Abstract
NAD(P)H-dependent oxidoreductases are valuable tools for synthesis of chiral compounds. The expense of the cofactors, however, requires in situ cofactor regeneration for preparative applications. We have attempted to develop an enzymatic system based on phosphite dehydrogenase (PTDH) from Pseudomonas stutzeri to regenerate the reduced nicotinamide cofactors NADH and NADPH. Here we report the use of directed evolution to address one of the main limitations with the wild-type PTDH enzyme, its low stability. After three rounds of random mutagenesis and high-throughput screening, 12 thermostabilizing amino acid substitutions were identified. These 12 mutations were combined by site-directed mutagenesis, resulting in a mutant whose T50 is 20°C higher and half-life of thermal inactivation at 45°C is >7,000-fold greater than that of the parent PTDH. The engineered PTDH has a half-life at 50°C that is 2.4-fold greater than the Candida boidinii formate dehydrogenase, an enzyme widely used for NADH regeneration. In addition, its catalytic efficiency is slightly higher than that of the parent PTDH. Various mechanisms of thermostabilization were identified using molecular modeling. The improved stability and effectiveness of the final mutant were shown using the industrially important bioconversion of trimethylpyruvate to l-tert-leucine. The engineered PTDH will be useful in NAD(P)H regeneration for industrial biocatalysis.
The potential of enzyme-catalyzed transformations in the pharmaceutical and fine-chemical industries has long been considered to have great promise (7, 10, 23). Despite the development of rapid and efficient methods for creating designer biocatalysts, inherent limitations in the chemistry that these enzymes perform still persist. The majority of biocatalysts used in industry are hydrolytic in action (∼65%) and perform rather simple chemistry (4). More complicated enzymatic reactions often require one or more costly cofactors, which, when added in stoichiometric quantities, make the process not economically feasible. Oxidoreductases, for example, catalyze a myriad of regio-, chemo-, and stereospecific reactions on a variety of functional groups, but often require either NADH or NADPH as a cofactor (8, 11).
Various in situ regeneration methods have been developed to allow the use of catalytic quantities of NAD(P)+ and NAD(P)H (20). The most successful and widely used enzymatic NADH regeneration system is based on the Candida boidinii formate dehydrogenase (FDH) (1). We have attempted to develop an NADH regeneration process using the enzyme phosphite dehydrogenase (PTDH) from Pseudomonas stutzeri (2). This enzyme catalyzes the nearly irreversible oxidation of phosphite to phosphate with the concomitant reduction of NAD+ to NADH. The kinetic and practical advantages of using PTDH for NADH regeneration have been recently reported (22).
In addition to NADH regeneration, the enzyme PTDH also has considerable potential in regenerating NADPH. The cost of NADPH is significantly higher than that of NADH, and no widely used NADPH regeneration system is currently available. To date, the most useful enzyme for NADPH regeneration is a mutant Pseudomonas sp. strain 101 FDH available from Jülich Fine Chemicals (18). Although wild-type PTDH has a preference for NAD+ over NADP+ by about 100-fold, a mutant PTDH has been created by rational design with relaxed specificity toward both nicotinamide cofactors (26). The catalytic efficiency of the PTDH double mutant with NADP+ (kcat/Km,NADP) is 33-fold higher than that of Pseudomonas sp. strain 101 FDH with a comparable turnover number.
As a biocatalyst for industrial applications, one of the main limitations of the wild-type PTDH is its low stability. Indeed, the wild-type enzyme shows fairly rapid inactivation under relatively mild temperatures (2). Thus, we sought to use directed evolution to improve its thermostability. Directed evolution has become a powerful tool for improving enzyme function and has been successfully used to improve the thermal stability of many different enzymes (9, 15, 19, 27). Here we report the directed evolution of a highly thermostable PTDH mutant for NAD(P)H regeneration. Three rounds of error-prone PCR and high-throughput screening coupled with site-directed mutagenesis yielded a PTDH mutant with higher thermostability than the most thermostable Candida boidinii FDH reported previously (17).
MATERIALS AND METHODS
Reagents.
Escherichia coli BW25141 and plasmid pLA2 were kindly provided by William Metcalf at the University of Illinois (Urbana, IL) (6). The pRW2 plasmid was created from pLA2 as described elsewhere (26). Escherichia coli BL21(DE3) and plasmid pET15b were purchased from Novagen (Madison, WI). Taq DNA polymerase was obtained from Promega (Madison, WI) and cloned Pfu Turbo DNA polymerase was obtained from Stratagene (La Jolla, CA). The DNA-modifying enzymes NdeI, BamHI, PciI, and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). PCR-grade deoxynucleotide triphosphates and DNase I were obtained from Roche Applied Sciences (Indianapolis, IN). l-(+)-Arabinose was purchased from Fluka (St. Louis, MO). Ampicillin, kanamycin, lysozyme, nitro blue tetrazolium (NBT), phenazine methosulfate, and NAD+ were purchased from Sigma (St. Louis, MO). Phosphorous acid was obtained from Aldrich (Milwaukee, WI). Other salts and reagents were purchased from either Sigma-Aldrich or Fisher Scientific (Pittsburgh, PA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). The QIAprep spin plasmid mini-prep kit, QIAEX II gel purification kit, and QIAquick PCR purification kit were purchased from QIAGEN (Valencia, CA). BD Talon metal affinity resin was purchased from BD Biosciences Clontech (Palo Alto, CA). Amicon Ultra-15 centrifugal filter devices were purchased from Fisher Scientific (Pittsburgh, PA). Purified formate dehydrogenase (FDH) from Candida boidinii was purchased from Jülich Fine Chemicals (Jülich, Germany).
