Abstract
The DNA sequence of the flaA short variable region (SVR) was used to analyze a random population of Campylobacter isolates to investigate the weakly clonal population structure of members of the genus. The SVR sequence from 197 strains of C. jejuni and C. coli isolated from humans, bovine, swine, and chickens identified a group of 43 strains containing disparate short variable region sequences compared to the rest of the population. This group contains both C. jejuni and C. coli strains but disproportionately consisted of bovine isolates. Relative synonymous codon usage analysis of the sequences identified two groups: one group typified C. jejuni, and the second group was characteristic for C. coli and the disparate alleles were not clustered. The data show that there is significant differentiation of Campylobacter populations according to the source of the isolate even without considering the disparate isolates. Even though there is significant differentiation of chicken and bovine isolates, the bovine isolates did not show any difference in ability to colonize chickens. It is possible that disparate sequences were obtained through the lateral transfer of DNA from Campylobacter species other than C. jejuni and C. coli. It is evident that recombination within the flaA SVR occurs rapidly. However, the rate of migration between populations appears to limit the distribution of sequences and results in a weakly clonal population structure.
Campylobacter is a leading cause of enteritis in the United States, accounting for an estimated 2.4 million cases per year (19). Symptoms are usually acute and are characterized by abdominal cramps, nausea, diarrhea, and fatigue. The illness is typically self-limiting, and treatment is contraindicated in all but the most severe cases. Sequelae are rare, but there is evidence that Campylobacter infection is associated with the autoimmune disorders Miller-Fisher syndrome and Guillain-Barré syndrome (24). Most Campylobacter infections are considered to be food-borne and are associated with contaminated meat, especially poultry (2). However, sporadic cases are common and the source of infection often goes undetermined (1).
Prevention of Campylobacter infections in humans through consumption of contaminated foodstuffs is dependent upon a good epidemiological understanding of this organism. To this end, many methods for typing and subtyping Campylobacter species have been developed. Phenotypic typing schemes such as Penner serotyping have been useful but suffer notable drawbacks such as cross-reactivity of antigens and the presence of nontypeable strains (28). Several genotype-based methods have been introduced, including restriction fragment length polymorphism (25), randomly amplified polymorphic DNA-PCR (27), multilocus enzyme electrophoresis (26), pulsed-field gel electrophoresis (31), and DNA sequence analysis (21). Payne et al. (29) also used a combination of randomly amplified polymorphic DNA-PCR and detection of the 23S rRNA intervening sequence to differentiate Campylobacter strains taken from poultry. Genotyping is generally more discriminatory than phenotypic typing, but each method has its own limitations and may show differing relationships.
Sequencing of a single locus, the flaA short variable region (SVR), has also been shown to be a useful typing scheme (8, 21). Due to the small size of the SVR (321 bp), sequence determination is quick and reliable. Sequence comparison of the flaA SVR is nearly as discriminating as the complete flaA sequence and can be more discriminatory than serotyping or PCR-restriction fragment length polymorphism of the flaA gene (8, 21). SVR comparisons have been used to study the spread of Campylobacter populations within the poultry industry, including differentiating between poultry and environmental isolates (4, 13, 14).
All of the subtyping methods have shown that Campylobacter is a highly diverse genus. Methods in which linkage analyses can be performed have demonstrated that C. jejuni is weakly clonal (5, 33). To be weakly clonal is to have significant linkage of alleles at different loci in a population while not demonstrating tree-like phylogenetic relationships, as predicted by the clonal model of bacterial population structure. However, the genesis of the weakly clonal structure is not clear but could be because of one or more of the following (20): (i) the frequency of recombination in the population may be insufficient to randomize genetic variation that leads to a panmictic population structure, which may be a simple rate-related phenomenon or due to the isolation of subpopulations; (ii) some subsets of the population may not be participating in recombination, resulting in some lineages that are evolving clonally; (iii) some subpopulations may be more successful, leading to their recent rapid expansion, so-called epidemic clones (32a); and (iv) some portions of the genome may not participate in recombination, leaving linkage islands within the genome that evolve clonally.
In this study, we ascertained whether alleles of the flaA SVR sequence show any linkage to subpopulations of Campylobacter isolates that were stratified by source. Isolates of Campylobacter jejuni and Campylobacter coli from poultry, bovine, swine, and human sources in the United States were compared, along with a subset of the European CampyNet strain collection (12). A subset of isolates was tested for the ability to colonize chicks to determine whether the stratification may be due to host adaptation.
