Abstract
The presence of Anaplasma phagocytophilum, a tick-transmitted zoonotic pathogen, was investigated in Sardinia using a molecular approach. Phylogenetic analysis revealed that Sardinian strains are genetically distinct from the two lineages previously described in Europe and are closely related to strains isolated in different areas of the United States.
Anaplasma phagocytophilum (formerly Ehrlichia phagocytophila), a tick-transmitted bacterium able to infect bone marrow-derived cell lines of several animal species, including humans, is the causative agent of a tick-borne fever of ruminants, as well as equine granulocytic anaplasmosis, canine granulocytic anaplasmosis, and human granulocytic anaplasmosis (HGA) (5).
To date, only two HGA cases have been reported in continental Italy (14). However, serological and molecular findings demonstrated the occurrence of A. phagocytophilum infections in dogs and Ixodes ricinus ticks in this country (9, 15, 17). An average rate of 13.4 tick bite-related disease cases/year/100,000 inhabitants was reported for the island of Sardinia between 1992 and 1996, compared to the national average value of 2.1 cases/year/100,000 inhabitants. Moreover, 117 cases of tick bite-related disease whose etiology remains obscure were registered between 1995 and 2002.
Indirect immunofluorescence is commonly used for A. phagocytophilum diagnosis, but it should take into account coinfection, cross-reactivity among different pathogens (5, 7, 13, 16), and seroconversion (2). The diagnostic strategy has therefore focused on molecular methods based on PCR approaches (10). PCR methods should be highly specific, and a recently developed PCR assay for amplification of the A. phagocytophilum 16S rRNA gene resulted in coamplification of the Anaplasma platys gene (6).
In order to establish the presence of A. phagocytophilum in an area of Sardinia characterized by a high incidence of tick bite-related diseases in dogs and humans and to compare Sardinian strains with American and European isolates, symptomatic dogs and horses were tested by a molecular method able to distinguish A. phagocytophilum from A. platys. To accomplish this, a strategy based on groEL PCR-restriction fragment length polymorphism and sequence analysis, an alternative to the 16S rRNA-based PCR method, was used.
Between 2002 and 2004, veterinarians based in central Sardinia were instructed to collect blood samples when a suspected case of tick bite-related disease was found in their clinics. A total of 40 blood samples were collected from 40 dogs showing tick infestation and symptoms consistent with tick bite-related disease, such as fever, anorexia, depression, anemia, myalgia, and a reluctance to move. Clinical hematology indicated thrombocytopenia and anemia. Moreover, 20 blood samples were obtained from 20 horses showing tick infestation, hyperthermia, anemia, anorexia, jaundice, myalgia, and a reluctance to move.
Genomic DNA was extracted from the buffy coat obtained by centrifugation of 2 to 4 ml of blood as previously described (12). A. phagocytophilum genomic DNA (NCH-1 strain) was extracted from FA substrate slides (VMRD, Inc.) and used as a positive control in canine granulocytic anaplasmosis-specific PCRs. A DNA preparation of a southern Italy A. platys isolate was used as a positive control in infectious cyclic thrombocytopenia-specific PCRs. Based on an alignment of the heat shock protein groEL gene sequences available in the GenBank database for species belonging to the family Anaplasmataceae, four primers were generated and used in combination in two heminested PCRs. Primers EphplgroEL(569)F (ATGGTATGCAGTTTGATCGC) and EphplgroEL(1193)R (TCTACTCTGTCTTTGCGTTC) anneal to nucleotide strings conserved in A. phagocytophilum and A. platys and were designed to