Random mutagenesis and library creation.
Random mutagenesis was carried out by error-prone PCR as described elsewhere (29). Plasmid pRW2 containing the parent gene was used as the template for the first generation of random mutagenesis. For the 1.0-kb PTDH parent target gene, 0.2 mM MnCl2 was used to obtain the desired level of mutagenesis rate (∼1 to 2 amino acid substitutions). Forward (5′-TTTTTGGATGGAGGAATTCATATG-3′, the NdeI restriction site is underlined) and reverse (5′-CGGGAAGACGTACGGGGTATACATGT-3′, the PciI restriction site is underlined) primers were used to amplify the gene. PCR-mutated genes were digested with NdeI and PciI and ligated into the plasmid pRW2 digested with the same two restriction enzymes. Ligation reactions (10 μl total volume) contained ∼100 ng inserts, ∼100 ng vector, 1X T4 DNA ligase buffer, and 0.5 U T4 DNA ligase, and were incubated at 16°C for 16 h. The resulting plasmids were transformed into freshly prepared electrocompotent E. coli WM1788 cells, which were then plated on Luria-Bertani (LB) agar plates containing 50 μg/ml kanamycin.
Thermostability screening.
For identification of thermostable PTDH variants, a 96-well plate thermostability screening assay was developed based on a colorimetric NBT-phenazine methosulfate assay (14). Library colonies were picked with sterile toothpicks and used to inoculate 100 μl of LB medium containing 50 μg/ml kanamycin in 96-well plates. The plates were incubated at 37°C for 5 h, and protein expression was induced with arabinose (final concentration, 10 mM) followed by incubation at 30°C for ∼14 to 16 h. Cells were lysed by adding lysozyme (1 mg/ml) and DNase I (4 U/ml) followed by freeze-thawing. The plates were centrifuged at 3,200 × g for 15 min at 4°C, and 50 μl of supernatant was transferred to two fresh plates. One plate was placed into a machined aluminum plate holder that had been incubated in an oven set at a desired temperature. After 10 min incubation, the plate was allowed to cool at room temperature for 15 min.
Initial and residual activities were determined by adding NBT assay solution (1mg/ml NBT, 0.5 mg/ml phenazine methosulfate, 15 mM NAD+, and 40 mM phosphite) and monitoring the change in absorbance at 580 nm for 5 min in a Spectramax 340PC microplate reader (Molecular Devices, Sunnyvale, CA). Thermostable mutants were identified by comparing their ratios of residual activity to initial activity with that of the parent enzyme. Only mutants that showed both higher ratio of residual activity to initial activity than the parent enzyme and similar initial activity compared to the parent enzyme were selected and characterized. The first-, second-, and third-generation libraries were incubated at 42°C, 57°C, and 62°C, respectively, for 10 min. Positives were confirmed by performing the identical assay in culture tubes (26).
DNA sequencing and analysis.
Plasmid DNA from E. coli BW25141 was isolated using QIAprep spin plasmid mini-prep kits. Sequencing reactions consisted of 100 to 200 ng of template DNA, 10 pmol each primer, sequencing buffer, and the BigDye reagent (Applied Biosystems, Foster City, CA). Reactions were carried out for 31 cycles of 96°C for 20 s, 50°C for 10 s, and 60°C for 3.5 min in an MJ Research (Watertown, MA) PTC-200 thermal cycler. Prepared samples were submitted to the Biotechnology Center at the University of Illinois for sequencing on an ABI PRISM 3700 sequencer (Applied Biosystems, Foster City, CA).
Site-directed mutagenesis.
A “megaprimer” method of site-directed mutagenesis was used to introduce site-specific mutations (16). Individual mutations identified from each round of screening were combined using the “megaprimer” method multiple times until all mutations were incorporated into the parent gene for a given round of screening. Combined site-directed mutant genes were cloned into the pRW2 vector to verify the increased thermostability followed by DNA sequencing. After verification, combined site-directed mutant genes were subcloned into pET15b as described elsewhere (26). The mutant genes were sequenced for a second time to ensure that no PCR-induced random mutations were incorporated into the final DNA construct. Plasmids containing the correct mutant genes were then transformed into E. coli BL21(DE3) and plated onto LB agar plates containing 100 μg/ml ampicillin.
PTDH overexpression and purification.