MATERIALS AND METHODS
Bacterial isolates.
Bacteria from two collections were used. The first was a subset of 69 isolates from the European CampyNet collection (12). Isolates that would not grow for us and isolates that were epidemiologically linked replicates were eliminated. A second group of 128 isolates was collected as part of the National Antimicrobial Resistance Monitoring System (34). All of the National Antimicrobial Resistance Monitoring System isolates were either geographically or temporally independent. The species were determined by using a commercially available PCR system (BAX; DuPont Qualicon, Wilmington, DE), and the isolates in the total test population (n = 197) were as follows: 141 C. jejuni, 52 C. coli, 1 C. lari, and 3 undetermined. The sources of the isolates were as follows: 66 from humans, 50 from chickens, 32 from cattle, 34 from swine, 6 from sheep, 4 from wild birds, 3 from canines, and 2 from water. For linkage analyses, the sheep, wild bird, canine, and water isolates were grouped together as “others.” A table with the complete list of isolates is available on request.
PCR and DNA sequencing.
The SVR of flaA was amplified as previously described (21). Forward and reverse sequence data were assembled using Sequencher version 3.1.2 (Gene Codes Corporation, Ann Arbor, MI) and aligned using ClustalX (16). No indels were introduced into the alignment, and no editing was necessary.
PAUP version 4.0b10 (Sinauer Associates, Inc., Sunderland, MA) was used to generate distance matrixes and perform cluster analyses by the neighbor-joining algorithm using HKY85 or absolute distances. Neither distance can be expected to give an accurate phylogenetic reconstruction because of the profound evidence for recombination among Campylobacter flagellin (11, 22, 35). Reticulograms showing the relationships of sequences were constructed with SplitsTree version 4b6 (15) using the Neighbor-Net algorithm (3). Correspondence analysis of the relative synonymous codon usage (RSCU) was performed using CodonW (J. Peden; http://www.molbiol.ox.ac.uk/cu/). An allele-based test of genotypic differentiation of populations was performed using GENEPOP (30). Chi-square analysis was performed with Statistica (Statsoft, Inc., Tulsa, OK).
Colonization of chickens.
Day-of-hatch broiler chicks were purchased from a commercial hatchery. The birds were sorted into groups of five, and each group was inoculated by oral gavage with one dose of a serial 10-fold dilution of between 103 and 106 bacterial cells from a culture that had been started the day before. The chicks were housed in isolation chambers; 7 days later, they were sacrificed and their ceca were pulled for direct plating on Campy-Cefex agar (23). Forty-eight hours later, the plates were examined for growth of Campylobacter, which was confirmed with phase-contrast microscopy and latex agglutination assays (Microscreen Campylobacter; Microgen Bioproducts, Ltd., Camberley, United Kingdom).
Nucleotide sequence accession numbers.
All sequences were submitted to GenBank and are available under accession numbers AY845438 through AY845634.
RESULTS
SVR sequences.
A total of 197 flaA SVR sequences were determined from Campylobacter spp. isolated from humans, bovine, swine, and chickens and other source populations. A total of 321 base pairs of DNA were sequenced for each isolate, of which 115 sites were polymorphic, with an average number of nucleotide differences between each pair of sequences being 25.86.
