Abstract
Although ibuprofen [2-(4-isobutylphenyl)-propionic acid] is one of the most widely consumed drugs in the world, little is known regarding its degradation by environmental bacteria. Sphingomonas sp. strain Ibu-2 was isolated from a wastewater treatment plant based on its ability to use ibuprofen as a sole carbon and energy source. A slight preference toward the R enantiomer was observed, though both ibuprofen enantiomers were metabolized. A yellow color, indicative of meta-cleavage, accumulated transiently in the culture supernatant when Ibu-2 was grown on ibuprofen. When and only when 3-flurocatechol was used to poison the meta-cleavage system, isobutylcatechol was identified in the culture supernatant via gas chromatography-mass spectrometry analysis. Ibuprofen-induced washed-cell suspensions also metabolized phenylacetic acid and 2-phenylpropionic acid to catechol, while 3- and 4-tolylacetic acids and 2-(4-tolyl)-propionic acid were metabolized to the corresponding methyl catechols before ring cleavage. These data suggest that, in contrast to the widely distributed coenzyme A ligase, homogentisate, or homoprotocatechuate pathway for metabolism of phenylacetic acid and similar compounds, Ibu-2 removes the acidic side chain of ibuprofen and related compounds prior to ring cleavage.
Ibuprofen [2-(4-isobutylphenyl)-propionic acid] is a pharmaceutical drug used for its analgesic, antipyretic, and anti-inflammatory properties. It is the third most consumed drug in the world, with an estimated annual production of several kilotons (3). Approximately 10% of the ibuprofen consumed by humans is excreted unmodified or as the glucuronide conjugate (17, 25). Wastewater treatment processes have been found to remove ibuprofen with varying success (3, 28), which may explain why Kolpin et al. (13) detected ibuprofen in 9.5% of the bodies of water that they surveyed. Environmental concentrations of ibuprofen have been found to range from low part-per-trillion (3, 9, 13, 28, 30) to low part-per-billion levels (3, 9).
Little information exists regarding how ibuprofen is oxidatively metabolized by environmental microbes. Side chain hydroxylation has been reported (13, 31), along with the formation of carboxyhydratropic acid [2-(4-carboxyphenyl)propionic acid] (3, 31) and ibuprofenol [2-(4-isobutylphenyl)-propanol] under anaerobic conditions (4).
The most similar compounds whose bacterial metabolisms have been described include 4-isopropylbenzoate (cumate), phenylacetic acid, and 2-phenylpropionic acid (2PPA). Cumate is dioxygenated at the 2,3 position by Pseudomonas putida F1 carrying the cmt operon and subsequently meta-cleaved (5-8). Other routes for the metabolism of phenylacetic acids include the well-characterized homoprotocatechuate (27) and homogentisate pathways (29), as well as the more recently described coenzyme A-ligase pathway (12). Additionally, Streptomyces rimosus has been shown to convert 2PPA to 4-hydroxy-2PPA (15). Finally, Pseudomonas cepacia has been shown to metabolize 2PPA (2) and tropic acid (21) (2-phenyl-3-hydroxypropionic acid) via a pathway involving decarboxylation to phenylacetaldehyde, followed by oxidation to phenylacetic acid (2).
In this study, we report the isolation of a bacterium capable of utilizing ibuprofen as a sole carbon and energy source. We also provide evidence of an apparently novel metabolic pathway for the degradation of ibuprofen and related phenylacetic acids.
MATERIALS AND METHODS
Materials and strains.
NAD+ was purchased from Sigma-Aldrich (St. Louis, MO). Dextrin 10 was purchased from Fluka BioChemika (Buchs, Switzerland). All other chemicals were purchased from Acros (Morris Plains, NJ). A 50/50 enantiomeric mixture of R/S-ibuprofen was used unless stated otherwise. Mineral salts medium (MSM) was prepared as previously described (20).
Isolation of Ibu-2 via enrichment of sewage sludge.