Protein purification of the parent and mutant PTDHs was carried out using a protocol described elsewhere (26) with some modifications. Small-scale spin columns containing approximately 0.5 ml of BD Talon resin were used to purify multiple enzymes in parallel. The columns were equilibrated in start buffer A (0.5 M NaCl, 20% glycerol, and 20 mM Tris-HCl, pH 7.6). The following method was used to purify PTDH (with His6 tag): ∼10 ml of clarified supernatant (∼1 to 5 g of cell paste) loaded onto the column, column washed 10 times with start buffer A, column washed five times with start buffer A plus 10 mM imidazole, and proteins eluted with elution buffer (0.5 M imidazole, 0.5 M NaCl, 20% glycerol, and 20 mM Tris-HCl, pH 7.6). Elution fractions were collected and each fraction was checked for activity using NBT-phenazine methosulfate assay. Pooled active fractions were concentrated using an Amicon Ultra-15 filter device and washed four times with 20 ml of 50 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.25). The enzyme was then stored at a concentration of >2 mg/ml in 20% glycerol at −80°C.
Enzyme kinetics.
The kinetic rate constants for the parent and mutant PTDHs were determined as described elsewhere (26). Enzyme concentration was determined by measuring A280 (ɛ = 30 mM−1 cm−1). The data represent an average of all statistically relevant data with a standard deviation of less than 10%.
Half-lives of thermal inactivation.
Purified enzymes (0.2 mg/ml) were incubated in an MJ Research PTC-200 thermocycler (±0.3°C) to study enzyme inactivation. PTDH enzymes were incubated in 50 mM MOPS buffer (pH 7.25), whereas Candida boidinii FDH was incubated in 100 mM potassium phosphate buffer (pH 7.5). Aliquots were taken at specific time points and placed on ice before assaying. Typically, inactivation was monitored until >80% of the activity was lost. Half-lives of thermal inactivation were calculated using t1/2 = ln2/kinact where kinact is the inactivation rate constant obtained from the slope by plotting log (residual activity/initial activity) versus time.
T50.
Values of T50, the temperature required to reduce initial enzyme activity by 50% after a fixed incubation period, were determined as described (29). Briefly, purified enzymes (0.2 mg/ml) were incubated for 20 min at various fixed elevated temperatures. After incubation, samples were placed on ice for 15 min before being assayed using saturating substrate conditions. Residual activity was determined and expressed as a percentage of the initial activity.
Topt.
Topt was determined by assaying the purified enzyme (0.2 mg/ml) with 1 mM phosphite, 0.5 mM NAD+ in 50 mM MOPS (pH 7.25) at increasing temperatures using a recirculating water bath with a jacketed cuvette holder. The enzyme activity was determined by monitoring the absorbance increase at 340 nm as described (26).
Batch production of l-tert-leucine.
Small-scale reactions were conducted to demonstrate the improved stability and effectiveness of the 12x thermostable PTDH mutant. The final volume of each batch reaction was 300 μl. Each sample contained 300 mM ammonium trimethylpyruvate, 400 mM diammonium phosphite (400 mM ammonium formate in the case of FDH), 0.4 mM NAD+, 5.26 U/ml Bacillus cereus leucine dehydrogenase, and 100 μg of PTDH or FDH. Each reaction was mixed gently and incubated at 40°C. At fixed time intervals, small aliquots were removed from the reaction and immediately frozen at −20°C. The conversion of l-tert-leucine was monitored using a high-pressure liquid chromatograph (Shimadzu) equipped with an evaporative light-scattering detector. Substrates and products were separated on an Alltech C-18 Prevail column with an isocratic elution of 94% water, 4.5% acetonitrile, and 1% acetic acid. The concentration of each sample was determined by using a standard curve prepared with authentic l-tert-leucine.
RESULTS
Generation of thermostable PTDH mutants.
The thermostability of PTDH was improved by combining error-prone PCR and site-directed mutagenesis approaches. Error-prone PCR was used to introduce an average of 1 to 2 amino acid substitutions per PTDH gene. This low mutation rate made it possible to identify the contribution of individual amino acid substitutions on enzyme thermostability. Three rounds of error-prone PCR mutagenesis were used to generate libraries of PTDH variants. After each round of error-prone PCR, site-directed mutagenesis was used to combine thermostabilizing mutations into one template and the resulting template served as the parent for the next round of error-prone PCR mutagenesis.
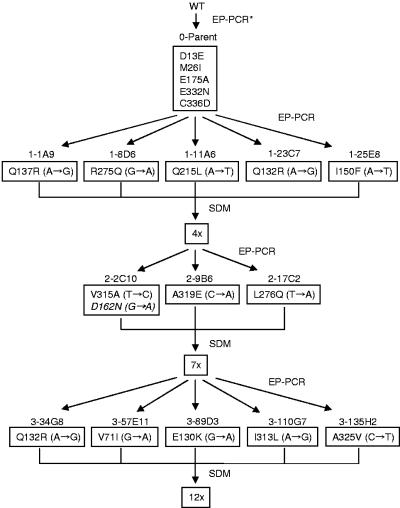
The first-generation mutant library was constructed using a mutant PTDH that we recently engineered (25). This PTDH mutant differs from wild-type PTDH at five amino acid positions (D13E, M26I, E175A, E332N, and C336D). These mutations improved the enzyme solubility and activity, but showed little effect on the enzyme thermostability. At 45°C, the t1/2 of wild-type PTDH is 1.4 ± 0.3 min, whereas the t1/2 of the parent is 1.2 ± 0.2 min. After creating the first-generation mutant library using error-prone PCR, approximately 3,200 clones were screened for increased thermostability at 43°C. Five variants were identified and confirmed to have longer half-lives than the parent at 45°C (Table 1). Sequencing these variants revealed the following amino acid substitutions: 1-1A9 (Q137R), 1-8D6 (R275Q), 1-11A6 (Q215L), 1-23C7 (Q132R), and 1-25E8 (I150F) (Fig. 1). Site-directed mutagenesis was used to combine these thermostabilizing mutations onto the parental template to create 4x (Q137R, I150F, Q215L, R275Q) and 5x (Q132R, Q137R, I150F, Q215L, R275Q) mutants.