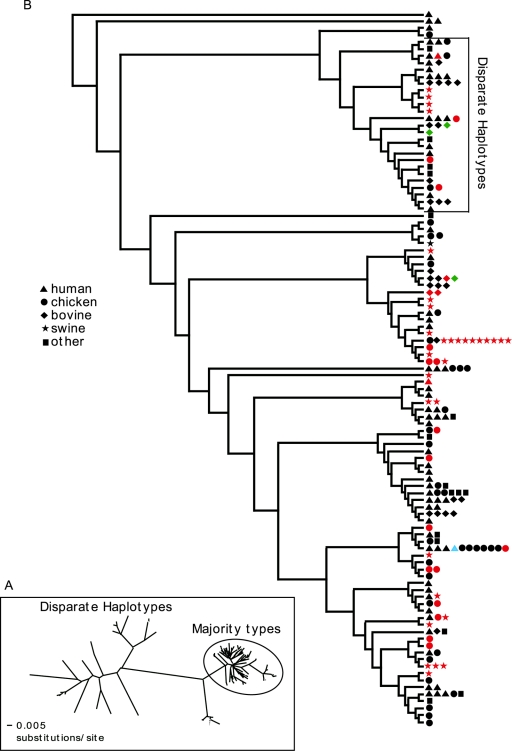
A cluster analysis using HKY85 distances of the flaA sequences (Fig. 1) revealed the presence of two distinct groups of sequences that we call the majority type (Fig. 1A, within the ellipse), and the remainder were called the disparate group based on their distance from most of the other sequences. The tree is not shown as an accurate representation of phylogeny because many of the branches have exceedingly low bootstrap values (not shown) but rather is shown to reveal clustering patterns that may relate to isolate source. However, the branch dividing the disparate from the majority-type groups had a bootstrap value of 99%. The majority-type group includes sequences from 154 isolates (78.2%), while the disparate group contains sequences from 43 isolates (21.8%). C. jejuni and C. coli are distributed in both groups in proportions (Fig. 1B) that are not significantly different (chi-square P = 0.73). The disparate group was significantly overrepresented by bovine isolates (chi-square P = 0.003), barely underrepresented by chicken isolates (chi-square P = 0.05), and underrepresented, but not significantly, by swine isolates (chi-square P = 0.12) (Table 1). The division of the isolates into majority and disparate types and the distribution of populations in those types are consistent with observations we have made with sequence analyses of all of the genes used in the multilocus sequence analysis of C. jejuni by using data in the Campylobacter jejuni and Campylobacter coli multilocus sequence typing home page (20).
FIG. 1.
Cluster analysis of SVR sequences. (A) Unrooted phylogram representation of neighbor joining of HKY85 distances. (B) From the same cluster analysis, a cladogram with the source of isolates indicated. The lengths of the horizontal lines in panel B are not proportional to genetic distance, and many of the branches had bootstrap values of less than 50%; thus, the tree does not represent an accurate phylogeny. Each haplotype is indicated once. The symbols indicate the sources of isolates within each haplotype, and the colors indicate C. jejuni (black), C. coli (red), C. lari (blue), or unknown species (green) within the indicated haplotype.
TABLE 1.
Percent of disparate type or majority type made up of isolates from each source
| Source | Disparate type | Majority type |
|---|---|---|
| Human | 37 | 34 |
| Bovine | 30a | 12a |
| Chicken | 14a | 29a |
| Swine | 9 | 19 |
| Other | 9 | 6 |
| Total percent | 99b | 100 |
Indicates that the disparate-type proportion is significantly different from the corresponding majority-type proportion.
Disparate types do not add up to 100 because of rounding errors.
Haplotype distribution.
There were a total of 103 haplotypes, unique flaA SVR sequences, in the test population (Fig. 1B). Thirty-nine haplotypes had multiple members; of these, 6 were found only in the European collection, 18 were found only in the U.S. collection, and 15 were found in both collections of isolates. The largest group containing strains with identical SVR sequences had 12 members. The haplotype diversity, which is the probability that a pair of samples randomly drawn from the test population will be different, for the entire population was 0.9869. With this haplotype diversity, there is a less than 2% chance that samples drawn randomly from the total population will have the same haplotype, yet it appears (Fig. 1B) that at least swine isolates are overrepresented in some haplotypes. The significance of this overrepresentation was tested with an exact test of genotypic differentiation of populations using GENEPOP (30). Not only was there evidence that the isolates from swine form a distinctly differentiated population, but isolates from bovine sources were also significantly differentiated (Table 2). In an effort to determine whether the differentiation of populations is a recent event, a series of analyses were performed in which haplotypes were progressively agglomerated into haplotypes that included isolates that were more and more distant. For instance, when a single haplotype was considered to be all the isolates that were within a 6-base-pair difference of any other individual within a group, isolates from bovine and swine sources continued to be significantly differentiated from the other populations (Table 2). If agglomeration of isolates were to continue to the point that there were only two haplotypes to consider, those two would be the majority-type and the disparate-type isolates, which were demonstrated to show significant differentiation of the bovine isolates by chi-square analysis (see above). To remove the influence of the strong differentiation of the disparate isolates, the analysis was repeated with isolates agglomerated by 6 base pairs and all the disparate alleles left out. The differentiation of bovine and swine isolates was still significant (Table 2).
TABLE 2.
