Abstract
Rhodococcus sp. strain DN22 can convert hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) to nitrite, but information on degradation products or the fate of carbon is not known. The present study describes aerobic biodegradation of RDX (175 μM) when used as an N source for strain DN22. RDX was converted to nitrite (NO2−) (30%), nitrous oxide (N2O) (3.2%), ammonia (10%), and formaldehyde (HCHO) (27%), which later converted to carbon dioxide. In experiments with ring-labeled [15N]-RDX, gas chromatographic/mass spectrophotometric (GC/MS) analysis revealed N2O with two molecular mass ions: one at 44 Da, corresponding to 14N14NO, and the second at 45 Da, corresponding to 15N14NO. The nonlabeled N2O could be formed only from -NO2, whereas the 15N-labeled one was presumed to originate from a nitramine group (15N-14NO2) in RDX. Liquid chromatographic (LC)-MS electrospray analyses indicated the formation of a dead end product with a deprotonated molecular mass ion [M-H] at 118 Da. High-resolution MS indicated a molecular formula of C2H5N3O3. When the experiment was repeated with ring-labeled [15N]-RDX, the [M-H] appeared at 120 Da, indicating that two of the three N atoms in the metabolite originated from the ring in RDX. When [U-14C]-RDX was used in the experiment, 64% of the original radioactivity in RDX incorporated into the metabolite with a molecular weight (MW) of 119 (high-pressure LC/radioactivity) and 30% in 14CO2 (mineralization) after 4 days of incubation, suggesting that one of the carbon atoms in RDX was converted to CO2 and the other two were incorporated in the ring cleavage product with an MW of 119. Based on the above stoichiometry, we propose a degradation pathway for RDX based on initial denitration followed by ring cleavage to formaldehyde and the dead end product with an MW of 119.
The two cyclic nitramine explosives hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) are powerful energetic compounds that are commonly used in conventional munitions and various military applications. Activities associated with manufacturing, training, waste disposal, and closures of bases have resulted in severe soil and groundwater contamination with the two explosives (9, 14, 21). RDX and HMX are both toxic (25, 29), thus necessitating their removal from the environment.
Previous studies have focused on the degradation of RDX by microorganisms under anaerobic conditions (1, 17, 18, 20, 24, 30). McCormick et al. (20) reported the formation of the three nitroso derivatives, hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine (MNX), hexahydro-1,3-dinitroso-5-nitro-1,3,5-triazine (DNX), and hexahydro-1,3,5-trinitroso-1,3,5-triazine (TNX), which after further reduction to the corresponding hydroxylamines undergo ring cleavage to produce hydrazine, dimethyl hydrazine, and methanol, respectively. No microorganisms or enzymes were identified in the study by McCormick et al. (20). Recently, Kitts et al. (19) reported the involvement of a type I nitroreductase in the degradation of RDX, but no products were identified. Using anaerobic sludge, Hawari et al. (11) reported that in addition to the occurrence of a ring cleavage via the nitroso route, other ring cleavage pathways, such as direct ring cleavage and/or denitration followed by ring cleavage, might be possible. In the latter study, several key intermediate ring cleavage products, including bis(hydroxymethyl)nitramine, methylenedinitramine (MEDINA), nitrous oxide, and formaldehyde, accumulated, but hydrazines were not detected (11, 12).
Several groups described RDX biodegradation under aerobic conditions, but little information was provided on the degradation pathway (5, 6, 16, 27). Products from biodegradation of cyclic nitramine explosives under aerobic conditions are poorly understood, particularly ring cleavage products (5, 10). Jones et al. (16) isolated a Rhodococcus sp. strain A from explosives-contaminated soil and demonstrated its potential for the degradation of RDX but did not report any products. Coleman et al. (6) reported the isolation and characterization of another Rhodococcus sp. strain DN22, which efficiently degrades RDX with the production of NO2−. No other products have been identified during RDX biodegradation with DN22. The aim of the present study was to identify key intermediates produced during aerobic degradation of RDX by Rhodococcus sp. strain DN22. In addition, uniformly labeled [U-14C]-RDX and ring-labeled [15N]-RDX were used in incubation mixtures with strain DN22 to determine the stoichiometry of the reaction and thus to provide insight about the RDX degradation pathway.
MATERIALS AND METHODS
Chemicals.
Commercial-grade RDX (purity of >99%) and ring-labeled [15N]-RDX (purity of >98%) (2) were provided by Defense Research Establishment Valcartier, Quebec, Canada. [U-14C]-RDX with chemical and radiochemical purities reaching 98 and 97% with a specific activity of 28.7 μCi/mmol (3) was also provided by Defense Research Establishment Valcartier (3). We determined the chemical purity of [U-14C]-RDX by high-performance liquid chromatography (HPLC)/UV analysis and radiochemical purity by collecting the HPLC fractions corresponding to RDX for subsequent radioactivity measurements. D2O (99% purity), 18O2 (97%), and [14C]HCHO (53-mCi/mmol specific activity as provided by the supplier) were from Aldrich, Oakville, Ontario, Canada. All other chemicals used were reagent grade.
