, PRA, and 16S) agreed. Strains were grown to log phase in Luria broth (LB) or 7H9-ADC and then mixed at a 1:1 ratio at an optical density of 0.001 at 650 nm in six 5-ml 7H9-ADC aliquots with the pH preadjusted to 4, 5, 6, 7, 8, or 9 using HCl or NaOH as appropriate (typical pH of the medium is 7.6). Cultures were incubated with shaking at 150 rpm and 37°C, and samples were taken at the time points indicated, serially 10-fold diluted in 7H9-ADC (control experiments show that pH is neutralized and free chlorine is eliminated following dilution), and plated in triplicate for viability on LB, 7H11-OADC, and selective 7H11.
For chlorine survival, log-phase strains were mixed at a 1:1 ratio at an optical density of 0.1 at 650 nm in four 5-ml aliquots of deionized water. After plating for initial viability as above, 1, 2, 4, or 8 ppm of chlorine was added. Sodium hypochlorite, 6% stock (Sigma, St. Louis, MO), was diluted to appropriate levels, with free chlorine levels standardized using a commercial diethyl-p-phenylene diamine system, the Mini Water Analyst (Orbeco Analytical Systems, Inc., Farmingdale, NY). Viability was measured at the indicated time points as above. The maximum sensitivity of viability detection for both strains was 1 CFU per ml.
Statistical analysis.
All statistical analysis was performed with SAS for Windows, version 7 (SAS Institute Inc., Cary, NC), in the Statistical Consulting Lab of the Border Biomedical Research Center by Julia Bader.
RESULTS
For water samples from the first month (February 2002), two isolation methods were utilized in parallel, a standard clinical method involving NaOH decontamination and the Neumann method described above. The NaOH clinical method isolated only 31% of the total environmental mycobacteria (site 1 had 25 acid-fast-positive colonies, site 2 had three, site 4 had one, sites 4 and 5 had zero, and site 7 had 22) isolated by the Neumann method (Table 1) from the same samples. The NaOH method also had a far higher contamination rate by fungi. Therefore, we considered the Neumann method superior for environmental water samples, and it was used for our entire study.
TABLE 1.
Number of environmental mycobacteria counted at seven study sites on the Rio Grandea
| Site | No. (CFU/10 ml)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Feb 02 | Mar 02 | Apr 02 | May 02 | Jun 02 | Jul 02 | Aug 02 | Sept 02 | Oct 02 | Nov 02 | Dec 02 | Jan 03 | Feb 03 | All | |
| 1 | 123 | 11 | 17 | AG | AG | AG | 0 | 3 | 14 | 22 | 1 | TNTC | 80 | 30 ± 42 |
| 2 | 6 | 5 | 14 | AG | AG | AG | * | 2 | 1 | 7 | 9 | TNTC | 15 | 7 ± 5 |
| 3 | 1 | 5 | 2 | AG | AG | AG | * | 1 | 27 | 6 | 4 | * | SNT | 7 ± 9 |
| 4 | 4 | 40 | 5 | AG | AG | AG | 3 | 4 | 24 | 30 | 11 | 100 | SNT | 25 ± 31 |
| 5 | 1 | 0 | 0 | AG | AG | AG | 0 | 3 | 4 | 10 | * | TNTC | SNT | 3 ± 4 |
| 6 | ND | ND | 6 | AG | AG | AG | * | 1 | 0 | 3 | ND | ND | SNT | 3 ± 3 |
| 7 | 31 | 4 | 0 | AG | AG | AG | 2 | 0 | 17 | 12 | * | 61 | 79 | 23 ± 28 |
Number given is CFU/10-ml water volume processed. ND, not determined because site was dry; *, not determined because plates were contaminated by fungi; AG, not determined because of alga bloom contamination; TNTC, too numerous to count; SNT, sample not taken. The column marked All contains the mean ± standard deviation for each site over all months, ignoring months without data.