Sewage sludge was enriched with ibuprofen according to standard protocols (14). A single colony was isolated and designated strain Ibu-2. A fragment of the 16S rRNA gene from Ibu-2 was PCR amplified using the universal primers 27F (AGAGTTTGATCMTGGCTCAG) and 1055R (CGGCCATGCACCACC) (16) and was sequenced using the 27F and 1055R primers.
Stereospecificity.
Ibu-2 was inoculated into 500 mg/liter R/S-ibuprofen, 500 mg/liter S-ibuprofen, or 250 mg/liter R/S-ibuprofen. Pure R-ibuprofen was not used because it is not commercially available. Maximal cell density was determined spectrophotometrically.
Chiral capillary electrophoresis (CE) was performed on the supernatants of ibuprofen-grown Ibu-2 cultures in order to determine if a difference existed in the rates at which the enantiomers were metabolized. Supernatants were harvested and filtered as described above. Samples were run on a Hewlett-Packard (HP) 3D CE using a 40-cm by 50-μm fused silica column from Agilent technologies (Palo Alto, CA) and a method adapted from Simo et al. (26). The cassette temperature was set at 25°C. The running buffer was composed of 6% dextrin 10 and 150 mM sodium borate at pH 9. Prior to each injection, the column was preconditioned for 1 min with water, 1 min with 0.1 M NaOH and 50 mM sodium dodecyl sulfate, and finally for 2 min with the running buffer. Injection was performed by applying 10 mbar pressure for 9 seconds, followed by running-buffer injection at 50 mbar for 5 seconds. Voltage was applied at +20 kV. A diode array detector was used with detection and reference wavelengths set at 194 ± 2 nm and 500 ± 80 nm, respectively.
Substrate specificity analysis with washed cells.
Ibu-2 was grown in MSM containing 500 mg/liter ibuprofen or 0.1% glucose. Washed-cell suspensions (WCS) were prepared according to standard protocols (10). Test compounds were added to a final concentration of 500 mg/liter in 500-μl aliquots of WCS, and yellow-color generation was monitored for 30 min. The yellow products were further characterized by determining their absorbance maxima via spectrophotometry. Yellow color was assumed to be indicative of meta-cleavage product (mcp) formation and was thus taken as a positive indication of metabolism of the test compound.
Chemicals tested included phenol, 2-, 3-, and 4-methylphenol; catechol; 3-methylcatechol; 4-methylcatechol; 4-tertbutylcatechol; benzoate; 4-methylbenzoate; phenylacetaldehyde; phenylacetic acid; R- and S-2-hydroxy-2-phenylacetic acid (mandelic acid); 2-, 3-, and 4-tolylacetic acid; 2- and 4-(hydroxyphenyl)acetic acid; 2- and 3-phenylpropionic acid; 2-(4-tolyl)propionic acid; 2-phenyl-3-hydroxypropionic acid (tropic acid); 3-phenyl-2-propenoic acid (cinnamic acid); 2-phenylbutyric acid; and 2,2-diphenylacetic acid.
Growth substrate analysis.
Growth of Ibu-2 was tested on compounds that gave a positive result in the substrate specificity analysis. Tests were performed in triplicate in test tubes. Five milliliters of an overnight Ibu-2 culture grown on ibuprofen was used to inoculate MSM containing 250 mg/liter or 500 mg/liter of the chemical of interest. The tubes were placed on a vertical rotor and monitored over the course of 1 week for growth via changes in optical density at 600 nm.
Compounds that did not support growth on their own were assayed for their ability to support growth in the presence of ibuprofen as an inducer. Ibu-2 was inoculated into mixtures of 250 mg/liter of these compounds plus 250 mg/liter ibuprofen in MSM.
Analysis of culture supernatants by GC-MS.