Significance of population differentiation
| Populations compared | Probabilitya | Probability agglomerated by 6-bp differenceb | Probability agglomerated by 6-bp majority- type differencec |
|---|---|---|---|
| Bovine and chicken | 0.00000 | 0.00000 | 0.00011 |
| Bovine and human | 0.00000 | 0.00000 | 0.00017 |
| Bovine and swine | 0.00000 | 0.00000 | 0.00001 |
| Bovine and other | 0.00001 | 0.00169 | 0.01337 |
| Chicken and human | 0.13559 | 0.51376 | 0.44640 |
| Chicken and swine | 0.00001 | 0.00245 | 0.00205 |
| Chicken and other | 0.05510 | 0.24609 | 0.21289 |
| Human and swine | 0.00000 | 0.00000 | 0.00000 |
| Human and other | 0.26126 | 0.57926 | 0.48877 |
| Swine and other | 0.00000 | 0.00075 | 0.00496 |
P values calculated from haplotype frequencies in each population using GENEPOP with the null hypothesis that the two populations are indistinguishable. A P value of <0.05 indicates significant differentiation of the two populations.
Probabilities calculated from haplotype data with isolates agglomerated by differences of up to 6 base pairs (i.e., a single haplotype includes all individuals that are different from any other member of the haplotype by six or fewer base pairs).
Probabilities calculated from haplotype data with isolates agglomerated by differences of up to 6 base pairs with all the disparate haplotypes left out of the calculations.
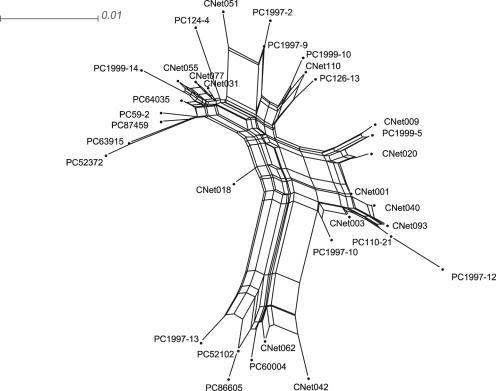
To determine the origin of the polymorphisms, mutation versus recombination, within an agglomerated group, analysis of recombination was done with sequences of members of agglomerated groups using the Neighbor-Net algorithm (3) in SplitsTree version 4b6 (15). It was seen that, even limiting groups to an agglomeration of members that were related to any other member by differences of six or fewer base pairs, SplitsTree produced a highly reticulated cluster that provides extensive evidence of recombination (Fig. 2).
FIG. 2.
Reticulate relationship of members of a superhaplotype in which all members are related to another by a difference of six or fewer base pairs. The relationship was calculated with the program SplitsTree using the Neighbor-Net algorithm.
Codon usage analysis.
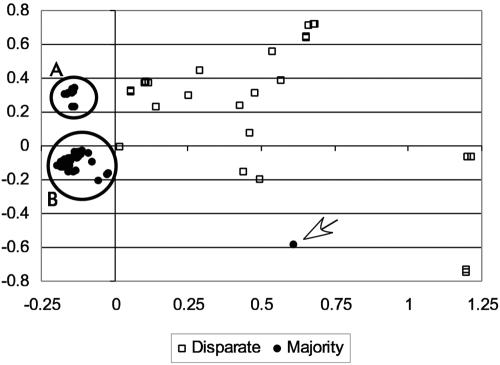
Correspondence analysis of the sequences for codon bias based on RSCU identified three subsets of the sequences (Fig. 3). A small cluster (Fig. 3, cluster A) consisted of 17 isolates of C. coli and 6 isolates of C. jejuni. A large cluster (Fig. 3, cluster B) consisted of 27 isolates of C. coli and 99 isolates of C. jejuni. The remaining isolates had correspondence vectors that were widely scattered without any apparent clustering. This last group consisted of all of the isolates identified in the disparate group (Fig. 1) plus one isolate that was a member of the majority-type group. Thus, the population of isolates that are isolated by this scattered group has the same biases as the disparate group (e.g., overrepresentation of bovine isolates, etc.). The changes in codon usage for the C. coli group are highlighted by the use of CGU in place of AGA for arginine, the use of GGU in place of GGC and GGG for glycine, and an increase in the use of GUU relative to GUG and GUA for valine (Table 3).
FIG. 3.
Correspondence analysis of RSCU comparisons of majority and disparate types. See the text for an explanation of the populations enclosed in circles. The arrow indicates the single isolate that was of the majority type outside of the two majority-type clusters.