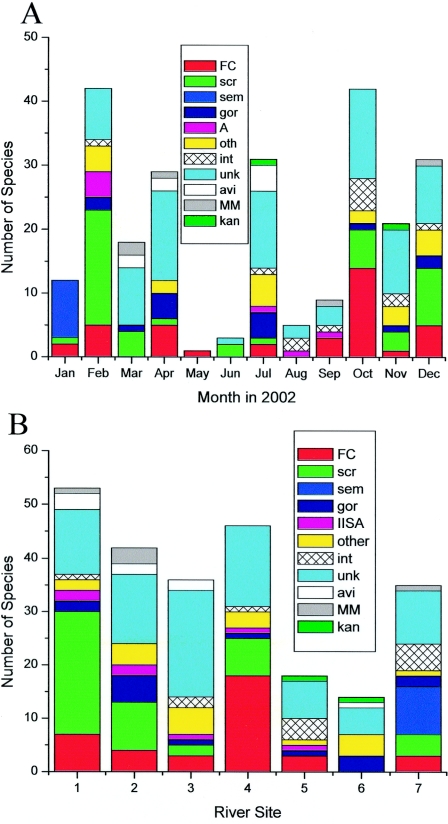
Mycobacteria were isolated from all seven sites in various numbers (Fig. 1B and Table 1). Sites 1, 4, and 7 consistently had the highest counts. Sites 1 and 7 are both agricultural, while 4 is effluent from a wastewater treatment plant. Chemical tests were not routinely performed on the effluent, though free chlorine was typically 3 to 4 ppm and coliform assays were usually negative and never higher than 10 CFU/100 ml. Sites 5 and 6 had the lowest environmental mycobacteria counts and were often dry or of very low water depth.
FIG. 1.
Number of species isolated by month and site. The raw number of each species group (colored sections of columns) isolated each month at all sites (panel A) and each site for all months (panel B). FC, M. fortuitum complex; scr, M. scrofulaceum; sem, M. semiae; gor, M. gordonae; IISA, a PRA pattern shared by M. avium, M. intracellulare, M. simiae, and M. intermedium; other, seven rare species combined (M. smegmatis, M.interjectium, M. lentiflavum, M. nonchromo, M. celatum, M. chitae, and M. pheli), int, M. intracellulare; unk, unknown; avi, M. avium; MM, M.malmoense or M. marinum (indistinguishable by PRA); and kan, M. kansasii.
The total number of representative acid-fast colonies that were subjected to identification procedures was 264 (only two of these were not identified as environmental mycobacteria). All isolates were subjected to five biochemical tests, but because of variability in the tests, 3.4 ± 1.2 reliable test results were generated on each isolate. PRA was also run on every isolate. As a quality control check on the PRA results, 37 isolates were selected that represented the most commonly observed PRA patterns and subjected to 16S ribosomal RNA gene sequencing. Five of the isolates (13.5%) matched a species in the MicroSeq database with 100% nucleotide sequence identity. Four out of these five had the same identification by PRA, with the exception of a PRA pattern being identified as M. smegmatis but matching the 16S sequence for M. mageritense, which does not have an entry in the PRA database. M.mageritense is a recently identified species in the M. fortuitum third biovariant complex (5, 11).
Twenty-three of the 37 isolates had a good match (≥99% homology to a database entry), and thus the most accurate genetic identification tool available was able to identify only 62% of the environmental isolates. This suggests that one-third of the mycobacteria isolated from the environment are novel species or subspecies. In a study of 83 clinical isolates, the same percentage (37%) were unidentifiable by 16S sequencing (37).
To examine mycobacterial ecology in this large natural river, the environmental data (Table 3) were compared to the total environmental mycobacteria counts for correlations (Table 2). Strong positive correlation was seen between total and fecal coliform counts and environmental mycobacteria counts. This was also supported by environmental mycobacteria correlations to the fecal coliform counts (coefficient = 0.719, P value = 0.0012) and E. coli counts (coefficient = 0.579, P = 0.019) independently performed by the IBWC. El Paso Community College (EPCC) fecal and total coliform counts correlate with each other as well, supporting the validity of the methods.
TABLE 2.