TABLE 1.
Thermostability data for the parent and mutant PTDHsa
| Generation | Variant | t1/2 (min, 45°C) | Fold Improvement | T50 (°C) | dT50 (°C) |
|---|---|---|---|---|---|
| 0 | Parent | 1.2 | 1 | 39.0 | 0 |
| 1 | 1-1A9 | 3.8 | 3.2 | 41.9 | 2.9 |
| 1-8D6 | 4.6 | 3.8 | 40.7 | 1.7 | |
| 1-11A6 | 8.7 | 7.3 | 42.5 | 3.4 | |
| 1-23C7 | 2.3 | 1.9 | 40.0 | 1.0 | |
| 1-25E8 | 7.0 | 5.8 | 42.2 | 3.2 | |
| 4x | 200 | 167 | 52.4 | 13.4 | |
| 5x | 161 | 134 | 53.4 | 14.4 | |
| 2 | 2-2C10 | 614 | 527 | 54.0 | 15.0 |
| 2-9B6 | 567 | 473 | 53.8 | 14.8 | |
| 2-17C2 | 437 | 364 | 54.5 | 15.5 | |
| 7x | 1,421 | 1,184 | 56.9 | 17.9 | |
| 3 | 3-57E11 | 2,350 | 1,958 | 57.9 | 18.9 |
| 3-89D3 | 2,000 | 1,667 | 58.3 | 19.3 | |
| 3-34G8 | 1,515 | 1,263 | 58.3 | 19.3 | |
| 3-110G7 | 1,765 | 1,471 | 57.5 | 18.5 | |
| 3-135H2 | 2,315 | 1,929 | 57.5 | 18.5 | |
| 12x | 8,440 | 7,033 | 59.3 | 20.3 |
Average of all statistically relevant data (standard deviation <10%). Wild type PTDH has a t1/2 of 1.4 ± 0.3 min at 45°C. All experiments were performed in 50 mM MOPS (pH 7.25).
FIG. 1.
Lineage and protein-level mutations in the evolved thermostable PTDH variants. Corresponding DNA-level mutations are shown in parentheses. The substitution shown in italics represents a mutation that does not increase PTDH thermostability. EP-PCR, error-prone PCR; SDM, site-directed mutagenesis; ∗, previous directed evolution work to improve activity and solubility (25).
The gene encoding the 4x mutant was chosen as a template for generating the second-generation mutant library using error-prone PCR (Table 2). The 4x mutant was chosen over the 5x mutant as the template for second-generation random mutagenesis based on a higher catalytic activity and longer half-life at 45°C. Approximately 6,000 clones from the 4x library were screened for increased thermostability at 57°C. Three clones were found to have longer half-lives than the parent at 45°C. After sequencing these three variants, the following additional amino acid substitutions were identified: 2-2C10 (D162N, V315A), 2-9B6 (A319E), and 2-17C2 (L276Q) (Fig. 1). Clone 2-2C10 had two amino acid substitutions. Both mutations were incorporated individually onto the 4x template using site-directed mutagenesis to determine the effect of each mutation separately on enzyme thermostability. Single mutant V315A shows a threefold increase in thermostability compared to the 4x mutant, whereas single mutant D162N shows a 1.8-fold decrease in thermostability. Using site-directed mutagenesis, the substitutions L276Q, A319E, and V315A were combined on the 4x template to create a 7x mutant.
TABLE 2.
Kinetic parameters of the parent and evolved PTDH variants in 50 mM MOPS (pH 7.25) at 25°Ca
| Generation | Variant | kcat (min−1) | Km (μM, NAD) | kcat/Km,NAD (μM−1 min−1) | Km (μM, Pt-H) |
|---|---|---|---|---|---|
| 0 | Parent | 262 | 66 | 4.0 | 57 |
| 1 | 1-1A9 | 285 | 66 | 4.3 | 48 |
| 1-8D6 | 244 | 70 | 3.5 | 78 | |
| 1-11A6 | 278 | 64 | 4.3 | 58 | |
| 1-23C7 | 238 | 60 | 4.0 | 45 | |
| 1-25E8 | 262 | 75 | 3.5 | 99 | |
| 4x | 218 | 74 | 2.9 | 144 | |
| 5x | 170 | 46 | 3.7 | 75 | |
| 2 | 2-2C10 | 171 | 65 | 2.6 | 155 |
| 2-9B6 | 198 | 52 | 3.8 | 92 | |
| 2-17C2 | 174 | 56 | 3.1 | 120 | |
| 7x | 174 | 66 | 2.6 | 96 | |
| 3 | 3-34G8 | 192 | 57 | 3.4 | 169 |
| 3-57E11 | 210 | 63 | 3.3 | 105 | |
| 3-89D3 | 216 | 75 | 2.9 | 123 | |
| 3-110G7 | 216 | 71 | 3.0 | 156 | |
| 3-135H2 | 213 | 50 | 4.3 | 135 | |
| 12x | 195 | 40 | 4.9 | 46 |
Average of all statistically relevant data (standard deviation <10%).
