Abstract
Autotrophic ammonia-oxidizing bacteria (AOB) are of vital importance to wastewater treatment plants (WWTP), as well as being an intriguing group of microorganisms in their own right. To date, corroboration of quantitative measurements of AOB by fluorescence in situ hybridization (FISH) has relied on assessment of the ammonia oxidation rate per cell, relative to published values for cultured AOB. Validation of cell counts on the basis of substrate transformation rates is problematic, however, because published cell-specific ammonia oxidation rates vary by over two orders of magnitude. We present a method that uses FISH in conjunction with confocal scanning laser microscopy to quantify AOB in WWTP, where AOB are typically observed as microcolonies. The method is comparatively simple, requiring neither detailed cell counts or image analysis, and yet it can give estimates of either cell numbers or biomass. Microcolony volume and diameter were found to have a log-normal distribution. We were able to show that virtually all (>96%) of the AOB biomass occurred as microcolonies. Counts of microcolony abundance and measurement of their diameter coupled with a calibration of microcolony dimensions against cell numbers or AOB biomass were used to determine AOB cell numbers and biomass in WWTP. Cell-specific ammonia oxidation rates varied between plants by over three orders of magnitude, suggesting that cell-specific ammonia oxidation is an important process variable. Moreover, when measured AOB biomass was compared with process-based estimates of AOB biomass, the two values were in agreement.
The quantification of microbial communities and populations is an invaluable aspect of microbial ecology. In principle, the autotrophic ammonia-oxidizing bacteria (AOB) are ideal candidates for the development of quantitative tools. AOB have a coherent phylogeny and defined nutritional requirements and are of profound practical importance in natural and engineered environments.
The number of individuals should be the ideal benchmark for quantitative studies. Individual counts can be converted to biomass, biovolume, or proportion of biomass, and results obtained by more indirect methods are typically compared to the number of cells per unit volume (15). Fluorescence in situ hybridization (FISH) represents the “gold standard” for quantification of specific bacterial cells in the environment, against which other methods should be compared. Classical (27) and immunological (20) methods are subject to methodological biases, while nonmicroscopic 16S rRNA-based methods (8, 34) or PCR-based methods (13, 14, 18, 19) deliver a proportion of total cell counts, copy number, or relative signal intensities rather than an absolute number of cells or biomass.
A quantitative method may be evaluated with respect to its precision and its accuracy. Wagner et al. (43) originally evaluated the accuracy of FISH counts of AOB by using cell specific oxidation rates, an approach previously used to show that most-probable-number-based methods underestimate AOB numbers (7, 41). Wagner et al. were able to show that the number of AOB detected could, in principle, account for the nitrification rates observed. However, cell-specific reaction rates are likely to be a crude method for corroborating a quantitative procedure, because the rate will vary with environmental conditions and possibly between taxa. For example, published cell-specific reaction rates in pure cultures of AOB vary by one and a half orders of magnitude (0.9 to 53 femtomoles/cell/hour) (7, 24, 39). Cell-specific ammonia oxidation rates estimated in situ are equally variable, but lower, and range from 0.22 to 2.3 femtomoles/cell/hour (reported values of 2.3 femtomoles/cell/hour [10], 0.63 femtomoles/cell/hour [17], 0.22 femtomoles/cell/hour [43], and 0.25 to 0.97 femtomoles/cell/hour [38]). It is impossible to know if the disparity between the rates measured in pure culture studies and rates estimated from in situ measurements is due to overestimation of the AOB community size in situ or to differences in environmental conditions (rates are likely to be a function of temperature, oxygen and ammonia concentrations, AOB taxa present, and the three-dimensional structure of biofilms or flocs). This critique is not new. When Knowles et al. (22a) first proposed the concept of estimating AOB numbers from cell-specific rates, in 1965, they believed that observed uptake rates could be normalized against known maximum specific uptake rates determined in culture. Writing in 1979, Belser (7) pointed out that this approach could be undermined by a discrepancy between the behaviors of pure cultures and AOB in the environment. Much of what we have learned about AOB in the intervening years would appear to confirm this suspicion.
The precision of AOB enumeration by FISH was not explicitly considered in the earliest literature. However, Schramm and colleagues (38) reported that the Shapiro-Wilks test (typically a test for a normal distribution) “showed an uneven distribution for all data,” and they expressed dissatisfaction with the exceptionally large standard deviations. They concluded that their results were only best estimates correct to an order of magnitude. However, high standard deviations and an uneven distribution would be expected if the data were not normally distributed (for example, if the data had a log-normal distribution). Log-normal distributions are associated with entities which grow and die (40).
To overcome, the apparent imprecision of AOB cell counts by FISH, image analysis tools have been used to measure the fluorescence from AOB as a proportion of the fluorescence from the Bacteria overall, and then this is converted to cell numbers by reference to Escherichia coli cells seeded at a known concentration (11). This elegant seven-step procedure yielded a coefficient of variation of 20%. However, the accuracy of the method was been evaluated on the basis of cell-specific ammonia oxidation rates in a single wastewater treatment plant.