selectively amplify, in a first PCR round, 624 bp of the groEL gene of both species from clinical samples. EphgroEL(1142)R (TTGAGTACAGCAACACCACCGGAA) and EplgroEL(1084)R (CATAGTCTGAAGTGGAGGAC) are specific for A. phagocytophilum and A. platys, respectively, and were alternatively coupled with primer EphplgroEL(569)F in two heminested PCRs in which the amplification products obtained from the first PCR round were used as DNA templates. The final 50-μl PCR mixture from the first PCR round contained 5 μl of the DNA extract, 1 μM primer EphplgroEL(569)F, 1 μM primer EphplgroEL(1142)R, and HotMaster Taq DNA polymerase (5 U/μl; Eppendorf) according to the manufacturer's basic protocols for cycle profiling and mixture composition. Five microliters of the PCR products obtained from the first PCR round was used as the template DNA in each of the two heminested PCRs. The heminested PCR profiles were the same as the profiles in the first PCR round, except for the reverse primer used [EphgroEL(1142)R for the A. phagocytophilum-specific heminested PCR and EplgroEL(1084)R for the A. platys-specific heminested PCR]. Amplicons were digested with restriction endonuclease HindIII. The HindIII restriction pattern of the A. phagocytophilum groEL gene amplified region (three fragments, 525 bp, 21 bp, and 27 bp) is different from the restriction pattern produced after digestion of amplicons of the A. platys groEL gene (two fragments, 199 bp and 316 bp). An ABI PRISM Big Dye terminator cycle sequencing Ready Reaction kit (Applied Biosystems) was used for direct cycle sequencing of the PCR products obtained according to the protocol supplied by the manufacturer. To ensure that polymorphisms did not represent PCR errors, all the amplicons obtained were cloned into the pCR2.1-TOPO vector (Invitrogen), and three clones per sample were sequenced using universal M13 primers. The sequences generated were edited with CHROMAS (Technelysium Pty. Ltd., Australia). Based on alignment of all the A. phagocytophilum groEL sequences available in the GenBank database with the sequences of A. phagocytophilum generated during this study (573 bp), 39 type sequences were identified (Table 1). The 39 groEL type sequences were used as operational taxonomic units (OTUs) for phylogenetic analyses, which were performed with MEGA, version 3.0 (8). Genetic distances among the OTUs were expressed as percent total nucleotide differences or by the Kimura two-parameter method and were used to construct neighbor-joining (NJ) trees. Statistical support for internal branches of the trees was evaluated by bootstrapping. Maximum-parsimony (MP) trees and consensus values were also generated.
TABLE 1.
Identity of the 39 groEL sequence types used as OTUs in phylogenetic analyses
| groEL sequence type | No. | Location(s) and host species | GenBank accession no. | Nucleotides at variable positionsb
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 593 | 613 | 660 | 662 | 674 | 700 | 708 | 726 | 732 | 774 | 840 | 849 | 933 | 939 | 969 | 972 | 979 | 996 | 1002 | 1022 | 1098 | 1113 | ||||
| Strains with proven pathogenicitya | |||||||||||||||||||||||||
| 1-HGA-Europe | 5 | Slovenia (human), Sweden (horse), Germany (tick), Switzerland (horse), Germany (horse) | AF033101, AY529490, AY281849, U96735, AF482760 | A | A | T | T | A | C | A | T | A | G | C | G | C | T | A | C | A | G | T | A | T | A |
| 2-HGA-USA | 11 | California (2 humans), New York (human), Minnesota (human), California (5 horses), California (woodrat), Missouri (dog) | AF172159/63, U96728, U72628, AF172158/60-62/U96727, AF173988, AY219849 | • | • | C | • | • | • | • | • | • | • | • | A | • | • | G | A | • | T | • | • | • | G |
| 3-Sweden.1 | 1 | Sweden (horse) | AY529489 | • | • | • | • | • | • | • | • | • | • | T | • | T | • | • | • | • | • | • | • | • | • |