Correlations between environmental parameters and microbesa
| Sample | TC | FC | CTX | HP | Fl | Temp (°C) | O2 | pH | Con | DS | Cl | Sl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Myco | 0.375 | 0.563 | (0.354) | 0.023 | (0.15) | (0.462) | 0.008 | (0.16) | 0.101 | 0.248 | 0.26 | 0.20 |
| 0.02 | 0.0002 | 0.029 | 0.90 | 0.63 | 0.023 | 0.97 | 0.44 | 0.64 | 0.34 | 0.32 | 0.45 | |
| 38 | 38 | 38 | 32 | 12 | 24 | 24 | 25 | 24 | 17 | 17 | 17 | |
| I-TC | 1.000 | 0.488 | (0.335) | 0.406 | (0.05) | (0.20) | (0.083) | (0.092) | 0.166 | 0.051 | (0.07) | (0.01) |
| <0.0001 | 0.01 | 0.003 | 0.86 | 0.23 | 0.66 | 0.57 | 0.31 | 0.80 | 0.71 | 0.95 | ||
| 60 | 60 | 59 | 52 | 15 | 39 | 31 | 40 | 40 | 27 | 30 | 30 | |
| I-FC | 0.488 | 1.000 | (0.494) | 0.12 | (0.22) | (0.21) | (0.12) | (0.17) | 0.16 | 0.09 | (0.02) | 0.08 |
| <0.0001 | <0.0001 | 0.41 | 0.44 | 0.20 | 0.53 | 0.30 | 0.34 | 0.65 | 0.90 | 0.66 | ||
| 60 | 60 | 59 | 52 | 15 | 39 | 31 | 40 | 40 | 27 | 30 | 30 |
The top, middle, and bottom values in each cell are the Pearson correlation coefficients, P value, and number of observations, respectively. Numbers in parentheses are negative. Myco, total counts of environmental mycobacteria. TC is total coliform counts from EPCC and FC is fecal coliform counts from EPCC, both in CFU/100 ml. CTX, chemical toxicity. HP, H. pylori antigen ELISA; Fl, water flow rate; O2, dissolved oxygen; Con, water conductivity; DS, dissolved solids; Cl, chloride; Sl, sulfates.
Environmental mycobacteria counts correlate negatively with chemical toxicity, while total coliforms and fecal coliforms share this correlation, and thus the toxicants have a general antimicrobial activity (Table 2).
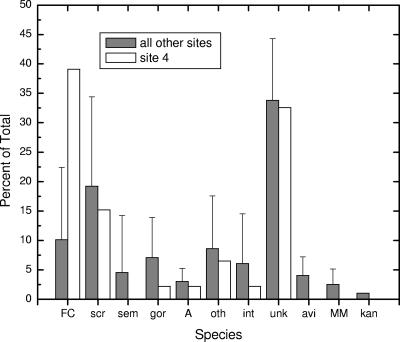
Comparison of the species of mycobacteria isolated from site 4 with the other sites (excluding 5 and 6 because of low numbers) shows the effect of water treatment and/or the municipal water distribution system (Fig. 2). For most species, the relative abundance was unchanged. However, with the M. fortuitum complex, abundance rose from 10.1 ± 12.3% to 39.1%. As M. fortuitum is the most common of the rapid growers clinically isolated from humans (1, 15), this result deserves further study. However, the Haskell secondary treatment plant utilizes activated sludge treatment, which is not commonly used in primary treatment plants responsible for potable water. Additionally, our findings contradict a previous report that secondary water treatment is adequate to prevent mycobacterial transmission (41).
FIG. 2.
Effect of treatment on mycobacteria. Data for site 4 are species plate counts from effluent from the Haskins Waste Water Treatment Plant. Data for all other sites are the mean ± standard deviation from the untreated river sites, excluding low-abundance sites 5 and 6. Abbreviations are the same as in Fig. 1.
There was a significant negative correlation between water temperature and environmental mycobacteria counts (Table 2). This is again in contrast with a previous study (29) in a different environment. In the Rio Grande, the water is cooler (Table 3; total mean temperature at all sites was 16.8 ± 6.9°C, n = 55) than in the swamp study, in which 36% of the samples were taken from water above 20°C and 20% were taken from water above 25°C.