Some investigators have reported FISH to be an inferior quantification method. Konuma et al. (23) compared the use of FISH (using Nso190 S-F-bAOB-0189-a-A-19) immunofluorescence and dot blot methods to enumerate AOB. They reported that quantitative FISH in activated sludge was confounded by weak signals, nonspecific binding, and autofluorescence and did not recommend its use. Rittmann and coworkers (34) used FISH and slot blot techniques to quantify AOB in a variety of activated sludge plants and compared their empirical and theoretical biomass estimates. Oligonucleotide probes Nso1225 (S-F-bAOB-1224-a-A-20) and Eub338 (S-D-Bact-0338-a-A-18) were used to detect AOB and Bacteria, respectively. The ratio of AOB to Bacteria obtained by slot blot analysis agreed with theoretical estimates. However, ratios of AOB biomass (obtained by FISH) to mixed-liquor volatile suspended solids (MLVSS) did not agree with theoretical predictions. Ratios obtained by FISH were much lower than predicted. No satisfactory explanation has been offered for this discrepancy. Rittmann et al. (34) tentatively suggested that the majority of AOB were not readily detectable by FISH, possibly because most of the biomass occurred as single cells rather than in microcolonies or failed to hybridize, perhaps due to permeabilization problems.
Rittman et al. (34) put forward an interesting methodology for assessing the amount of AOB that should be present in a system. In essence, they estimated the net production of AOB biomass from reduced ammonia by using the yield to convert ammonia consumed into biomass and simple mass balance concepts to account for losses. They used this technique to compare theoretical and measured biomasses in a number of plants. Thus, if measured and theoretical biomasses corresponded perfectly a plot of theoretical versus measured biomass would have a slope of 1, an r2 of 100, and an intercept of 0. Variation in the intercept would imply a systematic disagreement between the model of Rittman et al. and measured values. Low r2 values or a slope other than 1 would imply site-specific disagreement between theory and measurement. Unfortunately, in the original work the regression line was forced through the origin, and so the ability to assess systematic errors was lost. The accuracy of other published FISH studies cannot be retrospectively evaluated using the approach of Rittmann et al., because they typically focus on a single wastewater treatment plant and do not report the process variables required by the model.
We report a simple method for quantification of AOB by FISH. We explicitly consider the distribution of AOB microcolony sizes, which allows for the estimation of the proportion of AOB not detectable by FISH. The quantity of AOB per unit volume can be expressed as cells per unit volume or biomass. Using our methodology, we show that FISH and process-based estimates of AOB population size in several full- and lab-scale reactors are compatible and that cell-specific ammonia oxidation rates are very variable.
MATERIALS AND METHODS
Activated sludge plants.
Samples from five full-scale activated sludge plants in the United Kingdom (Wanlip, Stoke Bardolph, Preston, Hydburn, and Chorley) treating domestic wastewater and from one laboratory reactor treating artificial wastewater were collected to identify and quantify the AOB population by FISH. Relevant operational parameters of these plants are summarized in Table 1. The laboratory reactor is described in detail elsewhere (4, 5).
TABLE 1.
Typical operational parameters of the wastewater treatment plants
| Plant name | F/Ma (kg BOD/kg MLSS) | Avg flow (megaliters/day) | Aeration tank vol (m3) | Sludge age (days) | Observed hydraulic retention time (h) |
|---|---|---|---|---|---|
| Wanlip | 0.063 | 60.0 | 23,200 | 9.80 | 9.28 |
| Stoke Bardolph | 0.030 | 32.8 | 13,460 | 12.66 | 9.84 |
| Preston | 0.050 | 100.0 | 48,000 | 12.00 | 11.00 |
| Chorley | 0.070 | 27.9 | 7,587 | 6.7 | 6.50 |
| Hydburn | 0.070 | 66.0 | 28,100 | 9.00 | 5.28 |
| Lab reactor | 0.070 | 0.000005 | 0.005 | 20 | 24 |
F/M, food/microorganism ratio.
Culture.
Ralstonia eutropha (DSM 531T) was cultured in nutrient broth (Oxoid, United Kingdom) at 30°C in the dark. Nitrosospira sp. strain 40KI, Nitrosospira sp. strain B6, Nitrosospira sp. strain D11, Nitrosospira sp. strain GM4 (42), Nitrosospira sp. strain C_128, Nitrosospira sp. strain NpAV, Nitrosospira sp. strain Np22.2, and Nitrosomonas eutropha Nm57 were provided by the University of Liverpool culture collection. Nitrosospira multiformis N113 (NCIMB 1184) and Nitrosomonas europaea Nm50 (NCIMB 11850T) were obtained from the NCIMB; all were grown in the inorganic ammonia oxidizer growth medium of Watson and Mandel (45). For some experiments Nitrosomonas europaea (NCIMB 11850T) was grown in Skinner-Walker medium (39) modified to contain 50 μg/ml of ammonia (31). The growth of AOB was monitored by following the change in pH caused by the oxidation of ammonia to nitrite by means of a pH indicator (phenol red) present in the growth medium. When the medium changed color from pink to yellow, filter-sterilized sodium bicarbonate was added to neutralize the growth medium. Cells were harvested after three to five rounds of neutralization.
Sampling.