| 4-Norway.1 | 1 | Norway (sheep) | AF548385 | • | • | C | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | C | • | • | • |
| 5-Norway.2 | 2 | Norway (sheep), Scotland (sheep) | AF548386, U96730 | • | • | C | • | • | • | • | • | G | T | T | • | • | • | • | • | • | • | C | • | • | • |
| 6-Italy.1 | 1 | Italy (dog, Pippo) | AY848751 | • | • | C | C | • | • | • | C | • | • | • | A | • | • | G | A | • | T | • | • | C | G |
| 7-Italy.2 | 1 | Italy (horse, Quada) | AY848747 | • | G | C | • | • | • | • | • | • | • | • | A | • | • | G | A | • | T | • | • | • | G |
| 8-Italy.3 | 1 | Italy (horse, Dollaro) | AY848748 | • | • | C | • | T | T | G | • | • | • | • | A | • | • | G | A | G | T | • | • | • | G |
| 9-Italy.4 | 1 | Italy (dog, Stechi) | AY848750 | • | • | C | • | • | • | • | • | • | • | • | A | • | A | G | A | • | T | • | • | • | G |
| 10-Italy.5 | 1 | Italy (horse, Sogno) | AY848749 | T | • | C | • | • | • | • | • | • | • | • | A | • | • | G | A | • | T | • | • | • | G |
| 11-Italy.6 | 1 | Italy (dog, Perla) | AY848752 | • | • | C | • | • | • | • | • | • | • | • | A | • | • | G | A | • | T | • | G | • | G |
| Strains with unknown pathogenicitya | |||||||||||||||||||||||||
| 12-Slovenia.794 | 1 | Slovenia (roe deer) | AF478556 | ||||||||||||||||||||||
| 13-Slovenia.805 | 2 | Slovenia (roe deer), Germany | AF478555, AY281827 | ||||||||||||||||||||||
| 14-Slovenia.806 | 1 | Slovenia (roe deer) | AF478554 | ||||||||||||||||||||||
| 15-Switzerland.811 | 14 | Switzerland (roe deer), Slovenia (2 roe deer), Germany (11 ticks) | AF383226, AF478551/59, AF281810/12/14-17/19/21-22/46/50 | ||||||||||||||||||||||
| 16-Germany.X17 | 1 | Germany (tick) | AY281848 | ||||||||||||||||||||||
| 17-Scotland | 3 | Scotland (feral goat), Germany (2 ticks) | U96729, AY281844/31 | ||||||||||||||||||||||
| 18-Germany.L158 | 2 | Germany (2 ticks) | AY281832/20 | ||||||||||||||||||||||
| 19-Germany.145 | 2 | Germany (2 ticks) | AY281830/25 | ||||||||||||||||||||||
| 20-Germany.94 | 1 | Germany (tick) | AY281828 | ||||||||||||||||||||||
| 21-Germany.22 | 1 | Germany (tick) | AY281818 | ||||||||||||||||||||||
| 22-CzechRep.A41 | 2 | Czech Republic (roe deer), Germany (tick) | AY220468, AY281824 | ||||||||||||||||||||||
| 23-Germany.D20 | 1 | Germany (tick) | AY281815 | ||||||||||||||||||||||
| 24-Austria.A6 | 1 | Austria (tick) | AY220470 | ||||||||||||||||||||||
| 25-Switzerland.56 | 2 | Switzerland (roe deer), Germany (tick) | AF383225, AY28181 | ||||||||||||||||||||||
| 26-Austria.A4 | 1 | Austria (roe deer) | AY220469 | ||||||||||||||||||||||
| 27-Austria.A1 | 1 | Austria (roe deer) | AY220467 | ||||||||||||||||||||||
| 28-Switzerland.1 | 1 | Switzerland (bank vole) | AF192796 | ||||||||||||||||||||||
| 29-California | 1 | California (tick) | AF173989 | ||||||||||||||||||||||
| 30-Slovenia.478 | 2 | Slovenia (roe deer), Germany (tick) | AF478564, AY281836 | ||||||||||||||||||||||
| 31-Slovenia.474 | 1 | Slovenia (red deer) | AF478563 | ||||||||||||||||||||||
| 32-Slovenia.707 | 1 | Slovenia (red deer) | AF478557 | ||||||||||||||||||||||
| 33-Slovenia.473 | 1 | Slovenia (red deer) | AF478562 | ||||||||||||||||||||||
| 34-Slovenia.832 | 1 | Slovenia (red deer) | AF478553 | ||||||||||||||||||||||
| 35-Slovenia.812 | 1 | Slovenia (red deer) | AF478552 | ||||||||||||||||||||||
| 36-Slovenia.472 | 3 | Slovenia (roe deer), Germany (tick), Switzerland (roe deer) | AF478561, AY281813, AF383227 | ||||||||||||||||||||||
| 37-Slovenia.471 | 1 | Slovenia (roe deer) | AF478560 | ||||||||||||||||||||||
| 38-Slovenia.921 | 16 | Slovenia (roe deer), Germany (15 ticks) | AF478558, AY281823/26/29/33-35/37-43/45/47 | ||||||||||||||||||||||
| 39-Switzerland | 1 | Switzerland (tick) | AF202895 | ||||||||||||||||||||||
Pathogenicity based on the 530-bp region considered in this study.
