Abstract
Downward fluxes of nucleic acids adsorbed onto settling particles play a key role in the supply of organic phosphorus and genetic material to the ocean interior. However, information on pelagic-benthic coupling, diagenesis, and processes controlling nucleic acid preservation in deep-sea sediments is practically nonexistent. In this study, we compared nucleic acid fluxes, sedimentary DNA and RNA concentrations, and the enzymatically hydrolyzable fraction of DNA in a bathyal continental margin (North Aegean Sea) and an open-sea system (South Aegean Sea) of the Eastern Mediterranean. The two systems displayed contrasting patterns of nucleic acid fluxes, which increased significantly with depth in the North Aegean Sea and decreased with depth in the South Aegean Sea. These results suggest that in continental margin and open-ocean systems different processes control the nucleic acid supply to the sea floor. Differences in nucleic acid fluxes were reflected by nucleic acid concentrations in the sediments, which reached extremely high values in the North Aegean Sea. In this system, a large fraction of DNA may be buried, as suggested by the large fraction of DNA resistant to nuclease degradation and by estimates of burial efficiency (ca. eight times higher in the North than in the South Aegean Sea). Overall, the results reported here suggest that the preservation of DNA in deeper sediment layers may be favored in benthic systems characterized by high sedimentation rates.
Downward fluxes of particles exported from the ocean surface play a key role in the fuelling of organic material to the ocean interior, where it is mostly recycled (24). Although the intensity and temporal variability of vertical fluxes of organic matter to the deep sea can vary widely, previous investigations highlighted the intimate link between primary productivity in the euphotic zone and the particle rain (see reference 30 and citations therein). However, in topographically complex deep-sea marginal areas, mechanisms of particle transfer to the sea floor are dependent upon pathways that are different from those reported for open oceanic systems. These pathways include direct settling from the surface, down-slope transport, and rebound and/or recycling of bottom particles (3, 34, 41). In continental slopes, high sedimentation rates are typically observed, and in deep-sea areas adjacent to land, pelagic-benthic coupling is a more complex process than in open-ocean settings (21, 33). These factors, together with the slope topography, may lead to the formation of organic-matter hot spots characterized by fast degradation and burial rates. In this regard, previous studies reported carbon remineralization and burial rates for continental slope sediments that were 5 to 10 times higher than those for deep-sea sediments of open oceanic regions (1, 25, 36, 42). Organic matter buried in continental margin sediments may also be better preserved than that in open-ocean sediments (19, 20). All of these features suggest that marginal deep-sea areas and subsurface sediment layers may represent models for investigating the diagenesis and preservation of organic molecules (23, 35, 38, 39).
Recent studies have also shown that large amounts of extracellular DNA may reach the deep ocean floor adsorbed onto settling particles (up to ca. 10 mg DNA m−2 year−1 at a 4,800-m depth [12]). The nucleic acid supply may therefore contribute to the transfer of labile organic phosphorus from the water column to the sediment, with potential important implications both in phosphorus biogeochemical cycling and in benthic trophodynamics (9, 13). In this regard, previous studies reported that the regeneration of phosphates from the extracellular DNA pool in marine sediments may supply >40% of the daily benthic bacterial P requirement (14). At the same time, extracellular DNA escaping remineralization may be preserved in deeper sediment layers, thus representing a molecular marker for improving the reconstruction of past communities and related paleo-environments (7, 8). Despite the potential relevance of nucleic acids in benthic biogeochemical cycles and paleo-ecological reconstructions, information on pelagic-benthic coupling, diagenesis, and processes controlling nucleic acid preservation in marine sediments is practically nonexistent (12, 13).
In the present study, we compared nucleic acid fluxes, sedimentary DNA and RNA concentrations, and the enzymatically hydrolyzable fraction of DNA in a bathyal continental margin and an open-sea system of the Eastern Mediterranean. The contrasting features of the two systems are expected to influence the magnitude of nucleic acid supply to the deep-sea floor and related diagenetic processes, thus providing new insights into factors controlling pelagic-benthic coupling and the fate of nucleic acids in the deep sea.
MATERIALS AND METHODS
Study area and sampling.
Sediment sampling and trap deployments were carried out at two bathyal sites in the Eastern Mediterranean. The site located in the North Aegean Sea (40°14.85′N, 25°11.35′E; 1,250-m depth) is a continental margin ecosystem characterized by mesoeutrophic conditions (primary production values of 30 g C m−2 year−1), high terrigenous inputs from the continent (31), and high sedimentation and mixing rates (0.21 cm year−1 and 4.2 cm2 year−1, respectively [26]). In contrast, the site located in the South Aegean Sea (Cretan Sea; 35°44.7′N, 25°06.0′E; 1,550-m depth) is a typical oligotrophic open-sea system (primary production values of 15 gC m−2 year−1), characterized by a lack of terrigenous inputs (31) and relatively low sedimentation and mixing rates (0.02 cm year−1 and 0.022 cm2 year−1, respectively [26]). Organic carbon concentrations in the sediment were higher in the North than in the South Aegean Sea (31). A shift in the biochemical composition of the biopolymeric C fraction (as the sum of C equivalents of the protein, carbohydrate, and lipid pools) was observed, as the South Aegean was characterized by a dominance of carbohydrates, while the North Aegean was dominated by proteins (31).