Previous studies in swamps and brook sediments show that more acidic conditions favor higher environmental mycobacteria counts. In our study no significant correlation between pH and environmental mycobacteria counts was observed (Table 2). However, the Rio Grande is typically alkaline (total mean pH of 8.30 ± 0.35, n = 56) and the pH variation is narrow (Table 3), and thus this system is quite different from the ecosystems examined formerly. The alkaline sediments surrounding the El Paso region cause the basic water tone.
There was no significant correlation between water flow rate, dissolved oxygen, conductivity, chloride, or sulfate and total environmental mycobacteria counts. This is expected, as environmental mycobacteria can grow at a wide range of oxygen concentrations and are mostly halotolerant.
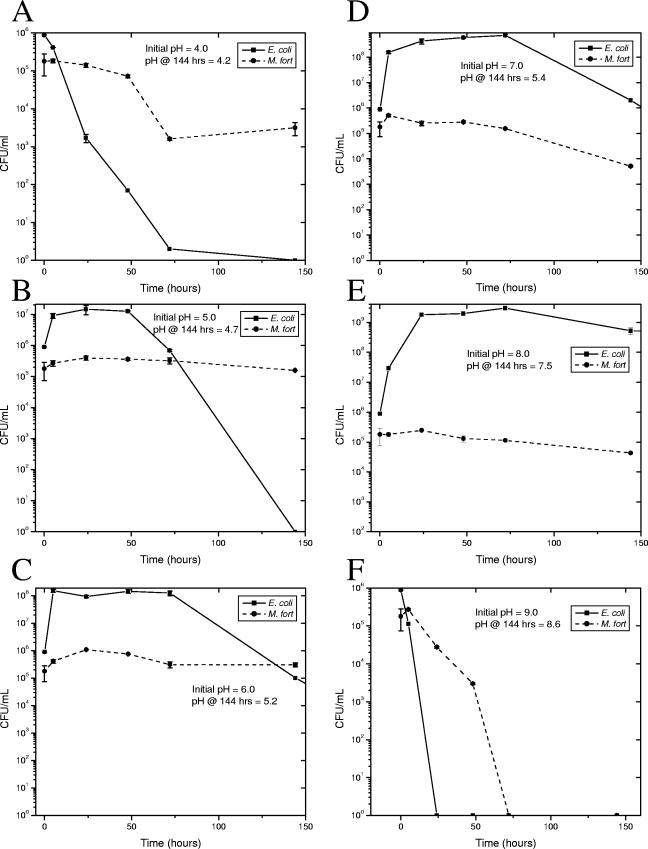
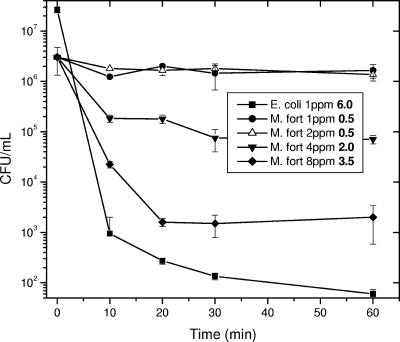
Since pH has been significant in other environmental studies and chlorine tolerance is important for persistence in municipal water, we have further examined these factors. Viability over time in combination cultures of the hardy E. coli O157:H7 strain Sakai (24) with a river isolate (M. fortuitum from site 4 in October, confirmed by 16S) was determined in 7H9-ADC medium with the pH set from 4 to 9 (Fig. 3). As predicted by environmental studies, the environmental mycobacteria survived much better than the E. coli strain at extreme pHs, both acidic and alkaline. Survival in combination was also examined in deionized water with increasing concentrations of chlorine (Fig. 4). E. coli lost 6 logs of viability after 1 h in 1 ppm of chlorine, while M. fortuitum lost only 45%. M. fortuitum lost only 3.5 log viability at 8 ppm, while no E. coli were recovered at chlorine concentrations above 1 ppm, and thus environmental mycobacteria are far more chlorine tolerant.