Numbering based on the accession number AF033101 sequence. Dots indicate nucleotides identical to nucleotides in the HGA-Europe sequence. The underlined nucleotides are nucleotides that are shared by the USA-HGA sequence type and the sequence types obtained in this study (Italy.1 to Italy.6).
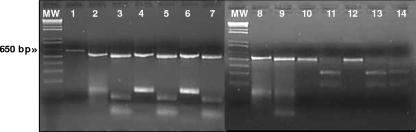
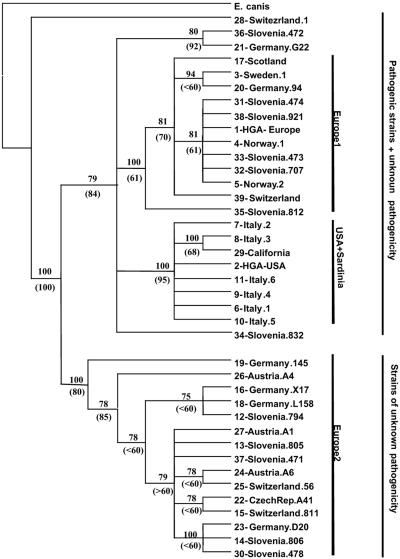
Of the 40 dog samples and 20 horse DNA samples, 3 dog samples and 3 horse samples tested positive for the presence of A. phagocytophilum, whereas only one dog sample (Lara) resulted in A. platys DNA amplification. HindIII digestion confirmed the PCR diagnosis (Fig. 1). Sample Lara generated the same HindIII restriction pattern as the A. platys DNA used as a positive control, and this pattern also coincided with the HindIII pattern predicted for A. platys. The six groEL A. phagocytophilum amplicons obtained in this study generated a pattern identical to that of A. phagocytophilum NCH-1 and to the HindIII pattern predicted for A. phagocytophilum. A total of seven new groEL sequence types were obtained from three horse samples and four dog samples. Alignment of the 530 bp investigated (not including the primer regions) of the groEL gene of the six A. phagocytophilum Sardinian genetic variants with all the groEL type sequences identified as described above (for a total of 39 sequence types) (Table 1) revealed a variability pattern characterized by nucleotide substitutions (60 variable sites, including 29 parsimony informative polymorphic sites and 31 singleton polymorphic sites). Six parsimony informative polymorphic sites (Table 1) are shared by Sardinian sequences, by a sequence obtained from a wood rat in California, by sequences of pathogenic A. phagocytophilum isolated from four human patients, five horses, and a dog in different states of the United States (type sequence 2-HGAUSA [Table 1]), and by a sequence obtained from a tick in California (type sequence 29-IxodesCali), but not by A. phagocytophilum isolated from symptomatic hosts in different regions of Europe (type sequences 1-HGAEurope and 39-HorseSweden). Coinciding and statistically supported NJ and MP trees (Fig. 2) allowed us to distinguish two main groups of groEL sequence types (MP consensus value, 100; NJ bootstrap value, 100). The first group (group Europe2) contained only strains with unknown pathogenicity isolated from ticks and roe deer in different European countries (Germany, Austria, Slovenia, Switzerland, and Czech Republic [Table 1]). The second group comprised pathogenic strains and strains with unknown pathogenicity isolated from various hosts in numerous European countries and the United States. Surprisingly, Sardinian groEL sequence types segregated with sequence type 2-HGAUSA isolated from human patients in California, Minnesota, and New York, from horses in California, and from a dog in Missouri and with sequence type 29-IxodesCali, a strain isolated from Ixodes pacificus in California. Indeed, 100% of the MP trees and 95% of the bootstrapped NJ trees generated during this study allowed us to identify a United States-Sardinia group that is genetically distinct from group Europe1, which contains groEL sequence types of European strains with unknown pathogenicity and pathogenic strains isolated from humans, horses, and sheep (Fig. 2 and Table 1). The average genetic distances calculated using MEGA highlight the low degree of genetic variation within groups (group Europe1, 0.005; group Europe2, 0.004; United States-Sardinia group, 0.006). Among groups, calculations of averages revealed greater genetic distances, especially between the two European lineages identified and between the United States-Sardinia group and group Europe2 strains (group Europe1 versus group Europe2, 0.031; United States-Sardinia group versus group Europe1, 0.013; United States-Sardinia group versus group Europe2, 0.029). None of the 39 sequence types identified was found in more than one group (Table 1 and Fig. 2), even when groups Europe1 and Europe2, which overlapped geographically and spatially, were considered.