Trap material was collected in each area by using two sediment traps (Technicap PPS3/3; surface, 0.125 m2) mounted on the same mooring line, placed at a 500-m depth and 35 m above the bottom, and armed with 12 collecting funnels. Trap samples were collected on a fortnightly basis from March to September 1997. Sediment trap samples were fixed in situ with buffered and prefiltered formalin (4% [vol/vol] final concentration) in order to minimize bacterial activity (18). Trap material was processed according to the method of Heussner et al. (22). The sediment trap material was split into 16 fractions, and subsamples were used for the analysis of nucleic acids.
Surface sediment samples (top 40 cm) were collected using a multiple corer (Maxicorer; internal diameter, 9.0 cm). This equipment allows the collection of undisturbed sediment samples and the sediment-water interface. Multiple corer deployments were carried out in March-April 1997 (referred to hereafter as April 1997) and September 1997. During each cruise, four to seven cores were taken from different independent deployments. All cores were sectioned vertically into five layers, i.e., 0 to 1, 1 to 2, 2 to 4, 4 to 6, and 8 to 10 cm, and frozen at −20°C until analysis. These layers were selected to highlight differences in the investigated variables within and below the sediment mixing depth (mixing depth was at 4 to 5 cm) (26).
Nucleic acid analysis of sediment trap material.
Subsamples of sediment trap material (5 to 10 ml) were filtered in duplicate under a vacuum (<100 mm Hg) onto GS-type membrane filters (0.22-μm pore size). Particulate nucleic acid extraction was carried out according to the procedure described by Bailiff and Karl (2). The filters were extracted in 100% acetone for 1 h at −20°C and then centrifuged, and the supernatants were discarded. The pellets were newly resuspended in cold acetone (100%, −20°C) and extracted for 30 min at −20°C until the filters were completely dissolved (usually two or three separate rinses were required). The samples were then washed once with 90% acetone (4°C), once with 10% trichloroacetic acid (4°C), and twice with 95% ethanol (4°C), and the resulting pellet was resuspended in NH4OH. In order to determine the amount of DNA and RNA in the same subsample, we combined spectrofluorometric DNA determination (using diaminobenzoic acid [27]) and spectrophotometric analysis for total nucleic acids (TNA; measured by absorbance at 260 nm [12]). Particulate DNA concentrations in trap samples were calculated by using a calibration curve of calf thymus DNA and then expressed as equivalents of absorbance at 260 nm in order to calculate (by difference) the absorbance due to RNA, as follows: ARNA = ATNA − ADNA.
RNA absorbance was then converted to a concentration using standard solutions of RNA type III from baker's yeast.
DNA and RNA concentrations determined for trap subsamples (5 to 10 ml) were converted to nucleic acid fluxes as follows: (i) DNA and RNA concentrations from subsamples were reported relative to the nucleic acid concentrations present in the total volume of the trap cups, (ii) total DNA and RNA concentrations were then divided by the surface area of the collecting trap (0.125 m2) to be reported relative to the surface area of 1 m2, and (iii) this value was divided by the number of days occurring at each deployment (i.e., the interval between an open and closed trap for each sampling period). DNA and RNA fluxes were then expressed in μg m−2 day−1.
Nucleic acid analysis of sediments.
Before analysis, large macroscopic organisms (i.e., macrofauna), if present, were removed from the sediment samples. Nucleic acid extraction and determinations from frozen sediment subsamples were carried out according to previously described spectrophotometric methods (15, 16). Briefly, 1 g of sediment (three replicates) was treated with 3.0 ml of 0.5 N perchloric acid, stirred for 3 min, and sonicated three times for 1 min (with intervals of 30 s). Nucleic acids were hydrolyzed at 75°C for 30 min with continuous stirring. After centrifugation (3,000 × g, 10 min), the absorbance of TNA content in the supernatant was measured at 260 nm. DNA absorbance was determined with a diphenylamine (2% in acetic acid) light-activated reaction (40 W, 12 h) at 598 nm and converted to a concentration using standard solutions of DNA type I from calf thymus. DNA concentrations were then expressed as equivalents of absorbance at 260 nm in order to calculate the absorbance due to RNA as described above.