The 7x mutant was used as the parent for the third round of random mutagenesis. From this library, ∼10,000 clones were screened for increased thermostability at 62°C. Five variants were identified and confirmed to have longer half-lives at 45°C than the 7x mutant. Sequencing these variants revealed the following changes: 3-34G8 (Q132R), 3-57E11 (V71I), 3-89D3 (E130K), 3-110G7 (I313L) and 3-135H2 (A325V) (Fig. 1). It is noteworthy that Q132R appeared twice, in the first and third-generations. This gave further evidence that Q132R is indeed a thermostabilizing mutation, and thus it was incorporated into the final template. Site-directed mutagenesis was used to incorporate all third round mutations into the 7x template to create the 12x mutant.
Thermostabilities of the evolved PTDHs.
Table 1 lists the t1/2 and T50 values of the parent and the evolved thermostable PTDHs accumulated during three generations of directed evolution. At 45°C, the t1/2 of the parent is 1.2 min and its T50 is 39°C. The first-generation variants have half-lives of thermal inactivation two- to sevenfold longer than the parent at 45°C and their T50s are 1.0 to 3.4°C higher. The half-lives of thermal inactivation of the combined site-directed mutants 4x and 5x are ∼130 to 170 times that of the parent and their T50s are ∼14°C higher.
A further round of random mutagenesis resulted in three variants that have half-lives of thermal inactivation 360 to 530 times longer than the parent at 45°C and their T50s are 14.8 to 15.5°C higher. The half life of the 7x mutant is ∼1,200 times that of the parent at 45°C and its T50 is increased by 17.9°C. The third-generation variants have half-lives of thermal inactivation ∼1,250 to 2,000 times longer than the parent at 45°C and their T50s are 18.5 to 19.3°C higher. The 12x mutant has a half-life at 45°C that is approximately 7,000 times longer than the parent or the wild-type enzyme. The T50 of the 12x mutant is 59.3°C, an increase of 20.3°C compared with the parent.
Activities of the evolved PTDHs.
The kinetic parameters of the parent and evolved PTDHs toward the substrates NAD+ and phosphite are listed in Table 2. All five first-generation variants show similar kcat and Km,NAD+ compared to the parent. First-generation variants 1-8D6 and 1-25E8 exhibit a slight increase in Km,Pt-H. The 4x and 5x mutants both have lower activities than the parent, and the 4x mutant exhibits about a 2.5-fold increase in Km,Pt-H. All three second-generation variants and the 7x mutant have similar kinetic parameters to the 4x mutant. All five third-generation variants are slightly more active than the 7x mutant, while their Km,NAD+ remains unchanged and their Km,Pt-H increased slightly. The final 12x mutant retains approximately 75% of the parent enzyme's turnover number and has a slightly higher catalytic efficiency (kcat/Km,NAD+). The 12x mutant exhibits a Km,Pt-H similar to that of the parent. The kinetic parameters of the 12x mutant toward nicotinamide cofactor NADP+ were also determined. The evolved 12x mutant has similar kcat and Km,NADP+ compared to the parent: 12x mutant, kcat = 80 ± 6 min−1, Km,NADP+ = 49 ± 5 mM−1 min−1 versus the parent, kcat = 110 ± 1 min−1, Km,NADP+ = 65 ± 8 mM−1 min−1 (25).
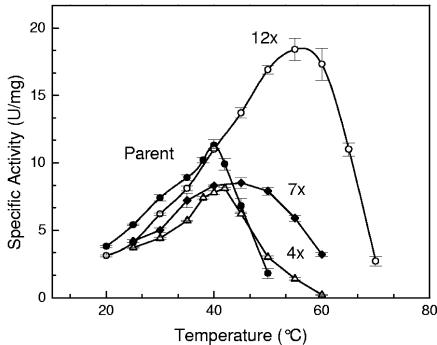
The activity-temperature profile for the parent and the combined mutants is shown in Fig. 2. For each successive round (parent→ 4x→ 7x→ 12x), the activity-temperature profile broadens and the specific activities of PTDH enzymes increase with increasing temperature until the enzyme denatures. The temperature optimum, Topt, of the 12x mutant is 59°C, which is ∼20°C higher than that of the parent, which is in good agreement with the observed increases in T50 (Table 1).
FIG. 2.
Activity-temperature profiles of the parent (•), 4x mutant (▴), 7x mutant (⧫), and 12x mutant (○).
Comparison with Candida boidinii formate dehydrogenase.