Grab samples of mixed liquor were preserved immediately in ethanol (sample/ethanol ration, 50:50 [vol/vol]), transported to the laboratory at 4°C, and stored at −20°C until analysis. Prior to FISH analysis, samples were fixed with 4% paraformaldehyde as described by Amann et al. (2). It has been reported in the literature (21, 26) that ethanol fixation can cause sufficient lysis of some gram-negative cells to affect the apparent proportional abundance of certain taxa. However, we did not observe lysis of AOB microcolonies when they were stored for up to 4 weeks.
Oligonucleotide probes.
Oligonucleotide probe nomenclature was based on the Oligonucleotide Probe Database protocol (1). Probes were labeled with fluorescein isothiocyanate, tetramethyl rhodamine isothiocyanate, or indocarbocyanine and were obtained commercially (Genosys, Cambridge, United Kingdom, or ThermoHybaid, Ulm, Germany). A calibration of cell numbers against microcolony dimensions was undertaken using Nso190 (S-F-bAOB-0189-a-A-19), Nsm156 (S-G-Nsm-0155-a-A-19), and Nsv443 (S-G-Nsp-0443-a-S-19) (29) and Nsm641 (S-*-Nsm-0641-a-A-23), a probe specific for the dominant AOB 16S rRNA gene sequence recovered from a laboratory-scale reactor (6). The probes used for counts were Nso1225 (S-F-bAOB-1224-a-A-20), Nso190 (S-F-bAOB-0189-a-A-19), Nsm641 (S-*-Nsm-0641-a-A-23), and Eub338 (S-D-Bact-0338-a-A-18). Negative control analyses using probe nonEub338 (S-D-Bact-0338-a-S-18) were conducted for all samples. It should be noted that Nso1225 has a single-base mismatch with the 16S rRNA of Nitrosococcus mobilis, which is common in reactors treating saline wastewaters, and studies of such systems should use a mixture of Nso1225 and probe NEU (22).
In situ hybridization.
All hybridizations were carried out as follows. Two hundred to 250 microliters of fixed activated sludge samples was placed in a 0.5-ml microcentrifuge tube. The samples were dehydrated in 60%, 80%, and 99.8% ethanol by successive suspension in 1 ml of the appropriate ethanol solution and centrifugation at 3,000 rpm in a microcentrifuge (MSE Microcentaur; MSE UK). After the final dehydration step, the supernatant was discarded and the pellet was resuspended in 38 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris HCl, 0.01% sodium dodecyl sulfate [SDS], X% formamide, where X is the amount of formamide optimal for each probe [Table 2 ]), and 2 μl of labeled probe (50 ng/μl) was added. Negative control hybridizations were done without a probe and with probe nonEub338. Samples were hybridized overnight under the appropriate conditions (Table 2). After hybridization, samples were washed twice in a 0.5 ml washing buffer (20 mM Tris HCl, 0.01% SDS, 5 mM EDTA, and X NaCl, where X is the optimal concentration for each probe [Table 2]) for 15 min at the hybridization temperature, followed by a brief wash in 0.5 ml molecular biology grade water. The samples were centrifuged, and the pellet was resuspended in 10 to 100 μl of filtered, distilled, deionized water. Ten microliters of sample was spotted onto a gelatin-coated slide and allowed to air dry (2). A drop of Citifluor (Citifluor, Kent, United Kingdom) was added to the sample and a coverslip placed over the preparation. The edges of the coverslip were sealed using nail varnish, and prepared slides were stored in the dark at 4°C before viewing. Hybridizations to establish the relationship between cell numbers and microcolony dimensions were conducted in a similar manner except that a complex hybridization buffer [0.9 M NaCl, 50 mM sodium phosphate (pH 7.0), 5 mM EDTA, 0.1% SDS, 0.5 mg of poly(A) per ml, 10× Denhardt's solution] was used (2).
TABLE 2.
Names, target positions, sequences, and specificities of oligonucleotide probes used during this study
| Oligonucleotide | Probe sequence, 5′→3′ | Specificitya | Hybridization/wash conditions (NaCl [mM]b, temp [°C], formamide [%]) | Reference |
|---|---|---|---|---|
| Nsm641 | TGC CGC ACT CTA GCT CTG CAG TT | Nitrosomonas 16S rRNA sequences recovered from lab-scale reactor | 900, 52, 45 | 5 |
| Nsv443 | CCG TGA CCG TTT CGT TCC G | 16S rRNA gene of β-Proteobacterial Nitrosospira spp. (444-462) | 32, 48, 30 | 29 |
| Nsm156 | TAT TAG CAC ATC TTT CGA T | 16S rRNA gene of β-Proteobacterial Nitrosomonas spp. (156-174) | 56, 48, 5 | 29 |
| Nso190c | CGA TCC CCT GCT TTT CTC C | 16S rRNA gene of ammonia-oxidizing β-Proteobacteria (190-208) | 900, 62, 55 | 29 |
| Nso1225 | CGCCATTGTATTACGTGTGA | 16S rRNA gene of β-subgroup ammonia-oxidizing bacteria (1224-1243) | 180, 51, 35 | 29 |
| Eub338 | GCT GCC TCC CGT AGG AGT | 16S rRNA gene of many eubacteria (338-355) | 180, 37, 30 | 2 |
| NonEub | ACT CCT ACG GGA GGC AGC | None (negative control; 355-338) | 180, 37, 30 | 25 |
The hybridization conditions for all the probes used during this study, except Nsm641, were optimized with reference to ammonia oxidizer cultures containing the appropriate target sequence. Nsm641 was optimized with aerobic sludge samples obtained from the DNB, as no reference organisms containing the target sequence for this probe were available. The hybridization temperature and/or formamide concentration in the hybridization buffer was successively increased until no fluorescent signal was observed from the reference cells. The optimal hybridization conditions were taken as the highest temperature and formamide concentration at which probe binding occurred. For Nso1225, Ralstonia eutropha was used in negative control hybridizations. Ralstonia eutropha DSM 531T has two mismatches with Nso1225 at the target site on the 16S rRNA. The relationship between the hybridization conditions and pixel intensity for our hybridization protocols is shown in Fig. 1 for pure cultures of target organisms (with no mismatches to the probe in their 16S rRNA) (N. europaea) and an organism with 16S rRNA with two mismatches with probe Nso1225 (Ralstonia eutropha). Hybridizations conducted using the protocol of Mobarry et al., (29) gave results comparable to those obtained with the protocol used in the current study (Fig. 1).