Ibu-2 was inoculated into 1 liter of MSM containing 500 mg of ibuprofen. When grown on ibuprofen, the culture accumulated a yellow color. The culture was allowed to continue growing until that color reached an apparent maximum level (48 to 60 h). At this point, the supernatant was harvested via centrifugation and filtered through a 0.22-μm filter. The supernatant was then acidified to pH 3 using 1 M HCl and extracted with 50 ml of ethyl acetate. The extract was concentrated to a volume of 2 ml under nitrogen at 35°C. The samples were then methylated with diazomethane by standard protocols (11). After 30 min at room temperature, the samples were evaporated to a minimal volume under a nitrogen stream and analyzed via gas chromatography-mass spectrometry (GC-MS) using an HP 6890 GC equipped with an HP-5MS column (5% phenyl methyl siloxane; 30 m by 0.25 mm; 0.25-μm film thickness) using helium as the carrier gas with a flow rate of 1 ml/min. The injector temperature was 250°C. The initial oven temperature of 40°C was held for 1 min and then ramped at a rate of 10°C/min to 250°C. The temperature was held at 250°C for 7 min and then ramped up at 30°C/min to 300°C. The detector was an HP 5973 MSD with the quadrapole and source set at 150°C and 230°C, respectively.
Analysis of catecholic ibuprofen intermediates.
3-Fluorocatechol, a meta-cleavage inhibitor, was added to a final concentration of 50 mg/liter in 100 ml of mid-log-phase Ibu-2 culture. After 30 min, the supernatant was removed and filtered. Potassium carbonate and acetic anhydride were added to final concentrations of 1.5% and 0.5%, respectively. This aqueous acetylation was performed to selectively acetylate aromatic hydroxyl groups (19). After 30 min, the samples were acidified to pH 3, extracted with ethyl acetate, and dried over a sodium sulfate column. The extracts were then methylated with diazomethane and analyzed as described above. A culture with ibuprofen but without 3-fluorocatechol was also analyzed.
Characterization of catechols produced from ibuprofen analogs.
Assays were performed with WCS and the 2-arylpropionic acids and phenylacetic acids that produced yellow metabolites. The assays were performed with and without the addition of 3-fluorocatechol. The supernatants were passed through a 0.22-μm filter and then analyzed via high-performance liquid chromatography (HPLC) for accumulation of catechols. The HPLC retention times of the metabolites were compared to those of catechol and methylcatechol standards. HPLC was performed using 70% methanol, 30% 40 mM acetic acid as the eluent. The sample was pumped at a rate of 1 ml/min using a Waters Model 590 pump through a Varian Microsorb-MV C18 column (250 mm by 4.6 mm). Samples were injected by a Shimadzu SIL-10AD AP autoinjector and detected with a Shimadzu SPD-10A VP UV-Vis detector by monitoring absorbance at 285 nm.
Cell extract assays.
Ibuprofen-grown Ibu-2 was washed with 0.9% NaCl to remove the exopolysaccharide matrix (24) and then resuspended in a minimal volume of sonication buffer (100 mM Tris, 2 μm phenylmethylsulfonyl fluoride, and 1 μm dithiothreitol at pH 8) (18). The cells were then lysed via sonication using a Branson Sonifer 450 set at 100% duty cycle using three cycles of 30 seconds each, separated by 1 min rest time. The cells were kept on ice at all times. The samples were then centrifuged at 21,000 × g at 4°C for 15 min, and the supernatant was retained. Extracts were assayed for activity by adding catechol to a small aliquot and monitoring it for yellow-product generation.
Assaying side chain oxidation.
Cell extracts from Pseudomonas sp. strain AT3 grown on tropic acid, prepared using the method described above, were used as a positive control for side chain oxidation (18). The assay, which monitors NAD+/NADP+ reduction, was performed with cell extracts from Ibu-2 or AT3 in sonication buffer with either 0.1 μM tropic acid or ibuprofen and 0.1 μM of either NAD+ or NADP+. The reaction mixtures were incubated at room temperature and monitored for reduction of NAD+ or NADP+ at 340 nm.