FIG. 3.
pH coincubation cultures. Exponentially growing M. fortuitum strain 4G and E. coli O157:H7 strain Sakai were mixed into 7H9-ADC with the pH adjusted to 4 (=A), 5 (B), 6 (C), 7 (D), 8 (E), or 9 (F). Culture pH after 144 h is also indicated. Cultures were incubated at 150 rpm and 37°C, and dilutions were plated in triplicate for CFU determination at the indicated time points. E. coli was determined by LB plates, and M. fortuitum was determined on selective 7H11. Square symbols, mean E. coli viability; circles, mycobacterial viability; error bars, standard deviation.
FIG. 4.
Chlorine combination survival. Exponentially growing M.fortuitum strain 4G and E. coli O157:H7 strain Sakai were mixed into deionized water with 1, 2, 4, or 8 ppm of free chlorine added. Viability was determined as in Fig. 3. E. coli is only indicated at 1 ppm because no colonies were obtained at higher chlorine concentrations.
DISCUSSION
Previous studies which correlate environmental parameters to isolation of environmental mycobacteria were performed in acidic, organic material-rich, stagnant water reservoirs. This study adds to the variety of environments studied, examining environmental mycobacteria in a free-flowing, alkaline, organic material-poor river. The previous concept of acidic ecosystems favoring environmental mycobacteria can be expanded to include alkaline as well, and thus any extreme promotes the hardy environmental mycobacteria.
The strong correlation between coliform and environmental mycobacteria levels in the river are presumably explained by conditions generally favoring bacterial replication or persistence. However, there was no significant correlation between H. pylori and coliforms or H. pylori and environmental mycobacteria. A potential explanation is that levels of H. pylori were determined by an antigen test, while coliform and environmental mycobacteria quantitations were culture based. Another possibility is that the coliforms and environmental mycobacteria share the same source, likely animal fecal material. Agricultural runoff is the major reason for high coliform counts in the Rio Grande (470 ± 800 CFU/ml fecal coliforms at all sites, n = 60). The coliform and environmental mycobacteria correlation observed in our study supports the finding that environmental mycobacteria correlate with total organism levels in brook sediments (26).
The Botsford cytotoxicity test used in this study (4) does not identify the source of toxicity. Often, biocides can selectively kill off less tolerant microorganisms, leaving hardier species, such as environmental mycobacteria. Mycobacteria are 100 to 330 times more chlorine tolerant than E. coli (32) (Fig. 4), and chlorine likely selects for environmental mycobacteria in treated water systems (35, 39), again attested to by the high counts and lack of coliforms from site 4 (chlorinated water effluent). The intrinsic tolerance of environmental mycobacteria compared to other bacilli results in a positive selective pressure in harsh environments.
Looking at the seasonal variation of environmental mycobacteria in the river, counts are higher in cooler months (Fig. 1A). However, this is complicated because water levels are also quite different. The seasonal variations cannot be substantiated without a multiyear study.
We have a systematic comparison of three identification methods for environmentally isolated mycobacteria. Biochemical tests are time-consuming and reagent intensive and involve significant variability even when performed correctly. Clearly, one must run a panel of more than five tests to correctly identify environmental isolates, however, more tests add more time and expense. PRA was just as effective as 16S sequencing but cheaper and more rapid. The percentage of isolates with unknown PRA patterns for each of the seven sites ranged from 22.6 to 55.6%, with a mean of 35 ± 10%. Thus, both PRA and sequencing could not identify one-third of the isolates. This suggests that the taxonomy of the mycobacteria is still quite incomplete, which is supported by the naming of >30 new species in the past decade. There is also a need to increase the number of entries in the PRA database.
Similar to previous large clinical studies, one-third of the environmental mycobacteria isolates in this study were not identifiable, even by 16S sequencing. We closely examined the PRA patterns of 53 unidentified isolates selected at random. Thirty percent of the isolates had a unique pattern (found only once within the group of 53). With an additional 42%, the pattern appeared in only two isolates. The most common pattern was displayed by only six isolates, and thus the unknown group appears very heterogeneous by PRA. This suggests that not a few but many new species of environmental mycobacteria are yet undiscovered. Well-demonstrated as a rapid, easy, and inexpensive method of identifying clinical isolates of environmental mycobacteria, PRA likewise was proven in this study to have accuracy equivalent to that of 16S sequencing. With expansion of the PRA database, this method shows great utility in the environmental field.