FIG. 1.
Fluorescence conferred by probe Nso1225 to whole fixed cells of Nitrosomonas europaea and Ralstonia eutropha at different formamide concentrations. Optimization was done under the protocol of Daims et al. (11) (squares and dashed line), under the protocol used in this paper (diamonds and solid line), and to the nontarget species Ralstonia eutropha (triangles and dotted line). Error bars indicate standard deviations among individual cells in a sample.
Microscopy.
Unless stated otherwise, slides were examined with a Bio-Rad MRC 600 confocal laser scanning microscope (CLSM) equipped with a Kr/Ar ion laser. All counting was undertaken at a magnification of ×600. Background fluorescence was accounted for by thresholding the images using data from hybridizations with the negative control probe (nonEub338). To calibrate microcolony dimensions against AOB cell numbers, optical sections were collected at 0.8-μm intervals and the number of cells in each microcolony was counted manually. The maximum and minimum diameters of each aggregate microcolony were used to determine a mean diameter, which was measured from stacked z-images (sections) for a given field of view. Comos (version 6.054; Bio-Rad) was used to record and analyze images. Conventional epifluorescence microscopy was done using an Olympus BX40 instrument fitted with an HBO 50W mercury lamp (Olympus, Tokyo, Japan) and an Olympus U-MWB filter set. The microcolony dimensions in this instance were determined by finding the focal plane for the maximum diameter for a given microcolony and then measuring the diameter to the nearest micrometer by using an eyepiece micrometer.
Chemical and physical properties of mixed liquor.
All physical parameters (mixed-liquor suspended solids [MLSS] and MLVSS) and chemical parameters (ammonia and biological oxygen demand [BOD]) were determined using standard methods (3).
Statistical analysis.
Probability distributions, Anderson-Darling normality tests, analysis of variance, and multiple comparisons of means were undertaken with MINITAB v11 (Minitab Inc., State College, PA). Other statistical analyses were used as described by Sokal and Rohlf (40).
Calculation of area under normal curve.
It is possible to calculate the area under a normal distribution curve based on mean (μ) and standard deviation (σ) values. However, if there are some data missing, both the mean and the standard deviation values are distorted and need to be corrected. This may be achieved by an iterative procedure described by Metcalfe (28). The procedure is as follows. The area of the unobserved proportion of the curve is calculated using the values for the “distorted” mean and standard deviation derived from the available data and the area under a standard normal distribution curve. The area calculated is then used to recalculate the values for the mean and standard deviation. The new mean and standard deviation are then used to recalculate the unobserved area under the curve. This procedure is iterated until the area of the recalculated mean and standard deviation reach a fixed value, and the corresponding unobserved area is determined using these values.
Sample size calculation.
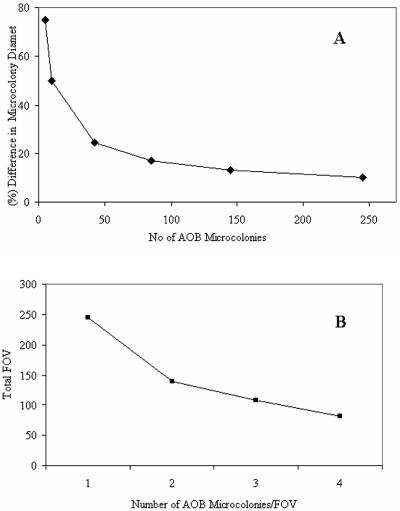
The sample size required to achieve a particular power of discrimination was determined using a method we have described previously (12), which was itself derived from a protocol suggested by Sokal and Rohlf (40). Briefly, nested analysis of variance was used to establish that virtually all the observed variation occurred from field of view to field of view (as opposed to sample to sample), and the mean microcolony diameter was identified as the most important variable in the estimation of biomass or cell counts (see below). On this basis we were able to calculate the number of microcolonies that must be counted if we were to have a 95% chance of detecting a given difference in size, significant at the 95% level (Fig. 2A). For example, counting 42 microcolonies ensures that there is a 95% chance of detecting a size difference of 25%, significant at the 5% level. The number of fields of view that must be counted, then becomes a function of the number of microcolonies per field of view (Fig. 2B). To give an 80% chance of detecting a difference of one AOB microcolony per field of view between two samples at the 5% level of significance, it was calculated that a sample size of 46 fields of view was required.