RESULTS AND DISCUSSION
Ibu-2 grew on ibuprofen as a sole source of carbon and energy (data not shown). When grown on ibuprofen in liquid media, a yellow color appeared in the supernatant. This yellow color disappeared upon acidification and reappeared upon neutralization, a phenomenon diagnostic of mcps. Sequencing and BLAST analysis (1) of a 16S rRNA gene fragment revealed that Ibu-2 was 98% identical to Sphingomonas species over 967 bp. Ibu-2 had yellow pigmentation and tended to develop an exopolysaccharide matrix, especially when grown on glucose. Both of these characteristics are common to Sphingomonas species (23).
Stereospecificity.
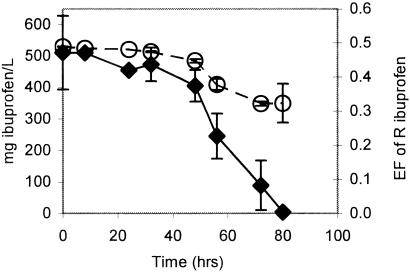
Ibu-2 grew to the same maximum cell density on 500 mg/liter R/S-ibuprofen as it did on 500 mg/liter S-ibuprofen. Both of these values were approximately twice that obtained using 250 mg/liter S-ibuprofen or R/S-ibuprofen. During growth on R/S-ibuprofen, the entantiomeric fraction dropped to less than 35% R-ibuprofen before both isomers were completely removed, suggesting that Ibu-2 may preferentially degrade the R enantiomer (Fig. 1).
FIG. 1.
The enantiomeric fraction (EF) of R-ibuprofen (○) and overall ibuprofen concentration (⧫) in a growing Ibu-2 culture in which ibuprofen was the sole carbon and energy source. The concentrations of both ibuprofen enantiomers were determined by chiral CE analysis. The error bars represent standard deviations.
GC-MS analysis of culture supernatant extracts.
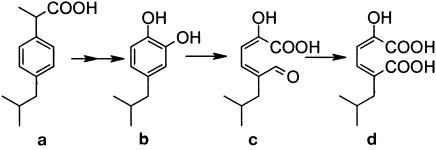
When ibuprofen-grown cultures were poisoned with 3-fluorocatechol, a metabolite accumulated in the supernatant whose mass spectrum was consistent with isobutylcatechol (metabolite b in Table 1). The mass spectrum of this compound had a molecular ion at m/z 250, which is consistent with diacetylated isobutylcatechol. The two acetyl groups, which were added during aqueous acetylation, are diagnostic of the presence of two aromatic hydroxyl groups (19). Acetyl groups give predictable losses of m/z 42, which in this case accounted for the peaks at m/z 208 and 166. The other large peak at m/z 123 represents a loss of 43, which is consistent with the loss of the isopropyl group from the base ion fragment. The combination of this mass spectral fragmentation pattern, the derivatizable nature of metabolite during aqueous acetylation, and its accumulation only in the presence of 3-fluorocatechol are strong evidence that the peak detected via GC-MS was indeed isobutylcatechol.
TABLE 1.
GC/MS retention times and major ionsa
| MS | Retention time (min) | Mass (relative abundance) | ||||
|---|---|---|---|---|---|---|
| b | 15.7 | 123 (99) | 166 (100) | 208 (17) | 250 (4) | |
| c | 14.7 | 226 (2) | 167 (100) | 151 (6) | 137 (1) | 123 (8) |
| d | 15.8 | 256 (10) | 225 (10) | 197 (100) | 139 (20) | |
Of isobutylcatechol (b) and two putative isobutylcatechol meta-deavage metabolites (c and d).