The RNA absorbance was then converted to a concentration using standard solutions of RNA type III from baker's yeast. The interference in TNA determination due to organic and inorganic compounds was tested on sediment subsamples which had previously been treated in boiling 0.5 N perchloric acid for 30 min (16). Data were normalized to the sediment dry weight after desiccation (60°C, constant weight).
Estimates of enzymatically hydrolyzable DNA fraction.
The analysis of enzymatically hydrolyzable DNA (HDNA) in sediment samples was carried out according to the method of Dell'Anno et al. (13). This method, based on extracellular DNA hydrolysis by means of commercial nucleases, mimics natural extracellular DNA degradation mediated by cell-associated and extracellular nucleases. Frozen sediment subsamples were stirred at 150 rpm in 2.5 volumes of 0.1 M Tris-HCl, 0.1 M NaCl, 1 mM CaCl2, and 10 mM MgCl2, pH 7.5. Triplicate sediment slurries were treated with DNase I (1.9 U ml−1), nucleases P1 and S1 (4.0 and 2.3 U ml−1, respectively), and exonuclease 3 (1.9 U ml−1); another set of replicates was added with an equal volume of buffer (but without enzymes) and utilized as a control. Samples were incubated at room temperature for 2 h with gentle agitation. After incubation, all samples were centrifuged at 2,000 × g for 5 min, and supernatants were utilized to determine the amount of DNA released from the sediments. Supernatants were dried under a vacuum and analyzed fluorometrically using diaminobenzoic acid. The fluorescence of hydrolyzed DNA was converted to concentrations using calibration curves obtained from standard solutions of calf thymus DNA. The amount of extracellular DNA hydrolyzed by nucleases was obtained by the difference between the nuclease-treated and untreated control samples. For all sediment types, control samples contained <5% of the extracellular DNA pool released by nucleases. HDNA concentrations were normalized to the sediment dry weight after desiccation (60°C, constant weight).
Statistical analysis.
Analyses of variance (ANOVA) followed by a post hoc Tukey test were carried out to test temporal and spatial differences of the investigated variables.
RESULTS
Nucleic acid fluxes.
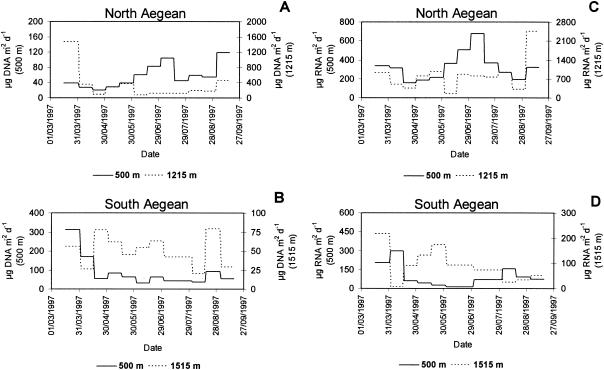
DNA and RNA fluxes in the North and South Aegean Sea at a 500-m depth and 35 m above the bottom are reported in Fig. 1A to D. Nucleic acid fluxes in the North Aegean Sea were significantly higher than those in the South Aegean Sea (P < 0.01), with the only exception being DNA fluxes measured at a 500-m depth, which did not show significant differences. In the North Aegean Sea, DNA and RNA fluxes increased significantly with the water column depth (on average, from 56.8 ± 8.9 to 316.3 ± 110.9 μg DNA m−2 day−1 and from 326.7 ± 42.8 to 814.8 ± 168.3 μg RNA m−2 day−1; for both variables, P < 0.01). Conversely, in the South Aegean Sea, nucleic acid fluxes decreased with increasing water depths (from 87.8 ± 23.2 to 49.6 ± 5.5 μg DNA m−2 day−1 and from 94.2 ± 24.8 to 87.5 ± 17.7 μg RNA m−2 day−1; for DNA, P < 0.01).
FIG. 1.
Temporal changes in nucleic acid fluxes. The data reported are DNA fluxes in the North (A) and South (B) Aegean Sea and RNA fluxes in the North (C) and South (D) Aegean Sea at a 500-m depth and 35 m above the bottom.
Nucleic acid fluxes in the North Aegean Sea were characterized by significant temporal changes (P < 0.01 for both traps). DNA and RNA fluxes in the trap at the 500-m depth were characterized by similar temporal patterns, with the highest values in July and the lowest in May. In contrast, temporal patterns of nucleic acid fluxes in the deepest trap were opposite, with the highest DNA fluxes in March and the highest RNA fluxes in September 1997. In the South Aegean Sea, significant temporal changes of DNA and RNA fluxes were only observed in the trap at the 500-m depth (for both variables, P < 0.01).
Nucleic acid concentrations in sediments.