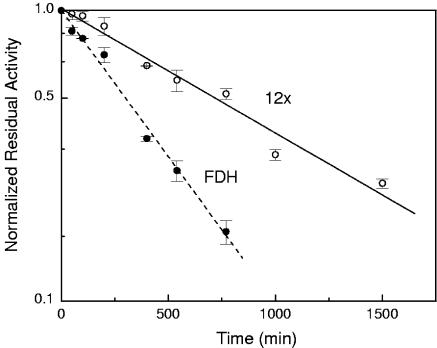
The half-lives of thermal inactivation of the 12x PTDH mutant and Candida boidinii FDH were measured at 50°C. Both enzymes exhibit first-order inactivation kinetics as shown in Fig. 3. At 50°C, the half-lives of thermal inactivation of the 12x PTDH mutant and FDH are 705 and 299 min, respectively. The t1/2 of FDH at 50°C is in good agreement with values reported elsewhere for the wild-type and recombinant FDH (t1/2 ≥300 min at 50°C in 100 mM potassium phosphate buffer, pH 7.5) (17). Slusarczyk and coworkers improved the oxidative stability of the wild-type FDH by replacing two cysteine residues using site-directed mutagenesis, but this resulted in several mutants with significantly lower thermostability (17). Thus, the 12x mutant is substantially more thermostable than these FDH variants.
FIG. 3.
Thermal inactivation of 12x PTDH and Candida boidinii FDH at 50°C.
NADH regeneration for l-tert-leucine production.
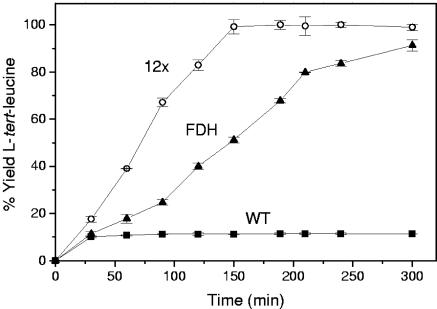
The 12x thermostable PTDH mutant was tested by coupling it with leucine dehydrogenase from Bacillus cereus for the production of l-tert-leucine in small-scale batch reactions. PTDH converts NAD+ into NADH with the conversion of phosphite to phosphate. The regenerated NADH is then used by leucine dehydrogenase to convert trimethylpyruvate and ammonia to l-tert-leucine. The 12x mutant was compared to wild-type PTDH and commercially available FDH from C. boidinii to demonstrate its improved stability and effectiveness. Figure 4 shows the production of l-tert-leucine over 300 min with the three different regeneration enzymes. The wild-type PTDH precipitates rapidly within the first 30 min of the reaction, whereas the 12x mutant retains its stability and reaches 100% conversion. The 12x mutant also has a ∼2-fold faster reaction rate than FDH under these conditions.
FIG. 4.
Production of l-tert-leucine from trimethylpyruvate with regeneration of NADH using wild-type PTDH (▪), the 12x mutant PTDH (○), and C. boidinii FDH (▴). The small-scale batch reaction contained 300 mM ammonium trimethylpyruvate, 400 mM diammonium phosphite or ammonium formate, 0.4 mM NAD+, and equal amounts (by mass) of either PTDH or FDH. Samples were incubated at 40°C.
DISCUSSION
Evolutionary strategy.
The evolutionary strategy implemented in this work consisted of random mutagenesis and high-throughput screening followed by site-directed mutagenesis to incorporate the best mutations into the parental template for the next round of directed evolution (Fig. 1). By introducing an average of only 1 to 2 amino acid substitutions per PTDH gene, it was possible to identify the contribution of individual amino acid substitutions on enzyme thermostability. The effects of thermostabilizing mutations are additive as found in many other enzymes (24, 28). Combined mutants (4x, 5x, 7x, and 12x) were significantly more thermostable than any single mutant in a given round. Similar to other directed evolution efforts, we found that the thermostability of PTDH could be increased significantly while retaining 75% of the enzyme's catalytic activity at lower temperatures (5, 29).
Structural analysis.
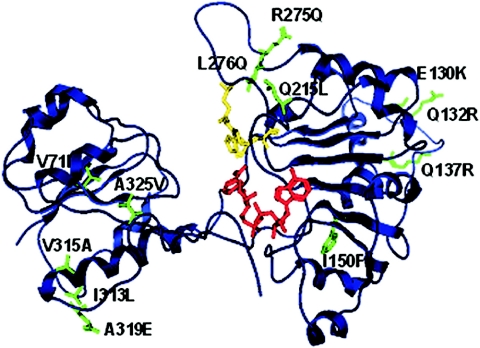
A three-dimensional homology model of the wild-type PTDH was created as described elsewhere (26), and the 12 thermostabilizing mutations were mapped into this model (Fig. 5). None of these mutations occurred near the three-active site residues (Arg237, Glu266, and His292), although one mutation (I150F) did occur in the Rossmann fold involved with NAD(P) binding. A mutation (R275Q) occurred in a loop region directly after the active site Glu266 in the sequence 267-DWARADRPR-275. It has been suggested that the three arginines in this region could be involved in binding substrate phosphite (26). Substituting arginine for glutamine at position 275 does not appear to affect the binding of phosphite according to the kinetic data (Table 2). Therefore, while the other two arginines may still be involved in binding phophite, it appears that Arg275 is not essential to phosphite binding. The mutations A319E and A325V occur in the highly flexible C-terminal region.
FIG. 5.