FIG. 2.
A. Number of microcolonies that must be counted to ensure having a 95% chance of detecting a given difference in microcolony diameter significant at the 5% level. The number of microcolonies can be decreased by accepting a marginally lower chance of detecting a given difference. B. Numbers of fields of view (FOV) that must be counted to give an 80% chance of detecting a difference of one AOB microcolony per field of view between two samples at the 5% level of significance. It was calculated that a sample size of 46 fields of view was required.
Calculation of AOB cell numbers and biomass from FISH data.
The basic methodology for the quantification of AOB was simple. The mean microcolony volume per unit volume of mixed liquor was determined and then converted to either biomass or cell numbers.
The number of AOB microcolonies per unit volume was determined on the basis of the mean number of microcolonies per field of view, the area covered by the sample spot, the area of one field of view, and correction factors to take account of sample dilution and concentration steps, including the initial dilution in alcohol. The diameters of the microcolonies observed were measured directly and found to be log-normally distributed. Ellipsoidal microcolonies were accounted for by taking the arithmetic mean of the longest and shortest axes of the ellipse. Average microcolony volume was calculated by assuming that AOB microcolonies are spherical and using the geometric mean radius of AOB microcolonies in the equation 4/3πr3. The measurements of mean microcolony abundance and volume were combined to give the mean microcolony volume per unit volume of sample.
The relationship between microcolony volume and cell numbers was determined empirically to provide a calibration curve relating microcolony volume to cell numbers. This relationship was used to convert measurements of microcolony volume per unit volume of activated sludge mixed liquor to cell counts per unit volume.
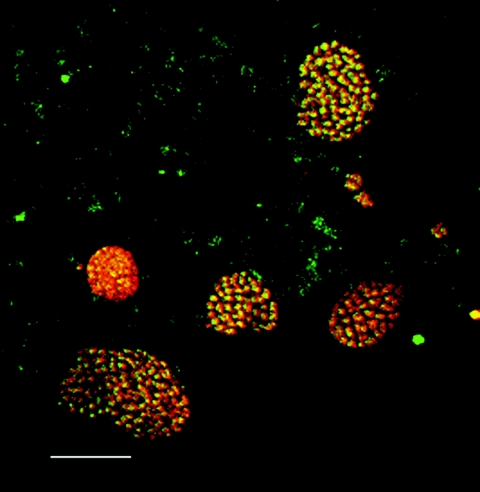
In principle, cell counts can also be converted to biomass by calculation of cell volume and density (15). In practice, biomass estimates may be obtained with less error by measuring microcolony dimensions. AOB biomass was therefore determined by multiplying the mean volume of AOB microcolonies per unit volume of mixed liquor and the dry weight of cells per unit volume of an AOB microcolony (0.49 g/cm3 [15]). This figure was obtained by using a consensus value of 318 fg of carbon/μm3 of cellular biomass (15) to convert volume to biomass. Carbon accounts for about 50% of cellular biomass (15), and therefore the total density of a bacterial cell is about 636 fg (dry weight)/μm3 of cellular material, which is equivalent to 0.636 g/cm3. However, this value cannot be applied directly to microcolonies, because they contain void spaces. Our observations indicated that most microcolonies appeared to be perfectly packed (Fig. 3). The maximum theoretical packing efficiency for a three-dimensional object (perfect packing) is 77% (for a sphere) to 76% (for an ellipsoid) (36), i.e., there is a 23 to 24% void volume. By allowing for a 23% void space, the biomass density in an AOB microcolony was determined to be no greater than 0.490 g/cm3 (i.e., 77% of 0.636 g/cm3). This is a maximum possible density, because lower packing efficiency in the microcolonies would yield lower densities.
FIG. 3.
Single optical slice through a section of an activated sludge floc (from Wanlip), showing Nso1225-labeled microcolonies of a range of diameters, most of which are of above average (see Fig. 4) for this plant. Bar, 10 μm. Although the cells are very close together, they will not all appear to touch, as even perfectly packed spheroids make contact with adjacent particles at only one point on any given side.
Calculation of theoretical AOB biomass.
Theoretical calculation of AOB biomass was undertaken as previously described (34) using the following equation:
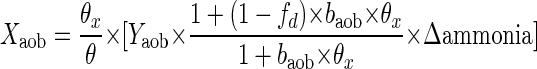 |
where θ is hydraulic retention time, θx is biomass residence time, X is biomass, Y is yield, b is the endogenous respiration rate, and fd is the fraction of newly synthesized biomass that is degradable by endogenous decay. The subscripts v and aob represent total bacteria and AOB, respectively. Values for Yaob (0.34 kg volatile suspended solids/kg N), fd (0.8), and baob (0.15 day−1) were taken from the literature (16, 34). Xv was measured directly and used to estimate sludge age as described previously (34). The ratio of active AOB to total MLVSS, Xaob/Xv, can be calculated using the equation above and employing known values for the parameters above and measured ammonia removal (Δ ammonia) and biomass concentration (Xv) (34) and was expressed as a percentage. This is a conservative estimate of the AOB biomass because it is based on ammonia alone; in reality, other forms of reduced nitrogen may become available for nitrification.