FIG. 1.
Results of groEL PCRs and HindIII digestion. Lane MW contained molecular weight standards. Lane 1 contained amplicon obtained after the first PCR round starting with DNA extracted from the buffy coat of a symptomatic dog. Lanes 2, 4, 6, and 8 show groEL heminested PCR results obtained using internal primer EPhGroR, specific for A. phagocytophilum, with DNA extracted from the reference strain of A. phagocytophilum and from samples Stechi, Sogno, and Perla, respectively. To the right of each PCR result, the corresponding HindIII digestion result is shown (lanes 3, 5, 7, and 9). Lanes 10 and 12 show groEL heminested PCR results obtained using internal primer EPlGroR, specific for A. platys, with DNA extracted from the control strain of A. platys and from sample Lara, respectively. To the right of each PCR result, the corresponding HindIII digestion result is shown (lanes 11 and 13). Lane 14 shows HindIII digestion of the amplicon obtained after the first PCR (lane 1) and identification of the sample as A. platys.
FIG. 2.
Coinciding NJ and MP trees generated by considering 39 A. phagocytophilum sequence types OTUs for phylogenetic analyses. The numbers in parentheses are statistically significant bootstrap values for the branches obtained by testing the robustness of NJ trees. The numbers not in parentheses are statistically significant consensus values for branches obtained by testing MP trees.
We successfully used a molecular approach to investigate the presence of A. phagocytophilum in an area characterized by a high rate of tick bite-related disease in humans and animal species. Our approach also allowed us to avoid the potential pitfalls caused by the high degree of genetic similarity between A. platys and A. phagocytophilum. Hancock and coworkers (6) addressed this problem when they discussed their A. phagocytophilum-specific 16S rRNA gene-based PCR that resulted in coamplification of A. platys DNA. Indeed, one of the dog buffy coats isolated during this study resulted in A. platys DNA amplification.
Two main groEL sequence types have been reported in Europe by von Loewenich and coworkers (18), in agreement with the findings of Petrovec and coworkers (11), who identified two genetic lineages of groEL sequences, one more closely related to strains isolated from humans and infecting mainly red deer and one infecting roe deer.
The phylogenetic analyses conducted during this study (Table 1 and Fig. 2) confirmed the presence of two distinct European lineages of A. phagocytophilum. Unexpectedly, both Sardinian equine and canine A phagocytophilum isolates do not fall in either of the two European lineages but are phylogenetically closely related to the American strains. The latter observation has implications for risk assessment, since A. phagocytophilum American strains seem to be associated with higher mortality rates in humans and HGA cases in Europe are less severe (1, 4). Statistical evaluation of the branches and group analysis strongly support the morphology of the trees. These observations could suggest that there was recent introduction via mammalian hosts and tick vectors that moved from the United States to Sardinia.
Alternatively, we cannot exclude the possibility of introduction from other regions of the Mediterranean area or from North Africa (for which information on the groEL gene type sequences are not yet available) through ticks transported by migratory birds. Indeed, 8 of 57 ticks (14%) collected on passerine migratory birds in the Baltic region of Russia were found to host A. phagocytophilum (3), thus demonstrating that Anaplasma exchange could occur between ticks cofeeding on animals without systemic infections. In conclusion, molecular analyses are required in order to characterize A. phagocytophilum strains circulating in hosts (especially humans) and vectors in the Mediterranean area and to confirm our observations.
Nucleotide sequence accession numbers.
The nucleotide sequences of A. phagocytophilum have been deposited in the GenBank database under the accession numbers shown in Table 1, and the accession number for the A. platys nucleotide sequence is AY848753.