An observation confirming the quality of the data is that conductivity correlates well with the measurement of ions chloride (coefficient = 0.962, P < 0.0001) and sulfates (coefficient= 0.919, P = <0.0001). Water temperature and dissolved oxygen have an inverse relationship (coefficient = −0.525, P = 0.0002). Water temperature exhibits significant negative correlation with conductivity, dissolved solids, chloride, and sulfates, while dissolved oxygen exhibits significant positive correlation with all of the same (data not shown). A potential explanation is that higher water temperatures allow more rapid microbial respiration, which in turn leads to lower dissolved oxygen. Additionally, gases dissolve better in liquids at lower temperatures.
The pathogenesis and treatment of mycobacterial infections are highly dependent on the species involved (1). Additionally, the epidemiology of infection by environmental mycobacteria is poorly understood due to a lack of data regarding the primary reservoirs of different mycobacterial species. The correlations between environmental parameters and prevalence of mycobacteria derived from this study may aid in future prediction and prevention of human exposure to environmental mycobacteria.
Acknowledgments
We thank Julia Bader for statistical analysis and interpretation (Statistical Consulting Lab, UTEP).
T.P.P. is funded by NIH K22 award AI01812-02 and grants from the Paso del Norte Health Foundation and the Lizanell and Colbert Coldwell Foundation. The Border Biomedical Research Center is funded by NIH RCMI award G12RR08124. C.S.B. and E.L. were supported by RISE grants R25GM069621-02 and 5R25GM060424-04, respectively.
REFERENCES
- 1. American Thoracic Society. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Official statement of the American Thoracic Society approved by the Board of Directors, March 1997. Medical Section of the American Lung Association. Am. J. Respir. Crit. Care Med. 156:S1-25. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, M. A., M. Nikulin, U. Koljalg, M. C. Andersson, F. Rainey, K. Reijula, E. L. Hintikka, and M. Salkinoja-Salonen. 1997. Bacteria, molds, and toxins in water-damaged building materials. Appl. Environ Microbiol. 63:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, T., A. Holtzman, N. Glover, M. Boian, S. Froman, O. G. Berlin, H. Hill, and G. Stelma, Jr. 1999. Comparison of large restriction fragments of Mycobacterium avium isolates recovered from AIDS and non-AIDS patients with those of isolates from potable water. J. Clin. Microbiol. 37:1008-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botsford, J. L. 2002. A comparison of ecotoxicological tests. Altern. Lab. Anim. 30:539-550. [DOI] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, L. A., L. A. Bland, L. B. Cusick, M. S. Favero, G. A. Bolan, A. L. Reingold, and R. C. Good. 1988. Prevalence of nontuberculous mycobacteria in water supplies of hemodialysis centers. Appl. Environ. Microbiol. 54:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cernoch, P. L., R. K. Enns, M. A. Saubolle, and R. J. Wallace. 1994. Laboratory diagnosis of the mycobacterioses. American Society for Microbiology, Washington, D.C.