Organisms and growth conditions.
Rhodococcus sp. strain DN22, previously isolated from a soil contaminated with RDX, 2,4,6-trinitrotoluene, 2,4-dinitrotoluene, and heavy metals, was kindly provided by Nicholas V. Coleman. The strain was grown in a mineral salt medium previously described by Coleman et al. (6). Unless specified otherwise, succinate (2.4 mM) was used as the carbon source and RDX (175 μM) (from a concentrated acetone stock solution) was added as the sole nitrogen source. Acetone was removed by evaporation prior to the addition of the aqueous medium. Growth was monitored spectrophotometrically at 530 nm (A530) (Thermo Spectronic, Rochester, N.Y.). Cultures were protected from light and agitated at 250 rpm at 25°C.
Mineralization of RDX by growing cultures of Rhodococcus sp. strain DN22 was determined as previously described (11). Briefly, the assays were performed with 120-ml serum bottles containing 10 ml of mineral salt medium and RDX (175 μM) containing [U-14C]-RDX (0.038 μCi). A 5-ml tube containing 1 ml of KOH (0.5 M) was placed in the serum bottle to act as a CO2 trap. The serum bottles were crimp sealed under a blanket of air with Teflon-coated rubber septa and incubated at room temperature under agitation (150 rpm). The KOH traps were regularly sampled for radioactivity determination (14CO2) using a Tri-Carb 4530 liquid scintillation counter (LSC) (model 2100 TR; Packard Instrument Company, Meriden, Conn.). After each sampling of the KOH tarp, a fresh volume (1 ml) of the original KOH solution was added to the trap to compensate for the withdrawn volume. When the stationary phase was reached, the microcosms were sacrificed to measure the remaining radioactivity in the culture supernatant and in the biomass.
Resting cells assays were performed using mid-log phase culture (A530 = 0.4 to 0.5). Cells were washed and resuspended in the mineral salt medium described above to an absorption equal to 1.2 without the addition of any carbon or nitrogen source apart from RDX. In some cases, (NH4)2SO4 (1 mM) was added to DN22 resting cells to prevent the uptake of NO2− produced during RDX degradation (6, 8). For some cell suspensions, we added ring-labeled [15N]-RDX] to DN22 cultures to determine which nitrogen atoms were incorporated into metabolites.
For experiments with 18O2 labels, the culture medium was first flushed with nitrogen to remove air and then 18O2 (97% pure) was added to the headspace with a gas-tight syringe followed by the addition of DN22 cells. The final oxygen concentration was roughly 20% (vol/vol) (O2/N2) as measured by a gas chromatograph (GC) connected to a thermal conductivity detector (TCD).
In experiments with deuterated water, RDX grown cells were harvested by centrifugation and then resuspended in pure D2O (5 ml) in the presence of RDX (175 μM) for subsequent analysis by liquid chromatography-mass spectrometry (LC-MS).
Analytical procedures.
Samples from the liquid culture medium of the above incubation mixtures were filtered through a 0.22-μm-pore-size membrane (Millex-GP; Millipore, Bedford, Mass.) and analyzed by reversed-phase HPLC connected to a photodiode array (PDA) detector. The system consisted of a W600 pump (Waters Associates, Milford, Mass.), a 717 plus auto-sampler, and a 996 PDA detector (λ = 254 nm). The filtered samples (50 μl) were injected into a Supelcosil LC-CN column (4.6-mm inner diameter [ID] by 25 cm) (Supelco, Bellafonte, Pa.), at 35°C. The methanol/water gradient was at a flow rate of 1.5 ml/min. The solvent composition was 30% methanol and 70% water held for 8 min, followed by a linear gradient from 30 to 65% methanol over 12 min. For the analysis of products from [U-14C]-RDX samples, a Waters HPLC system connected to a dual UV (254 nm) and radioactivity detector (In/Us ScintFlow System) was employed using chromatographic conditions similar to those described above.
A Micromass bench-top single quadrupole mass detector attached to a Hewlett Packard 1100 Series HPLC system equipped with a PDA detector was used for the analysis of RDX metabolites. The samples were injected into a 4-μm-pore-size SynergiPolar-RP column (4.6-mm ID by 15 cm) (Phenomenex, Torrance, Calif.) at 25°C. The solvent system consisted of a methanol/formic acid (200 μM) gradient (10 to 90% [vol/vol]) at a flow rate of 0.75 ml/min. For mass analysis, ionization was done in a negative electrospray ionization mode, ES(−), producing mainly the [M-H] mass ions. The mass range was scanned from 40 to 400 Da with a cycle time of 1.6 s, and the resolution was set to 1 Da (width at half-height).