Comparison and combination of errors.
Errors were combined using standard equations for the combination of errors and coefficients of variation (30). The combined error for the multiplication of the microcolonies per unit volume of sample, cell count per unit volume, and volume of a microcolony would be (CV12 + CV22)1/2 + 3(CV32), where CV1, CV2, and CV3 represent the coefficients of variation for the microcolonies per unit volume, the slope of the curve relating cell number to volume, and the radius, respectively. The errors in the radius are multiplied by 3 because this value is cubed to calculate the volume; this provides the most conservative estimate of the error.
Cell-specific ammonia oxidation rates.
Cell-specific ammonia oxidation rates were calculated by the method of Daims et al. (11).
RESULTS
Results of the chemical and physical analyses of the wastewater treatment reactors are summarized in Table 3. All of the plants appeared to be nitrifying.
TABLE 3.
BOD, ammonia, MLSS, and MLVSS in activated sludge samplesa
| Plant name | Concn (mg/liter) of:
|
|||||
|---|---|---|---|---|---|---|
| BODi | BODe | Ammoniai | Ammoniae | MLSS | MLVSS | |
| Wanlip | 150 | 20 | 32 | 4 | 2,400 | 1,800 |
| Stoke Bardolph | 81 | 14 | 15.8 | 4 | 3,678 | 2,705 |
| Preston | 243 | 5 | 18 | 0.97 | 2,805 | 2,580 |
| Chorley | 86 | 12.25 | 11.2 | 0.58 | 1,645 | 1,540 |
| Hydburn | 119.3 | 12.33 | 14.17 | 5.15 | 2,740 | 2,240 |
| Lab reactor | 410 | 40.3 | 22.2 | 0 | 5,400 | 3,240 |
BODi, influent BOD; BODe, effluent BOD; ammoniai, influent ammonia; ammoniae, effluent ammonia.
Probability distribution of AOB microcolony dimensions.
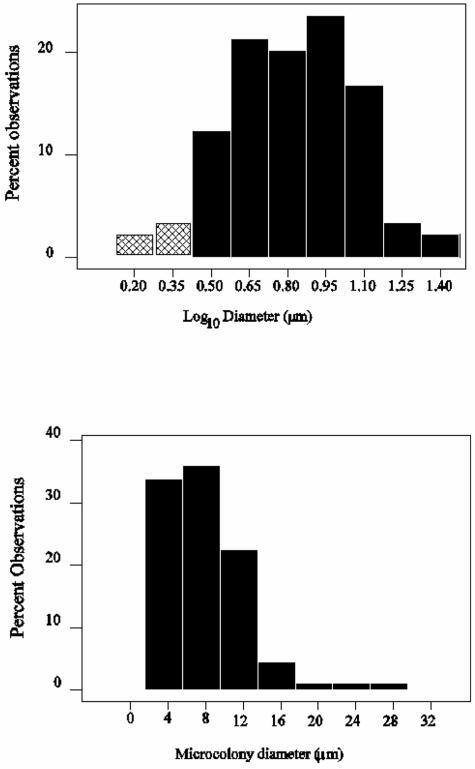
The detectable AOB occurred in characteristic microcolonies in all of the plants examined (Fig. 3). The diameters of AOB microcolonies were not normally distributed in any of the plants examined (Anderson-Darling normality test, P < 0.05). Log10-transformed microcolony diameter data, however, were normally distributed (Anderson-Darling probability values: Wanlip, 0.526; Stoke Bardolph, 0.211; Preston, 0.328; Chorley, 0.098; Hydburn, 0.191; and lab reactor, 0.305). Typical data are shown in Fig. 4. It is apparent that AOB microcolony diameters are log-normally distributed. These results have two practical implications: (i) we may use the finding of a normal distribution in log-transformed data to determine the proportion of the microcolonies that were not observed, and (ii) log-transformed data must be used when determining AOB biomass and cell numbers and associated errors, derived from microcolony dimensions.
FIG. 4.
Probability distributions of microcolony diameter for raw measurements (lower panel) and log-transformed measurements (upper panel) for 89 microcolonies from a full-scale wastewater treatment plant (Wanlip), using Nso1225. The putative unobserved fractions are shown as the shaded area in the log-transformed data and were calculated to be less than ca. 3.5% of the total possible observations.
Undetected fraction of AOB.
Since the AOB microcolonies have a characteristic distribution, we may calculate the proportion of the microcolonies that we have not observed because they are too small. The smallest observed microcolonies typically had a diameter of between 2 and 3 μm. The fraction of the AOB represented by small microcolonies and single cells may be represented by that proportion of the biomass lying between the smallest observed microcolony diameter and the diameter of a single cell. In the Wanlip wastewater treatment plant, the smallest microcolony diameter observed was 2.54 μm. Using a cell width of 1 μm, we found the undetected fraction of the AOB biomass to represent just 3.7% of the microcolonies and thus a very small proportion (∼0.02%) of the overall biomass. Since the microcolony diameter and volume were log-normally distributed in all the plants observed, we conclude that virtually all the AOB biomass was detected by FISH (Fig. 4).