Mass spectra were also obtained for two other compounds from extract of a culture that was not poisoned with 3-fluorocatechol and which had accumulated high levels of mcp (metabolites c and d in Table 1). These spectra were never detected in the extracts of cultures poisoned with 3-fluorocatechol. One spectrum was consistent with the mcp of isobutylcatechol: 5-formyl-2-hydroxy-7-methylocta-2,4-dienoic acid (metabolite c in Fig. 2), and the other was consistent with its formyl oxidized derivative, 2-hydroxy-5-isobutylhexa-2,4-dienedioic acid (metabolite d in Fig. 2).
FIG. 2.
Proposed pathway for the metabolism of ibuprofen by Ibu-2. Metabolites b to d were all detected via GC-MS. b, isobutylcatechol; c, 5-formyl-2-hydroxy-7-methylocta-2,4-dienoic acid; d, 2-hydroxy-5-isobutylhexa-2,4-dienedioic acid.
As expected, treatment with diazomethane methylated both the acidic hydroxyl and the alpha carbon hydroxyl groups of metabolite c, giving a molecular ion of m/z 226. The m/z 167 fragment represents a loss of 59 from the parent ion, which is consistent with the loss of a methylated carboxyl group and is a common loss from aliphatic esters. The m/z 137 fragment is consistent with the loss of CH2O from the m/z 167 fragment.
The major fragments of metabolite d (m/z 256/225/197/139) are consistent with the expected transformation product of metabolite c. After derivatization, this would be expected to have three additional methyl groups, one on each acidic hydroxyl group and one on the alpha hydroxyl group. A loss of 31 to give m/z 225 is consistent with loss of CH2OH from the parent ion. An alternative loss from the parent ion (m/z 256) yielded a fragment with m/z 197 and is consistent with the loss of a methylated carboxylic acid group (−59). Further impact of this fragment would be expected to result in a loss of 58, which would correspond to removal of the second methylated carboxylic acid group and yield the fragment with m/z 139.
Substrate specificity analysis.
An Ibu-2 washed-cell suspension was able to metabolize phenylacetic acid, 3- and 4-tolylacetic acids, 2-phenylpropionic acid, and 2-(4-tolyl)-propionic acid. However, it was not able to metabolize 2-phenylbutyric acid or 2,2-diphenylacetic acid, which may indicate that the nature of the aliphatic substitution on the alpha carbon could be important. Neither phenol nor any methylphenol was metabolized, making it less likely that a phenolic metabolite was involved. A washed-cell suspension could not metabolize either mandelic acids or tropic acid (2-phenyl-3-hydroxypropionic acid), implying that hydroxylation of the acid side chain was not an intermediate step in side chain removal. Furthermore, the washed-cell suspension was not able to metabolize benzoate or 4-methylbenzoate, suggesting that benzoic acids, which have been shown to be intermediates in the anaerobic degradation of phenylacetic acid by a Pseudomonas sp. and by Azoarcus evansii (21), were not likely to be intermediates in ibuprofen degradation. Other compounds that were not metabolized include 4-tertbutylcatechol, phenylacetaldehyde, 2-tolylacetic acid, 2- and 4-(hydroxyphenyl)acetic acid, 3-phenylpropionic acid, and 3-phenyl-2-propenoic acid (cinnamic acid). In all cases, it is possible that lack of metabolism was due to lack of transport into the cell.
Growth substrate analysis.