Analyses of nitrite (NO2−) and nitrate (NO3−) were performed by capillary electrophoresis using sodium borate (25 mM) and hexamethonium bromide (25 mM) electrolytic solution at pH 9.2 (23) and UV detection at 215 nm. The ammonium cation was analyzed using an SP 8100 HPLC system equipped with a Waters 431 conductivity detector and a Hamilton PRP-X200 (250 mm by 4.1 mm by 10 μm) analytical cation-exchange column as described earlier (11). Analysis of 15N14NO (45 Da) and 14N14NO (44 Da) was carried out with a GC/MS system as previously described (27).
Formaldehyde (HCHO) was analyzed as described by Summers (28) with a few modifications. Samples were derivatized with 2,4-pentanedione in the presence of ammonium acetate and glacial acetic acid for 1 h at pH 6.0, 40°C. The derivatives were then analyzed by HPLC using a 5-μm-pore-size Supelcosil LC-8 column (4.6-mm ID by 25 cm) (Supelco) maintained at 40°C. The mobile phase consisted of an acetonitrile gradient of 15 to 27%, at a flow rate of 1.5 ml/min for 6 min. The derivatives were detected and quantified with a fluorescence detector (excitation at 430 nm and emission at 520 nm).
RESULTS
Biotransformation of RDX with growing Rhodococcus sp. strain DN22 cells.
(i) Nitrogen-containing products.
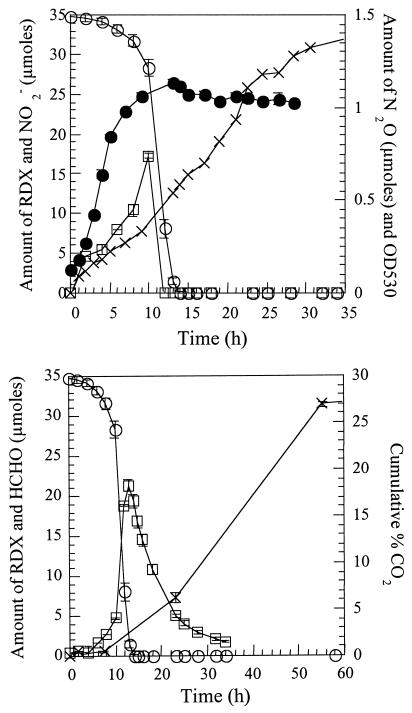
As already shown by Coleman et al. (6), we found that the growth of strain DN22 in a mineral salt medium containing RDX (35 μmol) as the N source and succinate (12 mM) as the carbon source was accompanied by RDX degradation and transient accumulation of nitrite (NO2−) (Fig. 1 top). There was no lag phase in the degradation of RDX because inocula were prepared with mid-log DN22 precultures grown on RDX. Maximum cell density (A530, 1.14) was attained after 14 h of incubation, during which RDX disappeared completely. In the uninoculated controls, no RDX removal was observed.
FIG. 1.
Time course study of RDX biodegradation with Rhodococcus sp. strain DN22. (Top) Removal of RDX (○), growth of cells (•), evolution of nitrite (□), and nitrous oxide (×). (Bottom) Removal of RDX (○), evolution of formaldehyde (□), and cumulative percentage of 14CO2 (×). Values are the averages and standard deviations of duplicate experiments.
The removal of RDX and the formation NO2− was accompanied by the formation of N2O, which continued to accumulate long after the disappearance of NO2− (Fig. 1 top). After 4 days of incubation, the amount of N2O attained a plateau of 4.5 μmol (not shown). We did not observe N2O in controls containing RDX in the absence of Rhodococcus. The presence of N2O was confirmed by comparison with a reference standard material and by measuring its molecular mass ion at 44 Da by GC-MS. Incubation of RDX grown cells with NaNO2 in the presence of succinate also lead to the accumulation of N2O also in trace amounts. When the above biodegradation experiment was repeated using ring-labeled [15N]-RDX as the nitrogen source for DN22, we detected a molecular mass ion at 45 Da, corresponding to 14N15NO, and another at 44 Da, corresponding to 14N14NO almost with the same intensity. The results indicate that trace amounts of N2O were produced from RDX or from RDX metabolites, such as NO2−.
We did not detect any of the RDX nitroso derivatives, such as MNX, DNX, and TNX, although such products have been frequently observed during biodegradation of RDX with anaerobic sludge (11, 20) and with P. chrysosporium (27).
(ii) Carbon-containing products.
Incubation of RDX with cultures led to the disappearance of RDX and the formation of formaldehyde (HCHO) (Fig. 1 bottom). Formaldehyde accumulated during the time of rapid RDX removal and was subsequently mineralized as determined by the accumulation of 14CO2 from [U-14C]-RDX (Fig. 1 bottom). In a separate experiment, incubation of H14CHO with RDX grown cultures under the same conditions led to the degradation of the aldehyde and the accumulation of 14CO2 in very high yield (88%).
(iii) Accumulation of a dead end product.