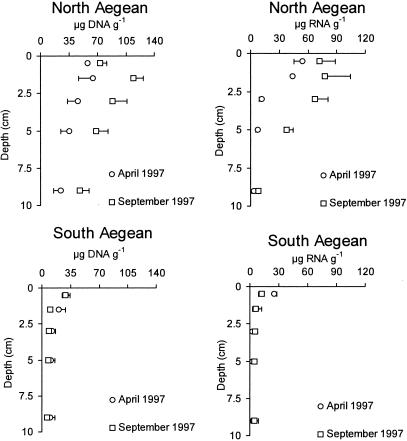
DNA concentrations in the sediment from the North Aegean Sea were about 4.5 times higher than those observed from the South Aegean Sea (integrated value for 0 to 10 cm for the two sampling periods, 55.5 ± 8.7 and 12.0 ± 3.8 μg DNA g−1, respectively; P < 0.01) (Fig. 2). Similarly, RNA concentrations were significantly higher in sediments from the North Aegean Sea than in those from the South Aegean Sea (28.7 ± 11.0 and 5.8 ± 2.9 μg RNA g−1, respectively; P < 0.01) (Fig. 2).
FIG. 2.
Vertical distribution of DNA concentrations in sediments from the North and South Aegean Sea (left panel) and RNA concentrations in sediments from the North and South Aegean Sea (right panel). Standard errors (n = 3) are reported.
Nucleic acid concentrations in the sediment from the North Aegean Sea were characterized by significant temporal changes, with higher values in September than in April 1997 (P < 0.01 for both DNA and RNA). In the South Aegean Sea, DNA concentrations showed a limited temporal variability (not significant by ANOVA), whereas RNA concentrations displayed significant differences between the two sampling periods (P < 0.01).
Sediments from the North Aegean Sea were characterized by subsurface maxima of DNA and RNA at the 1- to 2-cm sediment layer, whereas nucleic acid concentrations in the sediment from the South Aegean Sea decreased progressively from the surface down to the deepest sediment layer (P < 0.01).
Enzymatically hydrolyzable DNA in sediments.
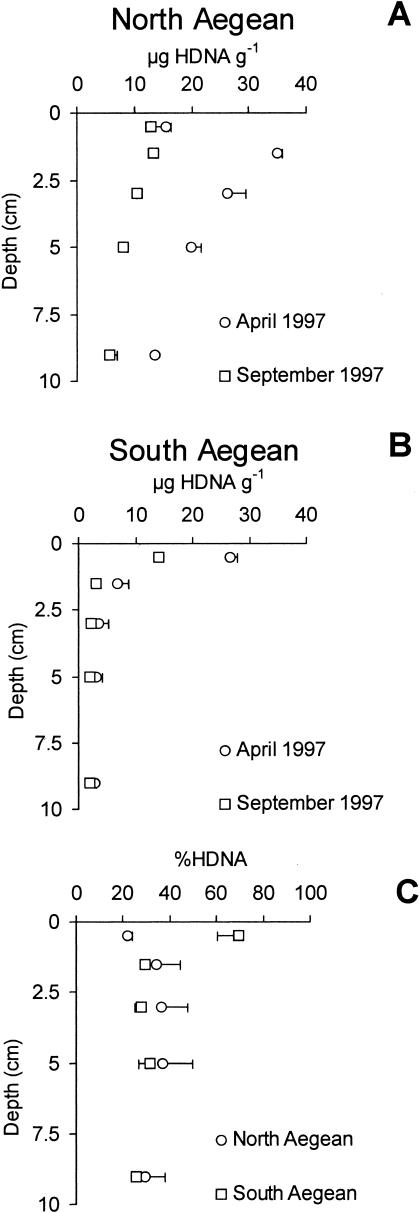
Enzymatically hydrolyzable DNA concentrations (HDNA) in sediments from the North Aegean Sea were about three times higher than those in sediments from the South Aegean Sea (integrated value, 14.6 ± 2.5 and 4.6 ± 3.5 μg HDNA g−1, respectively; P < 0.01) (Fig. 3). In the North Aegean Sea, HDNA concentrations were about double in April (20.4 ± 3.9 μg HDNA g−1) compared to those in September 1997 (8.9 ± 1.4 μg g−1; P < 0.01) (Fig. 3A). Similar temporal changes were observed in the sediments from the South Aegean Sea (5.8 ± 1.1 and 3.3 ± 0.1 μg HDNA g−1 in April and September 1997, respectively; P < 0.05) (Fig. 3B).
FIG. 3.
Vertical distribution of enzymatically hydrolyzable DNA (HDNA) in sediments from the North (A) and South (B) Aegean Sea and average contribution of the enzymatically hydrolyzable DNA fraction (C) to the total DNA pools (HDNA, expressed as a percentage). Standard errors (n = 3) are reported.
HDNA concentrations in the North Aegean Sea displayed the highest values at the 1- to 2-cm sediment horizon and the lowest values at the 8- to 10-cm sediment layer (P < 0.01). HDNA concentrations in the South Aegean Sea decreased significantly with increasing depths of the sediment (P < 0.01).