Ribbon representation of the monomeric structural model of PTDH from Pseudomonas stutzeri. NAD+ cofactor is shown in red, and active-site residues (His292, Arg237, and Glu266) are shown in yellow. The thermostabilizing mutations (green) are shown in stick representation.
Based on this structural model, the molecular basis for improved thermostability was further probed. The thermostabilizing mutations are all distributed over the surface of the enzyme except for mutations V71I and I150F. This finding underscores the importance of protein surface on stability as found in many other enzymes with improved thermostability (3, 29). It is consistent with the notion that the initial steps in protein unfolding during the irreversible thermal denaturation primarily involve surface-located parts of the protein (3). The two nonsurface mutations are located in β-sheets and both are buried in extremely hydrophobic regions within the protein. The isoleucine to phenylalanine substitution (I150F) in β5 is located in the Rossmann fold. Surprisingly, the incorporation of the large hydrophobic phenylalanine residue into the GxxGxGxxG (where x represents any type of amino acids) nucleotide binding motif does not seem to affect the enzyme's ability to bind the nicotinamide cofactor NAD+. The Km,NAD+ values for the parent and the I150F mutation (1-25E8) are essentially the same (66 and 75 μM, respectively). More importantly, based on the homology model, the substitution of isoleucine for phenylalanine (I150F) seems to increase the hydrophobic interaction between residue 150 and residues Met160, Leu171, Leu189, and Leu205, which enhances the thermostability of the enzyme. The stabilizing effect of V71I may be attributed to increased hydrophobic interactions with residues Met49, Val57, Leu93, and Phe81.
Only two mutations (I313L and V315A) are located in α-helices. These substitutions may increase stability by increasing helical propensity since Leu and Ala have nonbranched side chains compared to Ile and Val, respectively. Three mutation sites, E130K, Q132R, and Q137R, are located in the loop region between α6/β5. Mutations Q132R and Q137R introduce positively charged residues which may increase protein stability by creating a more favorable surface charge distribution, as found in the engineered thermostable cold shock proteins (13). Mutation E130K is located at the dimer interface and may increase enzyme stability by creating beneficial electrostatic interactions between charged residues of the two subunits (E130K is within ∼8Å of D272 of the other subunit).
The three mutations Q215L, R275Q, and L276Q are located in the loop region between β10/α13. Although it is surprising that substituting an uncharged hydrophilic residue for a nonpolar hydrophobic residue (Q215L) at the protein surface increases protein stability, and this effect has been observed in other thermostability studies (12, 13). Mutation R275Q substitutes a positively charged residue for an uncharged residue which may increase protein stability by influencing the surface charge distribution. The mechanism by which L276Q enhances thermostability is not clear. The two mutations on the highly flexible C-terminal region, A319E and A325V, most likely increase protein thermostability by anchoring the C terminus and this would make the protein more resistant to unfolding.
It should be noted that the above-proposed mechanisms for thermostabilization have been identified in many naturally occurring thermophilic and hyperthermophilic proteins as well as engineered thermostable proteins (3, 21). Further confirmation of these mechanisms should be greatly facilitated by the determination of the crystal structure of the 12x mutant and additional mutagenesis and stability analysis. However, since there are so many different structural routes to improved thermostability and the contribution of each route to the total free energy of stabilization is quite small, it seems very difficult to predict the specific thermostabilizing mutations for a protein of interest based on its structure, let alone a protein without a crystal structure such as PTDH.
In conclusion, this study shows that the thermostability of phosphite dehydrogenase can be rapidly improved by directed evolution without a loss of its catalytic efficiency. Various stabilization mechanisms have been uncovered, many of which are difficult to predict a priori, thus underscoring the notion that directed evolution is a generally applicable and highly effective approach for protein stabilization (29). The improved stability and effectiveness of the thermostable PTDH mutant were demonstrated in the production of an important specialty chemical, l-tert-leucine. The 12x mutant was found to be a more effective regeneration catalyst than the most successful and widely used NADH-regenerating enzyme, C. boidinii FDH, under the conditions tested. The results of this work will hopefully enable us to reach our ultimate goal of developing a low-cost, highly efficient biocatalyst for NAD(P)H regeneration.
Acknowledgments
We thank Yan Gao, Ka-chun Lai, Mike Mclachlan, and Kyle Kloepper for their help in screening the mutant libraries.
This work was supported by the Biotechnology Research and Development Consortium (BRDC) (project 2-4-121).
REFERENCES
- 1.Bommarius, A. S., and K. Drauz. 1994. An enzymatic route to l-ornithine from arginine—activation, selectivity and stabilization of l-arginase. Bioorg. Med. Chem. 2:617-626. [DOI] [PubMed] [Google Scholar]
- 2.Costas, A. M., A. K. White, and W. W. Metcalf. 2001. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 276:17429-17436. [DOI] [PubMed] [Google Scholar]
- 3.Eijsink, V. G. H., A. Bjork, S. Gaseidnes, R. Sirevag, B. Synstad, B. van den Burg, and G. Vriend. 2004. Rational engineering of enzyme stability. J. Biotechnol. 113:105-120. [DOI] [PubMed] [Google Scholar]
- 4.Faber, K. 2000. Biotransformations in organic chemistry, 4th ed. Springer Verlag, Berlin, Germany.