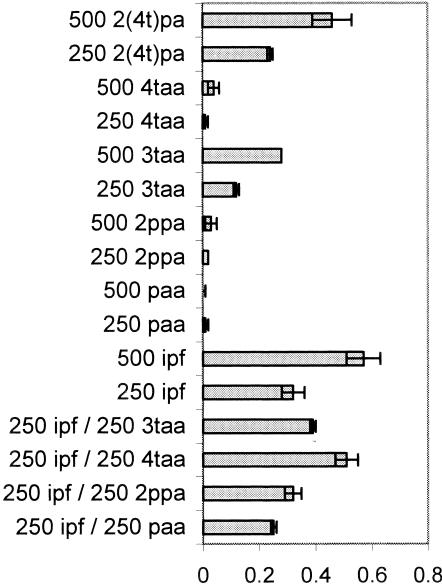
The only ibuprofen analogs that supported growth of Ibu-2 without the presence of ibuprofen as an inducer were 3-tolylacetic acid and 2-(4-tolyl)propionic acid. However, 4-tolylacetic acid was able to support growth when ibuprofen was also present in the medium. Phenylacetic acid and 2-phenylpropionic acid did not support growth under any conditions (Fig. 3). Surprisingly, 3-tolylacetic acid supported significantly less growth than was permitted by ibuprofen or 4-tolylacetic and 4-tolylpropionic acids (P < 0.02). This suggests the possibility of toxicity or incomplete metabolism. If the latter were true, accumulation of a metabolite might have been predicted. However, no dead-end metabolites were detected when the culture supernatant from 3-tolylacetic acid-grown cells was subjected to HPLC or GC-MS analysis (data not shown).
FIG. 3.
Average final culture density (n = 3) as measured by optical density (OD) at 600 nm when Ibu-2 was inoculated in liquid MSM culture containing ibuprofen or its analogs [ipf, ibuprofen; paa, phenylacetic acid; 2ppa, 2-phenylpropionic acid; 3taa, 3-tolylacetic acid; 4taa, 4-tolylacetic acid; 2(4t)pa, 2-(4-tolyl)propionic acid]. The error bars represent standard deviations.
Characterization of catechols produced from ibuprofen analogs.
When the supernatants of WCS that had been incubated with ibuprofen analogs and 3-fluorocatechol were examined via HPLC, each exhibited a novel peak. These peaks were present only after the addition of 3-fluorocatechol. In the phenylacetic acid and 2-phenylpropionic acid samples, a peak appeared whose retention time matched that of catechol (6.4 min). Within 2 h, almost 60% of the phenylacetic acid added was converted to catechol, and no other intermediates were detected, suggesting that the observed deacylation was not merely an unproductive side reaction. In the 3-tolylacetic acid sample, a peak appeared whose retention time matched that of 3-methylcatechol (13.5 min). In the 4-tolylacetic acid and 2-(4-tolyl)-propionic acid samples, a peak appeared whose retention time matched that of 4-methylcatechol (11.6 min).
Characterization of meta-cleavage products of ibuprofen analogs.
The mcps of phenylacetic acid, 2-phenylpropionic acid, and catechol all had the same maximum absorbance wavelength (378 nm). The mcps of 3-tolylacetic acid and 3-methylcatechol also had the same maximum absorbance wavelength (380 nm), as did 4-tolylacetic acid, 2-(4-tolyl)-propionic acid, and 4-methylcatechol (384 nm). These observations all suggest that deacylation of the acidic side chain occurred before ring cleavage.
Cell extract activities upon ibuprofen.
Ibu-2 cell extracts readily produced mcp from catechol, 3-methylcatechol, and 4-methylcatechol (data not shown). Ibu-2 cell extracts did not generate any yellow color when incubated with ibuprofen with or without the addition of NADH/ferrous iron, nor was any ibuprofen disappearance detected via HPLC under any conditions. The Ibu-2 cell extracts did not reduce NAD+ or NADP+ in the presence of either ibuprofen or tropic acid, although the positive control reduced NAD+ in the presence of tropic acid. Cofactor reduction would have been expected if Ibu-2 used a Pseudomonas sp. strain AT3-like mechanism to oxidize the side chain of ibuprofen.
Conclusions.
Unlike Variovorax sp. strain Ibu-1, which has been suggested to dioxygenate the aromatic ring of ibuprofen in the 2 and 3 positions (22), Ibu-2 appears to metabolize ibuprofen through a novel mechanism resulting in removal of the propionic acid moiety and dioxygenation of the ring at the 1,2 position, giving rise to isobutylcatechol. The accumulation of this compound when and only when a meta-cleavage inhibitor was added suggests that isobutylcatechol is further metabolized via meta-cleavage. The identification of compounds consistent with the meta-cleavage of isobutylcatechol only when a meta-cleavage inhibitor was not added lends further support to this conclusion.