- 8.Chi, J., J. A. VanLeeuwen, A. Weersink, and G. P. Keefe. 2002. Direct production losses and treatment costs from bovine viral diarrhoea virus, bovine leukosis virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum. Prev. Vet. Med. 55:137-153. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva, C. F., S. Y. Ueki, C. Geiger Dde, and S. C. Leao. 2001. hsp65 PCR-restriction enzyme analysis (PRA) for identification of mycobacteria in the clinical laboratory. Rev. Inst. Med. Trop. Sao Paulo 43:25-28. [DOI] [PubMed] [Google Scholar]
- 11.Domenech, P., M. S. Jimenez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1997. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. [DOI] [PubMed] [Google Scholar]
- 12.dos Santos, N. M., A. do Vale, M. J. Sousa, and M. T. Silva. 2002. Mycobacterial infection in farmed turbot Scophthalmus maximus. Dis. Aquat. Org. 52:87-91. [DOI] [PubMed] [Google Scholar]
- 13.du Moulin, G. C., K. D. Stottmeier, P. A. Pelletier, A. Y. Tsang, and J. Hedley-Whyte. 1988. Concentration of Mycobacterium avium by hospital hot water systems. JAMA 260:1599-1601. [DOI] [PubMed] [Google Scholar]
- 14.Environmental Protection Agency. 1993. Methods for chemical analyses of water and wastes. Document no. EPA-600/4-79-020. Environmental Protection Agency, Washington, D.C.
- 15.Falkinham, J. O., 3rd. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkinham, J. O., 3rd. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23:529-551. [DOI] [PubMed] [Google Scholar]
- 17.Falkinham, J. O., 3rd, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, O. A., L. Matlova, L. Dvorska, P. Svastova, and I. Pavlik. 2003. Nymphs of the Oriental cockroach (Blatta orientalis) as passive vectors of causal agents of avian tuberculosis and paratuberculosis. Med. Vet. Entomol. 17:145-150. [DOI] [PubMed] [Google Scholar]
- 19.Fox, C., B. Smith, O. Brogan, A. Rayner, G. Harris, and B. Watt. 1992. Non-tuberculous mycobacteria in a hospital's piped water supply. J. Hosp. Infect. 21:152-154. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, J., N. Nanki, K. Negayama, S. Tsutsui, T. Taminato, and T. Ishida. 2002. Nosocomial contamination by Mycobacterium gordonae in hospital water supply and super-oxidized water. J Hosp. Infect. 51:65-68. [DOI] [PubMed] [Google Scholar]
- 21.Gebo, K. A., A. Srinivasan, T. M. Perl, T. Ross, A. Groth, and W. G. Merz. 2002. Pseudo-outbreak of Mycobacterium fortuitum on a human immunodeficiency virus ward: transient respiratory tract colonization from a contaminated ice machine. Clin. Infect. Dis. 35:32-38. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths, P. A., J. R. Babb, and A. P. Fraise. 1999. Mycobactericidal activity of selected disinfectants using a quantitative suspension test. J. Hosp. Infect. 41:111-121. [DOI] [PubMed] [Google Scholar]
- 23.Gruft, H., A. Loder, M. Osterhout, B. D. Parker, and J. O. Falkinham 3rd. 1979. Postulated sources of Mycobacterium intracellulare and Mycobacterium scrofulaceum infection: isolation of mycobacteria from estuaries and ocean waters. Am. Rev. Respir. Dis. 120:1385-1388. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Ichiyama, S., K. Shimokata, and M. Tsukamura. 1988. The isolation of Mycobacterium avium complex from soil, water, and dusts. Microbiol. Immunol. 32:733-739. [DOI] [PubMed] [Google Scholar]
- 26.Iivanainen, E., P. J. Martikainen, P. Vaananen, and M. L. Katila. 1999. Environmental factors affecting the occurrence of mycobacteria in brook sediments. J. Appl. Microbiol. 86:673-681. [DOI] [PubMed] [Google Scholar]
- 27.Iivanainen, E., J. Northrup, R. D. Arbeit, M. Ristola, M. L. Katila, and C. F. von Reyn. 1999. Isolation of mycobacteria from indoor swimming pools in Finland. APMIS 107:193-200. [DOI] [PubMed] [Google Scholar]
- 28.Kazwala, R. R., C. J. Daborn, L. J. Kusiluka, S. F. Jiwa, J. M. Sharp, and D. M. Kambarage. 1998. Isolation of Mycobacterium species from raw milk of pastoral cattle of the Southern Highlands of Tanzania. Trop. Anim. Health Prod. 30:233-239. [DOI] [PubMed] [Google Scholar]