Estimating cell numbers.
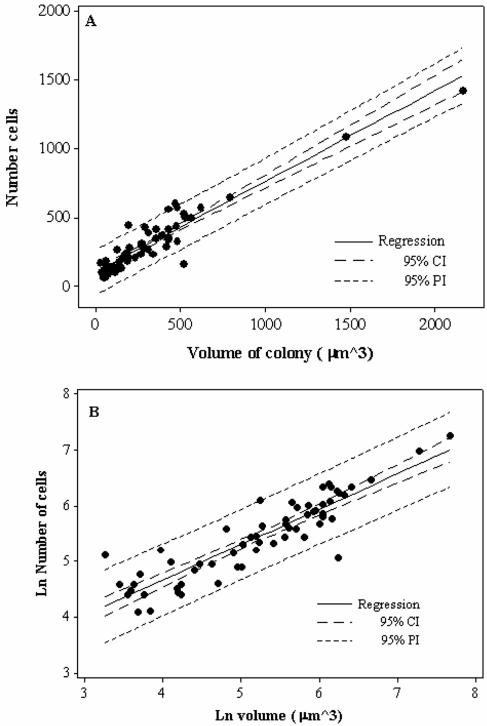
Total AOB cell numbers may be determined by using optical sections obtained with a confocal microscope. It is evident that that there is a relationship between cell numbers and microcolony volume (Fig. 5). However, both cell numbers and microcolony volume were log-normally distributed and a Box-Cox analysis showed that a natural log transformation was appropriate for describing the relationship between microcolony volume and cell numbers. Because log-log transformations can be used to force linear relationships, both raw and transformed data are presented(Fig. 5). For the raw data, the r2 value was 0.89 and both the slope and the intercept were significant (P < 0.001). For the transformed data, the r2 value was 0.81 and both the slope (0.64 [standard error, 0.04; coefficient of variation, 7%]) and the intercept (2.1) were statistically significant (P < 0.001); the residuals were normally distributed, suggesting that the scatter observed is random. The intercept is greater than zero. This could imply some systematic error in the estimation of either cell numbers or microcolony volume. Removing the three obvious outliers (those outside the 95% prediction interval) in the transformed data set changed the r2, slope, and intercept to 0.90, 0.70 and 1.7, respectively.
FIG. 5.
Relationship between microcolony size and AOB cell numbers in activated sludge for the raw (A) and natural-log-transformed (B) data. For the raw data, the r2 value was 0.89 and both the slope (0.66) and the intercept (101) were statistically significant (P < 0.001). However, both sets of raw data were log-normally distributed, and a Box-Cox analysis showed that a natural log transformation was appropriate. For the natural-log-transformed data, the r2 value was 0.81 and both the slope (0.64) and the intercept (2.1) were statistically significant (P < 0.001). We recommend the use of the log-transformed data. CI, confidence interval; PI, prediction interval.
Cell numbers and cell specific ammonia oxidation rates.
Cell numbers in a sample can be determined from measurement of the number and diameter of microcolonies per unit volume. The mean microcolony volume per unit volume of sample can then be calculated. The regression line (Fig. 4) may then be used to estimate cell numbers from the geometric mean microcolony volume. Using this approach, we have determined the concentration of AOB cells in a variety of full-scale plants and a bench-scale reactor by using these values ranged over three orders of magnitude, ca. 105 to ca. 108 cells/ml (Table 4).
TABLE 4.
AOB cells counts from full-scale reactors
| Plant name | Cells/ml | Plus SE | Minus SE |
|---|---|---|---|
| Wanlipa | 2.17E+08 | 6.36E+06 | 6.01E+06 |
| Prestona | 1.99E+07 | 2.57E+05 | 2.45E+05 |
| Chorleya | 1.40E+07 | 1.07E+05 | 1.03E+05 |
| Hydburna | 2.47E+05 | 1.09E+04 | 8.86E+03 |
| Stoke Bardolpha | 3.80E+05 | 2.62E+04 | 2.20E+04 |
| Bench-scale reactorb | 4.05E+07 | 2.44E+06 | 2.12E+06 |
| Bench-scale reactorc | 4.01E+07 | 8.04E+06 | 6.16E+06 |
Probe Nso1225 was used.
Probe Nso190 was used.
Probe Nsm641, designed to detect the AOB corresponding to the predominant AOB sequences recovered in a 16S rRNA gene clone library obtained from the bench-scale reactor (6), was used.