As a similar side chain removal of related 2-phenylpropionic acids or phenylacetic acids has not been previously reported, the ability of Ibu-2 to metabolize other aromatic acids was determined. The production of catechols and their respective mcps from these compounds provides additional evidence for this unique pathway.
The exact mechanism whereby Ibu-2 accomplished this acid side chain removal remains to be determined. Although it is likely that other steps were required to activate the acid moiety prior to removal, we could not detect any evidence suggesting the involvement of other intermediates prior to the formation of catechols. Further work will be required before the mechanism whereby this is accomplished can be elucidated.
Acknowledgments
R.W.M. was supported in part by NIH-NIEHS Environmental and Molecular Toxicology training grant ES 07052-27.
Pseudomonas sp. strain AT3 was a kind gift from David J. Hopper.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andreoni, V., G. Baggi, S. Bernasconi, and M. Zongrossi. 1992. Metabolism of phenylpropanoid compounds by Pseudomonas cepacia. Ann. Microbiol. Enzimol. 42:261-266. [Google Scholar]
- 3.Buser, H. R., T. Poiger, and M. D. Muller. 1999. Occurrence and environmental behavior of the chiral pharmaceutical drug ibuprofen in surface waters and in wastewater. Environ. Sci. Technol. 33:2529-2535. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., and J. P. N. Rosazza. 1994. Microbial transformation of ibuprofen by a Nocardia species. Appl. Environ. Microbiol. 60:1292-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Defrank, J. J., and D. W. Ribbons. 1977. P-cymene pathway in Pseudomonas putida ring cleavage of 2,3-dihydroxy p-cumate and subsequent reactions. J. Bacteriol. 129:1365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Defrank, J. J., and D. W. Ribbons. 1976. The p-cymene pathway in Pseudomonas putida strain Pl isolation of a dihydrodiol accumulated by a mutant. Biochem. Biophys. Res. Commun. 70:1129-1135. [DOI] [PubMed] [Google Scholar]
- 7.Eaton, R. W. 1996. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farre, M., I. Ferrer, A. Ginebreda, M. Figueras, L. Olivella, L. Tirapu, M. Vilanova, and D. Barcelo. 2001. Determination of drugs in surface water and wastewater samples by liquid chromatography-mass spectrometry: methods and preliminary result including toxicity studies with Vibrio fischeri. J. Chromatogr. A 938:187-197. [DOI] [PubMed] [Google Scholar]
- 10.Focht, D. D. 1994. Microbiological procedures for biodegradation research, p. 405-426. In R. W. Weaver, J. S. Angle, and P. S. Bottomley (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. Soil Science Society of America, Madison, Wis.
- 11.Hecht, S. M., and J. W. Kozarich. 1972. A convenient method for the production of diazomethane. Tetrahedron Lett. 13:1501-1502. [Google Scholar]
- 12.Ismail, W., M. E.-S. Mohamed, B. L. Wanner, K. A. Datsenko, W. Eisenreich, F. Rohdlich, A. Bacher, and G. Fuchs. 2003. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270:3047-3054. [DOI] [PubMed] [Google Scholar]
- 13.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 36:1202-1211. [DOI] [PubMed] [Google Scholar]
- 14.Krieg, N. R. 1981. Enrichment and isolation, p. 112-142. In P. Gerhardt (ed.), Manual of Methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 15.Kuge, Y., K. Mochida, and T. Uwajima. 1991. Microbial hydroxylation of 2 phenylpropionic acid. Agr. Biol. Chem. Tokyo 55:1099-1104. [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. S. M. Goodfellow (ed.), Nucleic Acid Techniques in Bacterial Systems. Wiley, Chichester, United Kingdom.