TABLE 3.
RSCU within C. jejuni- and C. coli-type isolates
| Amino acid and codon | RSCU for clustersa of:
|
|
|---|---|---|
| C. jejuni | C. coli | |
| Glycine | ||
| GGU | 1.75 | 2.29 |
| GGC | 0.26 | 0 |
| GGA | 1.9 | 1.7 |
| GGG | 0.07 | 0 |
| Valine | ||
| GUU | 0.05 | 1.3 |
| GUC | 0 | 0 |
| GUA | 2.3 | 1.3 |
| GUG | 1.5 | 1.3 |
| Arginine | ||
| AGA | 2.9 | 1.5 |
| CGU | 1.44 | 2.73 |
| CGC | 1.55 | .26 |
| AGG | 0.03 | 1.4 |
Clusters were defined by RSCU analysis and are shown in Fig. 3 as clusters A (C. coli) and B (C. jejuni).
Colonization ability.
The differentiation of chicken and bovine isolates and the underrepresentation of chicken isolates among the disparate types may correspond to differences in fitness of the isolates in the different hosts. To test this, isolates from four groups (majority-type human isolates, majority-type bovine isolates, disparate-type human isolates, and disparate-type bovine isolates) were selected and tested to determine their colonization dose for chickens. It turned out that all the isolates except for two from the disparate-type human isolates readily colonized chickens (Table 4).
TABLE 4.
Determination of colonization doses of selected isolates in chickens
| Type and isolate | ID50a (log 10) |
|---|---|
| Disparate | |
| Bovine | |
| 140-16 | 3.8 |
| 106-19 | 3.8 |
| 129-25 | <3 |
| 115-17 | 4.5 |
| Human | |
| 1997-11 | 3 |
| 1997-7 | <3 |
| 1998-5 | >6 |
| 1998-13 | >6 |
| 1999-9 | <3 |
| Majority | |
| Bovine | |
| 110-21 | <3 |
| 126-18 | 3.9 |
| 69-30 | 4 |
| 117-2 | <3 |
| 107-24 | 4 |
| Human | |
| 1997-13 | 5 |
| 1997-6 | <3 |
| 1999-6 | <3 |
| 1999-12 | 5 |
| 1997-4 | 3.8 |
ID50, 50% infective dose.
DISCUSSION
Isolation of populations will lead to differentiation when mutations are events that are independent from one another. However, horizontal gene transfer through recombination between subpopulations will obscure the differentiation. But this is a rate-dependent phenomenon, and a population can appear to be truly panmictic only if the migration rate between subpopulations is greater than or equal to the recombination rate. This is because horizontal gene transfer is much more likely to occur between nearest neighbors and migration is needed to mix the neighbors. The results presented here demonstrate that, even in the face of a high recombination rate, there is isolation of subpopulations of Campylobacter that results in a weakly clonal population structure.
Our laboratory and others have used the SVR of the flaA gene to discriminate between Campylobacter species. While only 321 bp in length, this segment of the flaA gene has been shown to be hypervariable and is useful for discriminating even closely related Campylobacter strains (6, 13, 14, 21, 32). However, the Campylobacter flaA locus is known to be active in both intragenomic recombination between the flaA and flaB genes within the same strain (22) as well as intergenomic recombination of the flaA gene between two different C. jejuni strains (11, 35). So the evidence we have shown for recombination is an expected finding.
The haplotype diversity that we found with the fla SVR sequence was quite high, with a less than 2% chance that two isolates selected at random are likely to be the same. Thus, the finding of multiple isolates from the same subpopulation with the same haplotype on several occasions appeared to be a clear departure from randomness, and this was shown to be the case with the test for population differentiation in GENEPOP. The technique we have used to agglomerate isolates into superhaplotypes may not be enclosing groups that have only a single most recent common ancestor because of the effects of recombination. However, recombination is more likely to bias the analysis toward showing a loss of differentiation. So we have confidence that the continued differentiation of Campylobacter populations with agglomeration of the haplotypes is indicative that the association of haplotypes with populations has some longevity. However, at this point, we are unable to apply a clock to determine how long that association may have been.