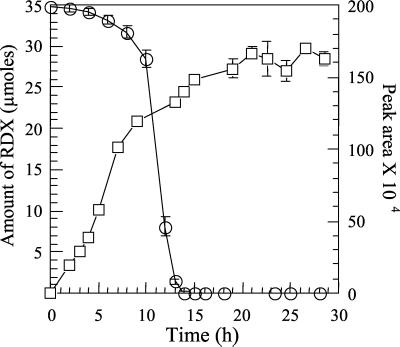
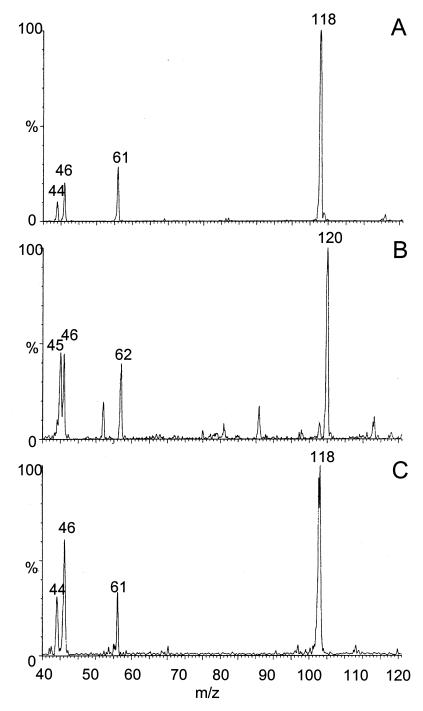
During the growth of strain DN22 on RDX (Fig. 2), there was a concurrent formation and accumulation of another soluble metabolite as detected by HPLC/UV analysis and LC-MS(ES−). The early chromatographic elution of the metabolite (retention time of 3.6 min) implies that the compound is more polar than RDX. The compound was not degraded by strain DN22. The metabolite yielded a deprotonated molecular mass ion [M-H] at 118 Da (MW, 119 Da) (Fig. 3A). The fragmentation pattern of the metabolite shows two other relevant mass ions at 61 and 46 Da representing fragments -NHNO2 and -NO2 (Fig. 3A). When ring-labeled [15N]-RDX was used, the [M-H] ion of the metabolite shifted to 120 Da (Fig. 3B), indicating that the metabolite contained two of the three ring-labeled 15N atoms in RDX. Also, the mass ion fragment -NHNO2 shifted from 61 to 62 Da, confirming that the fragment still contains one atom of 15N. High-resolution MS of the metabolite yielded a [M-H] at 118.0331, matching a deprotonated molecular formula of C2H4N3O3. The calculated mass of the deprotonated mass ion was 118.0253, suggesting that the metabolite had a molecular formula of C2H5N3O3 (MW, 119 Da) containing three N atoms (half of the total N content of RDX), two C atoms (two third of the total C content of RDX), and five H atoms.
FIG. 2.
Accumulation of the metabolite with an MW of 119 (□) during RDX biodegradation with Rhodococcus sp. strain DN22 (○). Mass area was used to monitor the formation of the metabolite MW 119. Values are the averages and standard deviations of duplicate experiments.
FIG. 3.
LC/MS (ES−) spectra of the metabolite with an MW of 119 produced during RDX degradation using resting DN22 cells (A), DN22 cells and ring-labeled [15N]-RDX (B), and sodium hydroxide (pH 12) (C).
Biodegradation of RDX by resting cells resuspended in pure D2O yielded two products with deprotonated molecular mass ions [M-H] at 118 and 119 Da. The percentage of 119 to 118 Da was 18% (data not shown). We did not account for the water residue that were retained with biomass after centrifugation. The D-labeled experiment indicated the incorporation of only one deuterium in the metabolite based on the assumption that H rather than D was lost during ionization in the mass spectrometer. The suggested molecular formula of the above metabolite would be C2H4DN3O3.
Figure 3C shows that alkaline hydrolysis of RDX with NaOH at pH 12 produced a product which had the same chromatographic (retention time of 3.6 min) and mass ([M-H] at 118 Da) data as observed earlier for the MW 119 RDX metabolite. Alkaline hydrolysis of RDX (pH 12) in the presence of D2O also resulted in the formation of a product with [M-H] at 119 Da, indicating the inclusion of one D atom in the product.
When RDX was incubated with cells in the presence of 18O2, we did not observe any mass change in the MW 119 Da metabolite. It was very difficult to search for 18O in HCHO, but no 18O was detected in carbon dioxide when the latter was trapped in an alkaline medium and analyzed on LC-MS.
Stoichiometry.