- 17.Lee, E. J. D., K. Williams, R. Day, G. Graham, and D. Champion. 1985. Stereoselective disposition of ibuprofen enantiomers in man. Br. J. Clin. Pharmacol. 19:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long, M. T., B. A. Bartholomew, M. J. Smith, P. W. Trudgill, and D. J. Hopper. 1997. Enzymology of oxidation of tropic acid to phenylacetic acid in metabolism of atropine by Pseudomonas sp. strain AT3. J. Bacteriol. 179:1044-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mars, A. E., T. Kasberg, S. Kaschabek, M. Van Agteren, D. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCullar, M., R. Brenner, R. Adams, and D. Focht. 1994. Construction of a novel polychlorinated biphenyl-degrading bacterium; utilization of 3,4′-dichlorobiphenyl by Pseudomonas acidovorans M3GY. Appl. Environ. Microbiol. 60:3833-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed, M. E.-S., W. Ismail, J. Heider, and G. Fuchs. 2002. Aerobic metabolism of phenylacetic acids in Azoarcus evansii. Arch. Microbiol. 178:180-192. [DOI] [PubMed] [Google Scholar]
- 22.Murdoch, R. W., and A. G. Hay. 2002. Isolation of a bacterium capable of using S-ibuprofen as a sole carbon source, p. 405-406. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, D.C.
- 23.Pollock, T. 1993. Gellan-related polysaccharides and the genus Sphingomonas. J. Gen. Microbiol. 139:1939-1945. [Google Scholar]
- 24.Richau, J., J. Leitao, and I. Sa-Correia. 2000. Enzymes leading to the nucleotide sugar precursors for exopolysaccharide synthesis in Burkholderia cepacia. Biochem. Biophys. Res. Commun. 276:71-76. [DOI] [PubMed] [Google Scholar]
- 25.Rudy, A. C., P. M. Knight, D. C. Brater, and S. D. Hall. 1991. Stereoselective metabolism of ibuprofen in humans: administration of R-, S- and racemic ibuprofen. J. Pharmacol. Exp. Ther. 259:1133-1139. [PubMed] [Google Scholar]
- 26.Simo, C., A. Gallardo, J. San Roman, C. Barbas, and A. Cifuentes 2002. Fast and sensitive capillary electrophoresis method to quantitatively monitor ibuprofen enantiomers released from polymeric drug delivery systems. J. Chromotogr. B 767:35-43. [DOI] [PubMed] [Google Scholar]
- 27.Sparnins, V. L., and P. J. Chapman. 1976. Catabolism of l-tyrosine by the homoprotocatechuate pathway in gram-positive bacteria. J. Bacteriol. 127:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stumpf, M., T. A. Ternes, R. D. Wilken, S. V. Rodrigues, and W. Baumann. 1999. Polar drug residues in sewage and natural waters in the state of Rio de Janeiro, Brazil. Sci. Total Environ. 225:135-141. [DOI] [PubMed] [Google Scholar]
- 29.Van Den Tweel, W. J. J., J. P. Smits, and J. A. M. De Bont. 1988. Catabolism of dl-alpha phenylhydracrylic, phenylacetic, and 3 and 4-hydroxyphenylacetic acid via homogentisic acid in a Flavobacterium sp. Arch. Microbiol. 149:207-213. [Google Scholar]
- 30.Winkler, M., J. R. Lawrence, and T. R. Neu. 2001. Selective degradation of ibuprofen and clofibric acid in two model river biofilm systems. Water Res. 35:3197-3205. [DOI] [PubMed] [Google Scholar]
- 31.Zwiener, C., S. Seeger, T. Glauner, and F. H. Frimmel. 2002. Metabolites from the biodegradation of pharmaceutical residues of ibuprofen in biofilm reactors and batch experiments. Anal. Bioanal. Chem. 372:569-575. [DOI] [PubMed] [Google Scholar]