An interesting group of sequences is comprised of those the isolates found within the disparate group. The flaA alleles of these 43 isolates are set apart by their genetic distance from the flaA alleles of the majority of the strains. The disparate alleles in this study are different enough to suggest that their origin is from a species other than C. jejuni or C. coli. Recent multilocus sequence typing studies of C. jejuni have shown that disparate alleles also occur within housekeeping genes that presumably have selective pressure for conservation as well as genes that may be under selective pressure for diversification, such as the flaA gene (20). Identification of possible donor species is made difficult because the availability of sequences from other Campylobacter species is limited. Sequences may also have been acquired from other genera. While there was no sequence difference between C. coli and C. jejuni strains that stood out based on the distance analysis of nucleotide sequence differences, the covariant analysis of RSCU demonstrated clusters that apparently typify sequences from C. jejuni or C. coli (Fig. 3). This leads to the expectation that the RSCU is species specific and the wide broadcasting of the disparate isolate sequences (Fig. 3) may indicate numerous donor species.
Codon usage by the consensus group and the C. coli group was similar overall in its bias toward codons ending with either A or U. This result was similar to that in a previous study that identified optimal and rare codons based on 67 genes of C. jejuni strain ATCC 11168 (9). The differences noted between the consensus group and the C. coli subset are limited to the optimal codon usage for glycine, valine, and arginine. The flagellin gene is highly expressed, and it is known that genes predicted to be highly expressed are likely to have optimized codon usage (17). It might be expected, then, that isolates with disparate flagellin alleles may have a selective disadvantage in their ability to express the gene, resulting in their eventual extinction.
There is an overrepresentation of isolates from cattle and an underrepresentation of isolates from chickens within the group containing the disparate allele sequences (Table 1). Chi-square analysis did not show a statistically significant difference of disparate- versus majority-type alleles within the swine or human isolates. There is no clear reason as to why bovine isolates would be more likely to contain disparate alleles. It is possible that acceptable donor strains are more prominent in cattle than in humans, chickens, or swine or that these sequences represent the environmental Campylobacter strains to which cattle would have more exposure due to farming practices. It is also possible that the bovine enteric tract provides an environment that enhances the rate of horizontal gene transfer.
Another clear distinction that has been noted before is the association of swine with C. coli (for an example, see reference 10). However, this isolation is not complete since there are swine isolates of C. coli that have fla SVR sequences that not only are similar to isolates of C. jejuni from other sources but are identical.
Our findings are compatible with the conclusion of Falush et al. (7) that recombination can occur as frequently as mutation in Helicobacter pylori, an organism with a rate of recombination similar to that seen with C. jejuni. That is because horizontal gene transfer requires opportunity, i.e., the colocation of donor DNA with the recipient organism. Cells that share the same environment are more likely to share DNA, and if the migration of alleles between types of hosts is limited, linkage to the source can be maintained. The strength of the linkage will be dependent on the rate of migration as well as the rate of recombination. With a low rate of migration, recombination is more likely to occur with very close relatives. Also, a breakpoint is required between a minimum of two polymorphic sites in order for the progeny of recombination to not be identical to at least one of the parents, and recombination between identical parents will yield identical progeny. The SplitsTree analysis for reticulate evolution that we have performed shows that there is ample evidence of recombination between closely related strains of Campylobacter (Fig. 2).
The determination of colonization doses in chickens of a subset of isolates appears to demonstrate that bovine isolates are not restricted in their ability to colonize chickens (Table 4), so we cannot conclude that the strains are selectively adapted to colonize chickens or cattle. If we accept that the differentiation of the Campylobacter subpopulation is due to isolation because of migration differences, then the source of infection of cattle is different from that of chickens. This is unexpected since so many of the haplotypes with multiple members (15 out of 29) were found on both the American and European continents. Thus, geographic isolation does not seem to be a key factor. It remains that there are local environmental factors that restrict the migration of Campylobacter between cattle and chickens. The mixing of different populations of Campylobacter is not occurring at a rate that is fast enough to allow recombination to totally break down the clonal structure, and they therefore appear to be weakly clonal.