- 5.Giver, L., A. Gershenson, P. O. Freskgard, and F. H. Arnold. 1998. Directed evolution of a thermostable esterase. Proc. Natl. Acad. Sci. USA 95: 12809-12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huisman, G. W., and D. Gray. 2002. Towards novel processes for the fine-chemical and pharmaceutical industries. Curr. Opin. Biotechnol. 13:352-358. [DOI] [PubMed] [Google Scholar]
- 8.Hummel, W., and M. R. Kula. 1989. Dehydrogenases for the synthesis of chiral compounds. Eur. J. Biochem. 184:1-13. [DOI] [PubMed] [Google Scholar]
- 9.Kim, J. H., G. S. Choi, S. B. Kim, W. H. Kim, J. Y. Lee, Y. W. Ryu, and G. J. Kim. 2004. Enhanced thermostability and tolerance of high substrate concentration of an esterase by directed evolution. J. Mol. Catal. B 27:169-175. [Google Scholar]
- 10.Kirk, O., T. V. Borchert, and C. C. Fuglsang. 2002. Industrial enzyme applications. Curr. Opin. Biotechnol. 13:345-351. [DOI] [PubMed] [Google Scholar]
- 11.Krix, G., A. S. Bommarius, K. Drauz, M. Kottenhahn, M. Schwarm, and M. R. Kula. 1997. Enzymatic reduction of alpha-keto acids leading to l-amino acids, d- or l-hydroxy acids. J. Biotechnol. 53:29-39. [Google Scholar]
- 12.Martin, A., I. Kather, and F. X. Schmid. 2002. Origins of the high stability of an in vitro-selected cold-shock protein. J. Mol. Biol. 318:1341-1349. [DOI] [PubMed] [Google Scholar]
- 13.Martin, A., V. Sieber, and F. X. Schmid. 2001. In-vitro selection of highly stabilized protein variants with optimized surface. J. Mol. Biol. 309: 717-726. [DOI] [PubMed] [Google Scholar]
- 14.Mayer, K. M., and F. H. Arnold. 2002. A colorimetric assay to quantify dehydrogenase activity in crude cell lysates. J. Biomol. Screen. 7:135-140. [DOI] [PubMed] [Google Scholar]
- 15.Sakaue, R., and N. Kajiyama. 2003. Thermostabilization of bacterial fructosyl-amino acid oxidase by directed evolution. Appl. Environ Microbiol. 69:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkar, G., and S. S. Sommer. 1990. The megaprimer method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 17.Slusarczyk, H., S. Felber, M. R. Kula, and M. Pohl. 2000. Stabilization of NAD-dependent formate dehydrogenase from Candida boidinii by site-directed mutagenesis of cysteine residues. Eur. J. Biochem. 267:1280-1289. [DOI] [PubMed] [Google Scholar]
- 18.Tishkov, V. I., A. G. Galkin, V. V. Fedorchuk, P. A. Savitsky, A. M. Rojkova, H. Gieren, and M. R. Kula. 1999. Pilot scale production and isolation of recombinant NAD+- and NADP+-specific formate dehydrogenases. Biotechnol. Bioeng. 64:187-193. [PubMed] [Google Scholar]
- 19.Uchiyama, H., T. Inaoka, T. Ohkuma-Soyejima, H. Togame, Y. Shibanaka, T. Yoshimoto, and T. Kokubo. 2000. Directed evolution to improve the thermostability of prolyl endopeptidase. J. Biochem. Tokyo 128:441-447. [DOI] [PubMed] [Google Scholar]
- 20.van der Donk, W. A., and H. Zhao. 2003. Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 14:421-426. [DOI] [PubMed] [Google Scholar]
- 21.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vrtis, J. M., A. White, W. W. Metcalf, W. A. van der Donk. 2002. Phosphite dehydrogenase, a versatile cofactor regeneration enzyme. Angew Chem. Int. Ed. Engl. 41:3257-3259. [DOI] [PubMed] [Google Scholar]
- 23.Wandrey, C., A. Liese, and D. Kihumbu. 2000. Industrial biocatalysis: Past, present, and future. Org. Process Res. Dev. 4:286-290. [Google Scholar]
- 24.Wells, J. A. 1990. Additivity of mutational effects in proteins. Biochemistry 29:8509-8517. [DOI] [PubMed] [Google Scholar]
- 25.Woodyer, R., W. A. van der Donk, and H. Zhao. Directed evolution of a phosphite dehydrogenase mutant with enhanced activity and expression for NAD(P)H regeneration. Comb. Chem. High Throughput Screen., in press. [DOI] [PubMed]
- 26.Woodyer, R., W. A. van der Donk, and H. M. Zhao. 2003. Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry 42:11604-11614. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, N. Y., W. C. Suen, W. Windsor, L. Xiao, V. Madison, and A. Zaks. 2003. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein Eng. 16:599-605. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, X. J., W. A. Baase, B. K. Shoichet, K. P. Wilson, and B. W. Matthews. 1995. Enhancement of protein stability by the combination of point mutations in T4 lysozyme is additive. Protein Eng. 8:1017-1022. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, H. M., and F. H. Arnold. 1999. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 12:47-53. [DOI] [PubMed] [Google Scholar]