- 29.Kirschner, R. A., Jr., B. C. Parker, and J. O. Falkinham 3rd. 1992. Epidemiology of infection by nontuberculous mycobacteria. Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum in acid, brown-water swamps of the southeastern United States and their association with environmental variables. Am. Rev. Respir. Dis. 145:271-275. [DOI] [PubMed] [Google Scholar]
- 30.Kressel, A. B., and F. Kidd. 2001. Pseudo-outbreak of Mycobacterium chelonae and Methylobacterium mesophilicum caused by contamination of an automated endoscopy washer. Infect. Control Hosp. Epidemiol. 22:414-418. [DOI] [PubMed] [Google Scholar]
- 31.Kusnetsov, J., E. Iivanainen, N. Elomaa, O. Zacheus, and P. J. Martikainen. 2001. Copper and silver ions more effective against legionellae than against mycobacteria in a hospital warm water system. Water Res. 35:4217-4225. [DOI] [PubMed] [Google Scholar]
- 32.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 68:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, J. S., M. Christensen, R. W. Wilson, R. J. Wallace, Jr., Y. Zhang, D. R. Nash, and B. Shelton. 2000. Mycobacterial contamination of metalworking fluids: involvement of a possible new taxon of rapidly growing mycobacteria. AIHAJ 61:205-213. [DOI] [PubMed] [Google Scholar]
- 34.Neumann, M., R. Schulze-Robbecke, C. Hagenau, and K. Behringer. 1997. Comparison of methods for isolation of mycobacteria from water. Appl. Environ. Microbiol. 63:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pai, H. H., W. C. Chen, and C. F. Peng. 2003. Isolation of non-tuberculous mycobacteria from hospital cockroaches (Periplaneta americana). J. Hosp. Infect. 53:224-228. [DOI] [PubMed] [Google Scholar]
- 37.Pauls, R. J., C. Y. Turenne, J. N. Wolfe, and A. Kabani. 2003. A high proportion of novel mycobacteria species identified by 16S rDNA analysis among slowly growing AccuProbe-negative strains in a clinical setting. Am. J. Clin. Pathol. 120:560-566. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, M. S., and C. F. von Reyn. 2001. Nosocomial infections due to nontuberculous mycobacteria. Clin. Infect. Dis. 33:1363-1374. [DOI] [PubMed] [Google Scholar]
- 39.Primm, T. P., C. A. Lucero, and J. O. Falkinham 3rd. 2004. Health impacts of the environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ristola, M. A., C. F. von Reyn, R. D. Arbeit, H. Soini, J. Lumio, A. Ranki, S. Buhler, R. Waddell, A. N. Tosteson, J. O. Falkinham 3rd, and C. H. Sox. 1999. High rates of disseminated infection due to non-tuberculous mycobacteria among AIDS patients in Finland. J Infect. 39:61-67. [DOI] [PubMed] [Google Scholar]
- 41.Shuval, H. I. 1991. Effects of wastewater irrigation of pastures on the health of farm animals and humans. Rev. Sci. Technol. 10:847-866. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, E. D., G. E. Gillaspy, and J. O. Falkinham 3rd. 2001. Fluorescent acid-fast microscopy for measuring phagocytosis of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum by Tetrahymena pyriformis and their intracellular growth. Appl. Environ. Microbiol. 67:4432-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, R. H., J. O. Falkinham 3rd, C. D. Norton, and M. W. LeChevallier. 2000. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl. Environ. Microbiol. 66:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Texas Natural Resource Conservation Commission. 1999. Surface water quality monitoring procedures manual. Texas Natural Resource Conservation Commission, Austin, Tex.
- 45.Vestal, A. L. 1981. Procedures for the isolation and identification of mycobacteria. Centers for Disease Control and Prevention, Atlanta, Ga.
- 46.Zumla, A., and J. Grange. 2002. Infection and disease caused by environmental mycobacteria. Curr. Opin. Pulmon. Med. 8:166-172. [DOI] [PubMed] [Google Scholar]
- 47.Zumla, A. I., and J. Grange. 2002. Non-tuberculous mycobacterial pulmonary infections. Clin. Chest Med. 23:369-376. [DOI] [PubMed] [Google Scholar]