In the North Aegean Sea, the contribution of HDNA to the total DNA pool remained relatively constant with the depth of the sediment (average for the two sampling periods, 22% ± 2% and 37% ± 13% in the top 1-cm and the 4- to 6-cm sediment layers, respectively), whereas in the South Aegean Sea such a contribution decreased significantly (from 70% ± 9% to 26% ± 2% in the top 1-cm and the 8- to 10-cm sediment layers, respectively; P < 0.01) (Fig. 3C).
DISCUSSION
Pelagic-benthic coupling of nucleic acids.
Previous studies dealing with downward fluxes of nucleic acids reported that changes in the DNA supply to the ocean interior are controlled by changes in primary productivity (2, 40). Therefore, differences in primary production between the North and the South Aegean Sea were expected to influence the downward fluxes of nucleic acids. However, we observed at a 500-m depth a lack of significant differences in DNA fluxes between the two sites. The two systems were under different hydrodynamic conditions (i.e., there was stronger lateral advection in the northern site [31]), which might have influenced the efficiency of sediment traps in collecting settling material (28). However, this was not the case for the present study, as significant differences in organic carbon fluxes were measured from the same set of samples (with values about double for the North Aegean Sea compared to those for the South Aegean Sea) (31). Therefore, the most likely explanation for the uncoupling of particle export from the photic zone and nucleic acid fluxes is that DNA molecules are more rapidly degraded in the North than in the South Aegean Sea. This was further supported by the analysis of RNA fluxes, which at a 500-m depth were about four times higher in the North Aegean than in the South Aegean Sea.
Different temporal changes were observed between the two sites, with the highest nucleic acid fluxes in early spring in the South Aegean and the highest fluxes in summer in the North Aegean. Such different patterns may be related to the different compositions of sinking organic material. Indeed, direct microscopic observations revealed a fresh input of phytodetritus (i.e., mainly diatom remains) in the South Aegean Sea in early spring, whereas aged material (i.e., amorphous particles) characterized summer downward fluxes in the North Aegean Sea.
In the North Aegean Sea, DNA and RNA fluxes increased significantly with the water column depth, reaching values among the highest reported in the literature (up to ca. 1.5 mg DNA m−2 day−1 and 2.5 mg RNA m−2 day−1) (2, 12, 29). Conversely, nucleic acid fluxes in the South Aegean Sea decreased with depth. Such patterns are consistent with those of total mass and organic matter fluxes reported for the two sites (10, 31). These contrasting patterns reflect the typical features of continental margin versus open-ocean ecosystems, suggesting that different processes controlled the nucleic acid supply to the sea floor. In the North Aegean Sea, the nucleic acid supply appears to be mainly derived from lateral export from the continental shelf down slope. The lateral control of particle supply to the deep site of the North Aegean Sea is suggested by the highly significant relationship between total mass and DNA fluxes (n = 12; R2 = 0.907; P < 0.01) (data on total mass fluxes are summarized in reference 31). Conversely, the vertical patterns reported for the South Aegean Sea suggest that the nucleic acid supply depends on vertical fluxes of particles produced in the photic layer.
Differences in nucleic acid fluxes from the deepest traps were reflected by the DNA and RNA concentrations in the sediment. Nucleic acid concentrations in the North Aegean sediments were high and comparable with values reported for coastal sediments (5, 32), thus suggesting the presence of a depocenter of nucleic acids in bathyal sediments of marginal areas. Conversely, DNA and RNA concentrations in the sediment from the South Aegean Sea were typical of deep sites of open-ocean regions (11, 12, 17).
Benthic biomass contributes to DNA concentrations in the sediment. However, estimates of benthic bacterial abundance carried out on the same sediment cores utilized for this study (average for the two sampling periods, 4.97 × 108 ± 1.25 × 108 cells g−1 in the North Aegean Sea and 1.88 × 108 ± 0.06 × 108 cells g−1 in the South Aegean Sea [data are summarized from reference 10]) revealed that DNA associated with bacterial cells accounted for a very small fraction of total DNA concentrations (on average, ca. 2% for both sites). Since bacterial biomass accounts for up to 90% of the total benthic biomass, these results indicate that the majority of DNA pools in the sediment are extracellular (13, 15). Therefore, we conclude that DNA concentrations in surface sediments may depend, to a large extent, on the magnitude of DNA input to the deep-sea floor.