We used resting cells assays to calculate the distribution of nitrogen in RDX metabolites produced during incubation with DN22 in the presence of ammonium sulfate (1 mM). The latter was added to prevent the uptake of NO2− produced as an RDX metabolite (6, 8). In experiments supplemented with ammonium sulfate, the concentrations of ammonia as an RDX product was calculated as the difference between initial and final concentrations of NH4+. Table 1 summarizes the percentage of N-containing products produced following RDX removal. We found an N-mass balance of 91.2% distributed as follows: NO2− (30%), N2O (3.2%), NH3 measured as NH4+ (10%), and a metabolite with an MW of 119 (48%). Table 1 also summarizes the carbon stoichiometry in a 4-day culture. After 4 days of incubation, 97.6% of the original radioactivity in RDX was found as follows: 30% was CO2 and 67.6% was in the liquid phase. However, Table 1 shows a 94% carbon mass balance distributed as follows: CO2 (30%) and a metabolite with an MW of 119 (64%), leaving 3% of measured radioactivity unidentified.
TABLE 1.
Stoichiometry of nitrogen and carbon during RDX degradation calculated based on percentage of reacted RDX using total theoretical numbers on N and C atoms
| Element (total no. of atoms) | NO2− | N2O | NH3a | CO2 | MW 119 | % Recovered |
|---|---|---|---|---|---|---|
| Carbon (3) | 30.0b (0.8)e | 64.0c | 94.0d | |||
| Nitrogen (6) | 30.0 (1.7)e | 1.5 (8.5)e | 10.0 | 48.0f | 89.5 |
No ammonium salt was added to the resting-cell suspension.
Based on 14CO2 measured using [U-14C]-RDX.
Based on HPLC/radioactivity measurement using [U-14C]-RDX.
Total of the measured values was 97.6%, distributed as follows: 30% was CO2 and 67.6% was in the liquid phase, leaving 3% unidentified.
Value in parentheses represents relative standard deviation of a triplicate.
Calculated based on the empirical formula C2H5N3O3 found by LC/MS (ES-) and high-resolution MS.
The stoichiometry shown in Table 1 indicates that following the removal of RDX (C3H6N6O6), 30% of its C content (one atom) was used to produce 14CO2 and 64% (two C atoms) were incorporated in the dead end product with an MW of 119 (C2H5N3O3). Controls containing RDX without strain DN22 retained close to 99.5% of the original radioactivity in the form of unreacted RDX.
DISCUSSION
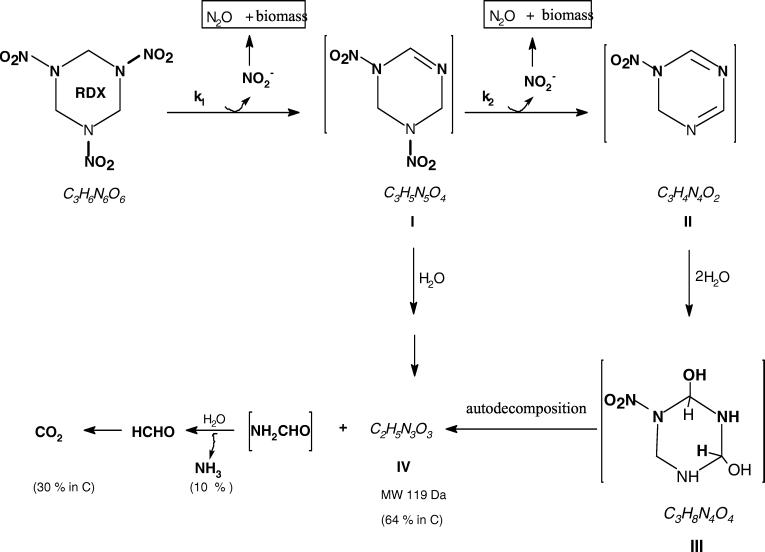
The formation of nitrite concurrent with RDX disappearance during incubation with growing DN22 cells (Fig. 1 top) clearly indicated the occurrence of an important early enzymatic denitration step in the degradation process (Fig. 4). The absence of any of the familiar nitroso products MNX, DNX, and TNX also supported the hypothesis that denitration was the most critical initial microbial (enzymatic) step in RDX degradation.
FIG. 4.
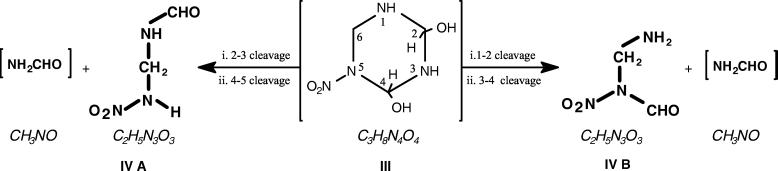
Schematic representation of potential steps involved during biodegradation of RDX with Rhodococcus sp. strain DN22. The scheme shows two steps of denitration to I and II prior to ring cleavage via the hypothetical hydroxylated product III. The presence of formamide as an RDX metabolite requires further confirmation.
Nitrite did not accumulate in the system and its disappearance was accompanied by microbial growth and the formation of N2O (Fig. 1), suggesting that nitrite was assimilated by the bacteria. Nitrogen assimilation via ammonia can be accompanied by the production of N2O (31).