REFERENCES
- 1.Aguero-Rosenfeld, M. E., H. W. Horovitz, G. P. Wormser, D. F. Mc-Kenna, J. Nowakowski, J. Munoz, and J. S. Dumler. 1996. Human granulocytic ehrlichiosis: a case series from a medical center in New York State. Ann. Intern. Med. 125:904-908. [DOI] [PubMed] [Google Scholar]
- 2.Aguero-Rosenfeld, M. E., F. Kalantarpour, M. Baluch, H. W. Horowitz, D. F. MCKenna, J. T. Raffalli, T. C. Hsieth, J. Wu, J. S. Dumler, and J. P. Wormser. 2000. Serology of culture-confirmed cases of human granulocytic ehrlichiosis. J. Clin. Microbiol. 38:635-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekseev, A. N., H. V. Dubinina, A. V. Semenov, and C. V. Bolshakov. 2001. Evidence of ehrlichiosis agents found in ticks (Acari: Ixodidae) collected from migratory birds. J. Med. Entomol. 38:471-474. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, J. R., and J. A. Oteo. 2002. Human granulocytic ehrlichiosis in Europe. Clin. Microbiol. Infect. 8:763-772. [DOI] [PubMed] [Google Scholar]
- 5.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 6.Hancock, S. I., E. B. Breitschwerdt, and C. Pitulle. 2001. Differentiation of Ehrlichia platys and E. equi infections in dogs by using 16S ribosomal DNA-based PCR. J. Clin. Microbiol. 39:4577-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kordick, S. K., E. B. Breitschwerdt, B. C. Hegarty, K. L. Southwick, C. M. Colitz, S. I. Hancock, J. M. Bradley, R. Rumbough, J. T. Mcpherson, and J. N. MacCormack. 1999. Coinfection with multiple tick-borne pathogens in a Walker Hound kennel in North Carolina. J. Clin. Microbiol. 37:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformat. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 9.Manna, L., A. Alberti, L. M. Pavone, A. Scibelli, N. Staiano, and A. E. Gravino. 2004. First molecular characterisation of a granulocytic Ehrlichia strain isolated from a dog in south Italy. Vet. J. 167:224-227. [DOI] [PubMed] [Google Scholar]
- 10.Massung, R. F., and K. G. Slater. 2003. Comparison of PCR assays for detection of the agent of human granulocytic ehrlichiosis, Anaplasma phagocytophilum. J. Clin. Microbiol. 41:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrovec, M., A. Bidovec, J. W. Sumner, W. L. Nicholson, J. E. Childs, and T. Avsic-Zupanc. 2002. Infection with Anaplasma phagocytophila in cervids from Slovenia: evidence of two genotypic lineages. Wien Klin. Wochenschr. 114:641-647. [PubMed] [Google Scholar]
- 12.Pusterla, N., J. Huder, C. Wolfensberger, B., Litschi, A. Parvis, and H. Lutz. 1997. Granulocytic ehrlichiosis in two dogs in Switzerland. J. Clin. Microbiol. 35:2307-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rikihisa, Y. 1991. Cross-reacting antigens between Neorickettsia helminthoeca and Ehrlichia species shown by immunofluorescence and Western immunoblotting. J. Clin. Microbiol. 29:2024-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruscio, M., and M. Cinco. 2003. Human granulocytic ehrlichiosis in Italy: first report on two confirmed cases. Ann. N. Y. Acad. Sci. 990:350-352. [DOI] [PubMed] [Google Scholar]
- 15.Sanogo, Y. O., P. Parola, S. Shpynov, J. L. Camicas, P. Brouqui, G. Caruso, and D. Raoult. 2003. Genetic diversity of bacterial agents detected in ticks removed from asymptomatic patients in northeastern Italy. Ann. N. Y. Acad. Sci. 990:182-190. [DOI] [PubMed] [Google Scholar]
- 16.Suksawat, J., C. Pitulle, C. Arraga-Alvarado, K. Madrigal, S. I. Hancock, and E. B. Breitschwerdt. 2001. Coinfection with three Ehrlichia species in dogs from Thailand and Venezuela with emphasis on consideration of 16S ribosomal DNA secondary structure. J. Clin. Microbiol. 39:90-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarello, W. 2003. Canine granulocytic ehrlichiosis (CGE) in Italy. Acta Vet. Hung. 51:73-90. [DOI] [PubMed] [Google Scholar]
- 18.von Loewenich, F. D., Baumgarten, B. U., K. Schröppel, W. Geißdörfer, M. Rollinghoff, and C. Bodgan. 2003. High diversity of ankA sequences of Anaplasma phagocytophilum among Ixodes ricinus ticks in Germany. J. Clin. Microbiol. 41:5033-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]