Temporal changes in nucleic acid concentrations in the sediment were generally coupled with changes in nucleic acid fluxes (i.e., March versus September 1997). In the South Aegean Sea, both DNA concentrations in the sediments and downward fluxes displayed a reduced temporal variability. In both areas, sedimentary RNA concentrations displayed significant temporal changes coupled with changes in RNA fluxes. The results presented here suggest that temporal changes in sedimentary nucleic acid concentrations may be detected when clear changes in nucleic acid fluxes occur. However, in the North Aegean Sea the temporal changes in sedimentary DNA concentrations revealed a pattern opposite to the one reported for particulate DNA fluxes, suggesting an uncoupling between DNA deposition and degradation rates. This was further confirmed by the analysis of temporal changes of enzymatically hydrolyzable DNA, which revealed a significant decrease in the degradable fraction with an increase in DNA pool size.
Diagenesis of nucleic acids in the sediment.
Once deposited on surface sediments, DNA undergoes complex biogeochemical transformations, which may strongly affect its diagenesis (13). However, the diagenetic processes influencing DNA degradation and burial in the sediment are still largely unknown (7, 14).
Previous studies carried out in deep-ocean systems reported a sharp decrease in nucleic acid concentrations along the sediment core (12, 17). Similarly, in the South Aegean, nucleic acid concentrations decreased exponentially with depth in the sediment core, whereas sediments from the North Aegean Sea displayed subsurface maxima. Such differences appear to be dependent on the strong differences in sedimentation and mixing rates reported for these bathyal sediments (26).
In order to explore the fate of nucleic acids, we determined the fraction of DNA that was hydrolyzable by nucleases in the different sediment layers. Our results indicated that, on average, ca. 80% of the total DNA pool in the top 1 cm of the sediment from the North Aegean Sea was represented by DNAs resistant to DNase degradation. Conversely, in the South Aegean the fraction resistant to nuclease degradation was ca. 30%. The different degradabilities of DNA pools are difficult to explain. However, it is known that mineral particles (e.g., clay particles) may strongly adsorb and stabilize DNA against DNase degradation (37). The large dominance of the lithogenic fraction of the material fuelling the deep North Aegean Sea (annual average, ca. 75% [31]) is likely responsible for such an effect (i.e., DNA stabilization against DNase attack). This was confirmed by analysis of the vertical profiles of the North Aegean Sea, which revealed a similar fraction of degradable DNA in surface sediment and deeper sediment layers.
Previous studies hypothesized that the accumulation of DNA in marine sediments might be dependent on an imbalance between DNA degradation and burial rates (14). To test this hypothesis, we estimated DNA decay rates based on a steady-state diagenetic kinetic model (4) and burial efficiencies by a mass balance approach (6). DNA decay rates were, on average, ca. five times higher in the sediment of the North Aegean than in that of the South Aegean Sea (0.14 ± 0.06 year−1 and 0.027 ± 0.003 year−1, respectively). As a consequence, the amounts of DNA remineralized in the sediments of the North Aegean (on average, 67.9 ± 3.2 mg DNA m−2 year−1) were much larger than those of the South Aegean Sea (on average, 4.6 ± 0.8 mg DNA m−2 year−1). These findings are consistent with synoptic in situ measurements of sediment community oxygen consumption, which in the North Aegean were ca. seven times higher than in the South Aegean Sea (119 and 18 μmol O2 m−2 h−1, respectively [31]). However, burial efficiency was much lower in sediments of the South Aegean than in those of the North Aegean Sea (7% ± 2% and 53% ± 17%, respectively). Therefore, although DNA was recycled at a higher rate in the North Aegean, ca. half of the DNA deposited on the sediment surface may escape remineralization and be buried.
Previous studies reported that extracellular DNA preservation in sedimentary deposits over geological time scales (from 1,000 to >100,000 years) is favored by low temperatures and permanent anoxic conditions (7, 8, 43). The results reported here suggest that large amounts of DNA may also be preserved in benthic systems characterized by a relatively high temperature (ca. 13°C) and high sedimentation rates.
Further studies are needed to clarify the relative importance of abiotic (e.g., oxygen availability, grain size and mineralogical composition, and pH) and biotic (e.g., DNase-mediated degradation rates) factors controlling the diagenesis and preservation of DNA in marine sediments.
Acknowledgments
This work was financially supported by the EU within the framework of the program MATER (“Mass Transfer and Ecosystem Response”; contract MAS3-CT-950018) and by HERMES (“Hotspot Ecosystem Research on the Margins of European Seas”; contract GOCECT-2005-511234-1) under the Sixth Framework Programme.
REFERENCES
- 1.Anderson, R. F., G. T. Rowe, P. F. Kemp, S. Trumbores, and P. E. Biscaye. 1994. Carbon budget for the mid-slope depocenter of the Middle Atlantic Bight. Deep Sea Res. II 41:669-703. [Google Scholar]
- 2.Bailiff, D. M., and D. M. Karl. 1991. Dissolved and particulate DNA dynamics during a spring bloom in the Antarctic Peninsula region, 1986-87. Deep Sea Res. I 38:1077-1095. [Google Scholar]
- 3.Bauer, J. E., and E. R. M. Druffel. 1998. Ocean margins as a significant source of organic matter to the deep ocean. Nature 329:482-485. [Google Scholar]
- 4.Berner, R. A. 1980. Early diagenesis. A theoretical approach. Princeton University Press, Princeton, N.J.