The continued formation of N2O (total yield, 3.6%) long after the complete removal of RDX and nitrite indicated that in addition to nitrite assimilation by DN22, the presence of other RDX intermediate(s) might have been directly responsible for its formation. Also, the detection of both 14N14NO (m/z, 44 Da) and 15N14NO (m/z, 45 Da) during biodegradation of ring-labeled [15N]-RDX indicated that in addition to -NO2− assimilation there must be another route for its production. The formation of 15N14NO requires the direct participation of an RDX nitramine group (15N-14NO2). Earlier, we reported the formation of N2O from the spontaneous decomposition of nitramide, NH2NO2, considered as a ring cleavage product of RDX biodegradation with anaerobic sludge (11). Regardless of which mechanism leads to the production N2O, its formation as a secondary product in a small yield (3.6%) does not provide insight about early steps in RDX metabolism.
The formation of 2 mol of NO2− per molecule of RDX that disappeared is consistent with the stoichiometry reported earlier by Coleman et al. (6). A plausible hypothesis would be that the first loss of NO2− produced the cyclohexenyl product (I) whereas the second denitration produced the cyclohexadienyl intermediate (II) (Fig. 4).
The transient accumulation of HCHO and the subsequent formation of CO2 clearly indicate the cleavage of the RDX ring following its denitration. The concurrent formation of carbon dioxide with the disappearance of HCHO (Fig. 1 bottom) might indicate the direct involvement of the aldehyde in the formation of CO2. The above conclusion is supported by the rapid and efficient degradation of H14CHO to 14CO2 (88%) under the same conditions. The total yield of CO2 indicated that roughly one carbon atom in each RDX molecule mineralized.
We tentatively identified the molecular structure of the detected C2H5N3O3 metabolite as the aldehyde (IVA) or the amine (IVB) (Fig. 5). We based our conclusion on the assumption that the denitrated intermediate II would first react with water to produce the hypothetical hydroxylated structure III. The subsequent spontaneous decomposition of the resulting α-hydroxylated product (III) would then produce both IV A and IVB (Fig. 5). The autodecomposition of the unstable α-hydroxylated product (III) would also produce NH3, HCHO in addition to the dead end product IV (C2H5N3O3) (Fig. 4). No nitrite assimilation occurred during resting-cell assays. Therefore, the formation of ammonia in these assays was presumed to be generated directly through the decomposition of the hydroxylated product III or via one of its ring cleavage intermediates possibly formamide (NH2CHO). Pseudomonas putida biodegrades NH2CHO to NH3 and CO2 (4). α-Hydroxylation of cyclic and acyclic dialkyl nitrosamines catalyzed by a mixed function oxidase can lead to the production of unstable carbinol products which decompose to nitrogen and cationic alkyl groups, R+ (26, 22).
FIG. 5.
Proposed chemical structures of the MW 119 (C2H5N3O3) metabolite and the hypothetical hydroxylated product (III) that lead to its formation.
Our hypothesis of the occurrence of a rapid hydrolytic ring cleavage (II to III to IV) following RDX initial denitration (RDX to I to II) was supported by our observation of deuterium in the MW 119 metabolite during RDX biodegradation in the presence of D2O. The evidence does not support a simple exchange with the solvent because when we added an HPLC-purified sample of C2H5N3O3 to D2O, we did not observe any change in the MW. Furthermore, we observed the MW 119 product during alkaline hydrolysis of RDX at pH 12. The absence of 18O in the metabolite produced during RDX transformation in the presence of 18O2 clearly supports the formation of MW 119 products via a hydrolytic step. Interestingly, we did not observe any 18O incorporation in CO2. However, further spectroscopic evidence on the identity of the 119-Da metabolites is needed to provide insight about the mechanism of its formation and also its subsequent reactions.
Previously, we reported that biodegradation of RDX with another isolated Rhodococcus strain produced the cyclohexenyl intermediate (I) which autodecomposed to produce the dead end product MW 119 (IV) that was tentatively identified as the aldehyde IV A (10). In fact, both mono denitration followed by ring cleavage (13) and successive elimination of two HNO2 molecules followed by ring cleavage (15) have been reported for the destruction of RDX under alkaline conditions.
Most of the products (NO2−, NH3, N2O, HCHO, and CO2) detected during RDX biodegradation with DN22 have also been detected during the alkaline hydrolysis of RDX, suggesting a resemblance in the degradation mechanisms of both reactions. RDX hydrolyzes in an alkaline solution (pH 12) via a bimolecular elimination of HNO2 to initially produce a 1,3,5-triaza-3,5-dinitrocyclohex-1-ene intermediate (I) (7). The intermediate (I) decomposes at a rate 105 times faster than the initial rate of RDX degradation by E2 (13). As Fig. 3C shows, we detected a product with an MW of 119 with the same retention time (3.6 min) as observed during incubation with Rhodococcus. It is possible that intermediate I was also formed by strain DN22, but its rapid decomposition would have prevented detection.