REFERENCES
- 1.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evans. 1995. The Public Health Laboratory Service national case-control study of primary indigenous sporadic cases of Campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant, D., and V. Moulton. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 4.Cox, N. A., N. J. Stern, K. L. Hiett, and M. E. Berrang. 2002. Identification of a new source of Campylobacter contamination in poultry: transmission from breeder hens to broiler chickens. Avian Dis. 46:535-541. [DOI] [PubMed] [Google Scholar]
- 5.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duim, B., P. C. R. Godschalk, N. van den Braak, K. E. Dingle, J. R. Dijkstra, E. Leyde, J. van der Plas, F. M. Colles, H. P. Endtz, J. A. Wagenaar, M. C. J. Maiden, and A. van Belkum. 2003. Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curaçao, Netherlands Antilles. J. Clin. Microbiol. 41:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, C., L. T. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, S. A., and M. E. Konkel. 1999. Codon usage in the A/T-rich bacterium Campylobacter jejuni, p. 231-235. In P. S. Paul and D. H. Francis (ed.), Mechanisms in the pathogenesis of enteric diseases 2. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 10.Guevremont, E., R. Higgins, and S. Quessy. 2004. Characterization of Campylobacter isolates recovered from clinically healthy pigs and from sporadic cases of campylobacteriosis in humans. J. Food Prot. 67:228-234. [DOI] [PubMed] [Google Scholar]
- 11.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implicatons for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrington, C. S., L. Moran, A. M. Ridley, D. G. Newell, and R. H. Madden. 2003. Inter-laboratory evaluation of three flagellin PCR/RFLP methods for typing Campylobacter jejuni and C. coli: the CAMPYNET experience. J. Appl. Microbiol. 95:1321-1333. [DOI] [PubMed] [Google Scholar]
- 13.Hiett, K. L., N. A. Cox., R. J. Buhr, and N. J. Stern. 2002. Genotype analyses of Campylobacter isolated from distinct segments of the reproductive tracts of broiler breeder hens. Curr. Microbiol. 45:400-404. [DOI] [PubMed] [Google Scholar]
- 14.Hiett, K. L., N. J. Stern, P. Fedorka-Cray, N. A. Cox, M. T. Musgrove, and S. Ladely. 2002. Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68:6220-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 16.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 17.Karlin, S., J. Mrazek, A. Campbell, and D. Kaiser. 2001. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 183:5025-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinersmann, R. J., K. E. Dingle, and M. C. J. Maiden. 2003. Genetic exchange among Campylobacter species. Genome Lett. 2:48-52. [Google Scholar]
- 21.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinersmann, R. J., and K. L. Hiett. 2000. Concerted evolution of duplicate fla genes in Campylobacter. Microbiology 146:2283-2290. [DOI] [PubMed] [Google Scholar]
- 23.Musgrove, M. T., M. E. Berrang, J. A. Byrd, N. J. Stern, and N. A. Cox. 2001. Detection of Campylobacter spp. in ceca and crops with and without enrichment. Poultry Sci. 80:825-828. [DOI] [PubMed] [Google Scholar]
- 24.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nachamkin, I., J. Engberg, M. Gutacker, R. J. Meinersmann, C. Y. Li, P. Arzate, E. Teeple, V. Fussing, T. W. Ho, A. K. Asbury, J. W. Griffin, G. M. McKhann, and J. C. Piffaretti. 2001. Molecular population genetic analysis of Campylobacter jejuni HS:19 associated with Guillain-Barre syndrome and gastroenteritis. J. Infect. Dis. 184:221-226. [DOI] [PubMed] [Google Scholar]
- 27.Ono, K., T. Kurazono, H. Niwa, and K. Itoh. 2003. Comparison of three methods for epidemiological typing of Campylobacter jejuni and C. coli. Curr. Microbiol. 47:364-371. [DOI] [PubMed] [Google Scholar]
- 28.Patton, C. M., and I. K. Wachsmuth. 1992. Typing schemes: are current methods useful?, p. 110-128. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 29.Payne, R. E., M. D. Lee, D. W. Dreesen, and H. M. Barnhart. 1999. Molecular epidemiology of Campylobacter jejuni in broiler flocks using randomly amplified polymorphic DNA-PCR and 23S rRNA-PCR and role of litter in its transmission. Appl. Environ. Microbiol. 65:260-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond, M., and F. Rousset. 1995. An exact test for population differentiation. Evolution 49:1280-1283. [DOI] [PubMed] [Google Scholar]
- 31.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Smith, J. S., N. H. Smith, M. O’Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tollefson, L., P. J. Fedorka-Cray, and F. J. Angulo. 1999. Public health aspects of antibiotic resistance monitoring in the USA. Acta Vet. Scand. Suppl. 92:67-75. [PubMed] [Google Scholar]
- 35.Wassenaar, T. M., B. N. Fry, and B. A. M. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]