- 5.Boon, A. R., G. C. A. Duineveld, and A. Kok. 1999. Benthic organic matter supply and metabolism at depositional and non depositional area in the North Sea. Estuar. Coast. Shelf Sci. 49:747-761. [Google Scholar]
- 6.Burdige, D. J., and C. S. Martens. 1988. Biogeochemical cycling in an organic-rich marine basin: the role of amino acids in sedimentary carbon and nitrogen cycling. Geochim. Cosmochim. Acta 52:1571-1584. [Google Scholar]
- 7.Coolen, M. J., and J. Overmann. 1998. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coolen, M. J. L., G. Muyzer, W. I. C. Rijpstra, S. Schouten, J. K. Volkman, and J. S. Sinninghe Damsté. 2004. Combined DNA and lipid analyses of sediments reveal changes in Holocene haptophyte and diatom populations in an Antarctic lake. Earth Planet. Sci. Lett. 223:225-239. [Google Scholar]
- 9.Danovaro, R., A. Dell'Anno, A. Pusceddu, and M. Fabiano. 1999. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the Eastern Mediterranean: relationships with seasonal varying organic inputs and bacterial dynamics. Deep Sea Res. I 46:1077-1094. [Google Scholar]
- 10.Danovaro, R., D. Marrale, N. Della Croce, P. Parodi, and M. Fabiano. 1999. Biochemical composition of sedimentary organic matter and bacterial distribution in the Aegean Sea: trophic state and pelagic benthic coupling. J. Sea Res. 42:117-129. [Google Scholar]
- 11.Danovaro, R., M. Fabiano, and N. Della Croce. 1993. Labile organic matter and microbial biomasses in deep-sea sediments (Eastern Mediterranean Sea). Deep Sea Res. I 40:953-965. [Google Scholar]
- 12.Dell'Anno, A., M. Fabiano, M. L. Mei, and R. Danovaro. 1999. Pelagic-benthic coupling of nucleic acids in an abyssal location of the Northeastern Atlantic Ocean. Appl. Environ. Microbiol. 65:4451-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dell'Anno, A., S. Bompadre, and R. Danovaro. 2002. Quantification, base composition and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47:899-905. [Google Scholar]
- 14.Dell'Anno, A., and C. Corinaldesi. 2004. Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 70:4384-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dell'Anno, A., M. Fabiano, G. C. A. Duineveld, A. Kok, and R. Danovaro. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dell'Anno, A., and R. Danovaro. 2001. Nucleic acid turnover in aquatic environments. 1. Determination of total and extracellular DNA in marine sediments, p. 1-9. In A. D. L. Akkermans, J. D. van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Duineveld, G., M. Lavaleye, E. Berghuis, and P. de Wilde. 2001. Activity and composition of the benthic fauna in the Whittard Canyon and the adjacent continental slope (NE Atlantic). Oceanol. Acta 24:69-83. [Google Scholar]
- 18.Gardner, W. D., K. R. Hinga, and J. Marra. 1983. Observation on the degradation of biogenic material in the deep ocean with implication on accuracy of sediment trap fluxes. J. Mar. Res. 41:195-214. [Google Scholar]
- 19.Goni, M. A., K. C. Ruttenberg, and T. I. Eglinton. 1997. Sources and contribution of terrigenous organic carbon to surface sediments in the Gulf of Mexico. Nature 389:275-278. [Google Scholar]
- 20.Hartnett, H. E., R. G. Keil, J. Hedges, and A. H. Devol. 1998. Influence of oxygen exposure time on organic carbon preservation in continental margin sediments. Nature 391:572-575. [Google Scholar]
- 21.Herman, P. M. J., K. Soetart, J. J. Middelburg, C. Heip, L. Lohse, E. Epping, W. Helder, A. N. Antia, and R. Peinert. 2001. The seafloor as the ultimate sediment trap—using sediment properties to constrain benthic-pelagic exchange processes at the Goban Spur. Deep Sea Res. II 48:3245-3264. [Google Scholar]
- 22.Heussner, S., C. Ratti, and J. Carbonne. 1990. The PPS3 sediment trap and trap sample processing techniques used during the ECOMARGE experiment. Cont. Shelf Res. 10:943-958. [Google Scholar]
- 23.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahnke, R. A. 1996. The global ocean flux of particulate organic carbon: areal distribution and magnitude. Global Biogeochem. Cycles 10:71-88. [Google Scholar]
- 25.Jahnke, R. A., C. E. Reimers, and D. B. Craven. 1990. Intensification of recycling of organic matter at the seafloor near ocean margins. Nature 348:50-54. [Google Scholar]
- 26.Kaberi, H., and V. Lykousis. 2004. Recent sedimentation rates in the Aegean Sea, p. 39. In 37th CIESM Congress Proceedings, Barcelona, Spain.