The nitrogen and carbon stoichiometry shown in Table 1 is consistent with the scheme in Fig. 4 in which we assumed that both RDX denitration steps occurred prior to ring cleavage. Figure 4 thus represents the best explanation for the detected RDX degradation products and their time course of production and stoichiometry. Future work should focus on the actual enzymes involved in the initial attack on RDX and the role of enzymes versus abiotic mechanisms in the subsequent complex reactions that take place following ring cleavage.
Acknowledgments
We thank Louise Paquet, Carl Groom, Alain Corriveau, and Chantale Beaulieu for their technical assistance and Sylvie Beaudet for mass analysis. We also thank Nick Coleman and Sandra Trott for providing the strain and for reviewing the manuscript. We are grateful to Sonia Thiboutot and Guy Ampleman from the Defense Research Establishment Valcartier, Quebec, Canada for providing us with the energetic chemicals.
Funding was provided by the U.S. DoD/DoE/EPA Strategic Environmental Research and Development Program (SERDP # 1213).
Footnotes
This is NRCC publication number 44634.
REFERENCES
- 1.Adrian, N. R., and T. Chow. 2001. Identification of hydroxylaminodinitroso-1,3,5-triazine as a transient intermediate formed during the anaerobic biodegradation of RDX. Environ. Toxicol. Chem. 20:9. [PubMed] [Google Scholar]
- 2.Ampleman, G., A. Marois, S. Thiboutot, J. Hawari, C. W. Greer, J. Godbout, G. I. Sunahara, C. F. Shen, and S. R. Guiot. 1999. Synthesis of 14C-labelled octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX), and 15N-isotopically labeled hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) for use in microcosm experiments. DREV-TR-1999-99. Defense Research Establisment Valcartier, Val Bélair, Québec, Canada.
- 3.Ampleman, G., S. Thiboutot, J. Lavigne, A. Marois, J. Hawari, A. M. Jones, and D. Rho. 1995. Synthesis of 14C-labelled hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), 2,4,6-trinitrotoluene (TNT), nitrocellulose (NC) and glycidylazide polymer (GAP) for use in assessing the biodegradation potential of these energetic compounds. J. Label. Compd. Radiopharm. 36:559–577. [Google Scholar]
- 4.Babu, G. R. V., O. K. Vijaya, V. L. Ross, J. H. Wolfram, and K. D. Cahpatwala. 1996. Cell free extract(s) of Pseudomonas putida catalyzes the conversion of cyanides, cyanates, thiocyanates, formamide, and cynide-containing mine waters into ammonia. Appl. Microbiol. Biotechnol. 45:273–277. [DOI] [PubMed] [Google Scholar]
- 5.Binks, P. R., S. Nicklin, and N. C. Bruce. 1995. Degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Stenotrophomonas maltophilia PB1. Appl. Environ. Microbiol. 61:1318–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, N. V., D. R. Nelson, and T. Duxbury. 1998. Aerobic degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol. Biochem. 30:1159–1167. [Google Scholar]
- 7.Croce, M., and Y. Okamoto. 1979. Cationic micellar catalysis of the aqueous alkaline hydrolyses of 1,3,5-triaza-1,3,5-trinitrocyclohexane and 1,3,5,7-tetraaza-1,3,5,7-tetranitrocyclooctane. J. Org. Chem. 44:2100–2103. [Google Scholar]
- 8.Ecker, S., T. Widmann, H. Lenke, O. Dickel, P. Fischer, C. Bruhn, and H.-J. Knackmuss. 1992. Catabolism of 2,6-dinitrophenol by Alcaligenes eutrophus JMP 134 and JMP 222. Arch. Microbiol. 158:149–154. [Google Scholar]
- 9.Haas, R., I. Schreiber, E. von Löw, and G. Stork. 1990. Conception for the investigation of contaminated munitions plants. 2. Investigation of former RDX-plants and filling stations. Fresenius’ J. Anal. Chem. 338:41–45. [Google Scholar]
- 10.Hawari, J. 2000. Biodegradation of RDX and HMX: from basic research to field application, p.277–310. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 11.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal sludge. Appl. Environ. Microbiol. 66:2652–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawari, J., S. Beaudet, A. Halasz, S. Thiboutot, and G. Ampleman. 2000. Microbial degradation of explosives: biotransformation versus mineralization. Appl. Microbiol. Biotechnol. 54:605–618. [DOI] [PubMed] [Google Scholar]
- 13.Hoffsommer, J. C., D. A. Kubose, and D. J. Glover. 1977. Kinetic isotope effects and intermediate formation for the aqueous alkaline homogeneous hydrolysis of 1,3,5-triaza-1,3,5-trinitrocyclohexane (RDX). J. Phys. Chem. 81:380–385. [Google Scholar]
- 14.Jackson, M., J. M. Green, R. L. Hash, D. C. Lindsten, and A. F. Tatyrek. 1978. Nitramine (RDX and HMX) wastewater treatment at the Holston Army Ammunition Plant. Report ARLCD-77013. U.S. Army Armament Research and Development Command, Dover, N.J.