- 27.Karl, D. M., and M. D. Bailiff. 1989. The measurement and distribution of dissolved nucleic acids in aquatic environments. Limnol. Oceanogr. 34:543-558. [Google Scholar]
- 28.Karl, D. M., G. A. Knauer, and J. H. Martin. 1988. Downward flux of particulate organic matter in the ocean: a particle decomposition paradox. Nature 332:438-441. [Google Scholar]
- 29.Karl, D. M., and G. A. Knauer. 1984. Vertical distribution, transport, and exchange of carbon in the northeast Pacific Ocean: evidence for multiple zones of biological activity. Deep Sea Res. I 31:221-243. [Google Scholar]
- 30.Lampitt, R. S., and A. N. Antia. 1997. Particle flux in deep-seas: regional characteristics and temporal variability. Deep Sea Res. I 44:1377-1403. [Google Scholar]
- 31.Lykousis, V., G. Chronis, A. Tselepides, N. B. Price, A. Theocharis, I. Siokou-Frangou, F. Van Wambeke, R. Danovaro, F. S. Stavrakakis, G. Duineveld, D. Georgopoulos, L. Ignatiades, A. Souvermezoglou, and F. Voutsinou-Taliadouri. 2002. Major outputs of the recent multidisciplinary biogeochemical researches undertaken in the Aegean Sea. J. Mar. Syst. 33-34:313-334. [Google Scholar]
- 32.Manini, E., M. Fabiano, and R. Danovaro. 2000. Benthic response to mucilaginous aggregates in the Northern Adriatic Sea: biochemical indicators of eutrophication. Chem. Ecol. 17:171-179. [Google Scholar]
- 33.Monaco, A., P. Biscaye, J. Soyer, R. Pocklington, and S. Heussner. 1990. Particle fluxes and ecosystem response on a continental margin: the 1985-1988 Mediterranean ECOMARGE experiment. Cont. Shelf Res. 10:809-839. [Google Scholar]
- 34.Puig, P., A. S. Ogston, B. L. Mullenbach, C. A. Nittrouer, and R. W. Sternberg. 2003. Shelf-to-canyon sediment-transport processes on the Eel continental margin (northern California). Mar. Geol. 193:129-149. [Google Scholar]
- 35.Ransom, B., D. Kim, M. Kastner, and S. Wainwright. 1998. Organic matter preservation on continental slopes: importance of mineralogy and surface area. Geochim. Cosmochim. Acta 62:1329-1345. [Google Scholar]
- 36.Reimers, C. E., R. A. Jahnke, and D. C. McCorkle. 1992. Carbon fluxes and burial rates over the continental slope and rise off central California with implications for the global carbon cycle. Global Biogeochem. Cycles 6:199-224. [Google Scholar]
- 37.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1991. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 57:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulte, S., K. Mangelsdorf, and J. Rullkötter. 2000. Organic matter preservation on the Pakistan continental margin as revealed by biomarker geochemistry. Org. Geochem. 31:1005-1022. [Google Scholar]
- 39.Shippers, A., L. N. Neretin, J. Kallmeyer, T. G. Ferdelman, B. A. Cragg, R. J. Parkes, and B. B. Jørgensen. 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433:861-864. [DOI] [PubMed] [Google Scholar]
- 40.Vanucci, S., A. Dell'Anno, A. Pusceddu, M. Fabiano, R. Lampitt, and R. Danovaro. 2001. Microbial assemblages associated to sinking particles in the Porcupine Abyssal Plain. Progr. Oceanogr. 50:105-122. [Google Scholar]
- 41.van Weering, T. C. E., I. R. Hall, H. C. de Stigtera, I. N. McCaveb, and L. Thomsen. 1998. Recent sediments, sediment accumulation and carbon burial at Goban Spur, N.W. European Continental Margin (47-50°N). Progr. Oceanogr. 42:5-35. [Google Scholar]
- 42.van Weering, T. C. E., H. C. de Stigtera, W. Balzer, E. H. G. Epping, G. Graf, I. R. Hall, W. Helder, A. Khripouno, L. Lohse, I. N. McCave, L. Thomsen, and A. Vangriesheim. 2001. Benthic dynamics and carbon fluxes on the NW European continental margin. Deep Sea Res. II 48:3191-3221. [Google Scholar]
- 43.Willerslev, E., A. J. Hansen, J. Binladen, T. B. Brand, M. T. P. Gilbert, B. Shapiro, M. Bunce, C. Wiuf, D. A. Gilichinsky, and A. Cooper. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300:791-795. [DOI] [PubMed] [Google Scholar]