- 15.Jones Walter, H. 1954. Mechanisms of the homogeneous alkaline decomposition of cyclotrimethylenetrinitramine: kinetics of consecutive second-and first-order reactions. A polarographic analysis for cyclotrimethylenenitramine. J. Am. Chem. Soc. 76:829–835. [Google Scholar]
- 16.Jones, A. M., C. W. Greer, G. Ampleman, S. Thiboutot, J. Lavigne, and J. Hawari. 1995. Biodegradability of selected highly energetic pollutants under aerobic conditions, p.251–257. In R. Hinchee, R. E. Hoeppel, and D. B. Anderson (ed.). Third International In Situ and On-site Bioreclamation Symposium. Battelle Press, Columbus, Ohio.
- 17.Kaplan, D. L. 1998. Biotransformation and bioremediation of munitions and explosives, p.549–575. In S. K. Sikdar and R. L. Irvine (ed.), Bioremediation: principles and practice, vol. II. Biodegradation technology developments. Technomic Publishing Inc., Lancaster, Pa.
- 18.Kitts, C. L., D. P. Cunningham, and P. J. Unkefer. 1994. Isolation of three hexahydro-1,3,5-trinitro-1,3,5-triazine degrading species of the family Enterobacteriaceae from nitramine explosive-contaminated soil. Appl. Environ. Microbiol. 60:4608–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitts, C. L., C. E. Green, R. A. Otley, M. A. Alvarez, and P. J. Unkefer. 2000. Type I nitroreductase in soil enterobacteria reduce TNT (2,4,6-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can. J. Microbiol. 46:278–282. [DOI] [PubMed] [Google Scholar]
- 20.McCormick, N. G., J. H. Cornell, and A. M. Kaplan. 1981. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine. Appl. Environ. Microbiol. 42:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myler, C. A., and W. Sisk. 1991. Bioremediation of explosives contaminated soils (scientific questions/engineering realities), p.137–146. In G. S. Sayler, R. Fox, and J. W. Blackburn (ed.), Environmental biotechnology for waste treatment. Plenum Press, New York, N.Y.
- 22.Okada, M., M. Mochizuki, T. Anjo, T. Stone, Y. Wakabayashi, and E. Suzuki. 1980. Formation, deoxygenation, and mutagenicity of α-hydroperoxydialkylnitrosamines, p.71–79. In E. A. Walker et al. (ed.), N-nitroso compounds: analysis, formation and occurrence. Publication no. 31. International Agency for Research on Cancer, Lyon, France. [PubMed]
- 23.Okemgbo, A. A., H. H. Hill, S. G. Metcalf, and M. A. Bachelor. 1999. Determination of nitrate and nitrite in Hanford defense waste by reverse-polarity capillary zone electrophoresis. J. Chromatogr. A 844:387–394. [Google Scholar]
- 24.Regan, K. M., and R. L. Crawford. 1994. Characterization of Clostridium bifermentans and its biotransformation of 2,4,6-trinitrotoluene (TNT) and 1,3,5-triaza-1,3,5-trinitrocyclohexane (RDX). Biotechnol. Lett. 16:1081–1086. [Google Scholar]
- 25.Robidoux, P. Y., J. Hawari, S. Thiboutot, G. Ampleman, and G. I. Sunahara. 2001. Chronic toxicity of and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) in soil determined using the earthworm (Eisenia andrei) reproduction test. Environ. Pollut. 111:283–292. [DOI] [PubMed] [Google Scholar]
- 26.Rowland, R. 1988. The toxicology of N-nitroso compounds, p. 117–141. In M. J. Hill (ed.), Nitrosamines: toxicology and microbiology. Ellis Horwood Ltd., Chichester, England.
- 27.Sheremata, T. W., and J. Hawari. 2000. Mineralization of RDX by the white rot fungus Phanerochaete chrysosporium to carbon dioxide and nitrous oxide. Environ. Sci. Technol. 34:3384–3388. [Google Scholar]
- 28.Summers, W. R. 1990. Characterization of formaldehyde and formaldehyde releasing preservatives by combined reversed phase cation-exchange high performance liquid chromatography with postcolumn derivatization using Nash’s reagent. Anal. Chem. 62:1397–1402. [Google Scholar]
- 29.Talmage, S. S., D. M. Opresko, C. J. Maxwel, C. J. E. Welsh, F. M. Cretella, P. H. Reno, and F. B. Daniel. 1999. Nitroaromatic munition compounds: environmental effects and screening values. Rev. Environ. Contam. Toxicol. 161:1–156. [DOI] [PubMed] [Google Scholar]
- 30.Young, D. M., P. J. Unkefer, and K. L. Ogden. 1997. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a prospective consortium and its most effective isolate Serratia marcescens. Biotechnol. Bioeng. 53:515–522. [DOI] [PubMed] [Google Scholar]
- 31.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]