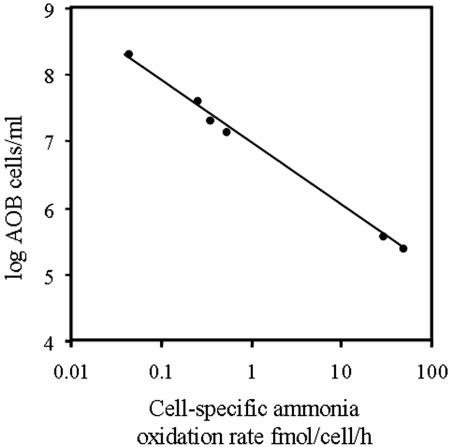
Cell-specific ammonia oxidation rates were found to range over nearly three orders of magnitude, from 43.00 to 0.03 femtomoles per cell per hour (Fig. 6). The plant with the highest number of AOB (Wanlip) had the lowest cell-specific ammonia oxidation rates, and Hydburn, a plant which the operators reported to be close to failure, had the highest cell-specific ammonia oxidation rates. Moreover, the range in ammonia oxidation rates was almost entirely driven by the differences in the numbers of AOB in different plants. Since there is a great difference in cell numbers between plants and this seems to lead to corresponding differences in cell-specific ammonia oxidation rates, it is logical to ask if we were finding more or less AOB biomass than theory would predict.
FIG. 6.
Variation of the cell-specific ammonia oxidation rate with the concentration of AOB. The plant with the lowest rate is Wanlip, while the plant with the highest rate is Hydburn.
Comparison of experimental and theoretical AOB biomasses in different plants.
The fraction of total biomass that AOB comprise was calculated using theoretical model of nitrification described by Rittmann et al. (34)and operating data from the wastewater treatment plants. These predicted values were compared with those determined using FISH (Fig. 7). Perfect correspondence between measured and predicted values would imply a slope of 1 and an intercept of 0. The regression line has a statistically significant (P = 0.003) slope of 1.28 (standard error of 0.20) and an intercept (−2.8) that is statistically distinguishable from 0 (P = 0.022). The regression line explained 89% of the variation between the two estimates which employed full-scale and bench-scale plants; one plant (Hydburn) was reported to be failing shortly before the time of sampling and contained fewer AOB than predicted (Fig. 7A). This data point was identified as an outlier (Dixon's test statistic, P < 0.05). Excluding this data point (Fig. 7B), we found that the slope is 1.23 (standard error of 0.16) and r2 rises to 94% and that the intercept (−2.47) is still significantly different from zero (although only marginally so) (P = 0.034).
FIG. 7.
A. Relationship between theoretical and measured AOB fractions in full-scale activated sludge reactors in the United Kingdom and a bench-scale reactor. The regression line has a statistically significant (P = 0.003) slope of 1.28 (standard error of 0.20) and an intercept (−2.8) that is statistically distinguishable from 0 (P = 0.022). The regression line explained 89% of the variation between the two estimates. Xaob/Xv is the proportion of the total biomass measured as MLVSS that is contributed by the AOB. Hydburn was identified as an outlier by using Dixon's test (P < 0.0.05). B. Relationship between theoretical and measured AOB fractions in full-scale activated sludge reactors in the United Kingdom and a bench-scale reactor, but with Hydburn removed. The slope is 1.23 (standard error of 0.16), r2 rises to 94%, and the intercept (−2.47) is still significantly different from zero (although only marginally so) (P = 0.034). CI, confidence interval.
Magnitude and importance of random errors.
The ostensibly satisfactory agreement between observed and estimated biomasses suggests that between 2 and 20% of the variation between sites is attributable to random error. In order to assess and improve this, one must determine the source and magnitude of these errors.
The combined site-specific error estimates for the number of cells per unit volume were 18% (Wanlip and Chorley), 19% (Preston), 31% (Stoke Bardolph), and 40% (Hydburn and bench-scale reactor). Not surprisingly the higher coefficients of variation were associated with lower cell counts. The number of cells per unit volume of mixed liquor was calculated by multiplying the number of microcolonies per unit volume of mixed liquor (coefficient or variation, < 5%), the mean microcolony volume based on measurement of the geometric mean diameter of the microcolony (mean coefficient of variation, 17%; range, 9 to 22%), and the number of cells per unit microcolony volume (from the slope in Fig. 5) (coefficient of variation, < 7%). Virtually all the error is attributable to the error in the microcolony mean radius, as this error is multiplied by 3 (as the radius is cubed).
The random error in the estimation of the biomass per unit volume is on the order of 39%. Again, much of the error is attributable to the error in the measurement of microcolony diameter, and the site-specific errors vary accordingly from 30% to 46% (30% for Wanlip and Chorley, 31% for Preston, 40% for Stoke Bardolph, and 46% for Hydburn and the bench-scale reactor). The biomass per unit volume was calculated by the multiplication of microcolonies per unit volume (coefficient of variation, < 5%), the mean microcolony volume based on measurement of the geometric mean diameter of the microcolony (mean coefficient of variation, 17%; range, 9 to 22%), and the estimate of AOB carbon per unit volume (coefficient of variation, ∼25%) (15). The errors due to the variation in the packing efficiency were not included in the analysis because the value employed in the calculations represented a fixed upper value, not a mean. However, since we know that the coefficient of variation in the number of cells per unit volume is about 11% in the size range used in biomass estimates, it appears that this is not a predominant source of error in biomass estimations.
Confocal microscopy versus conventional fluorescence microscopy.
To investigate whether image quality affects quantification, we counted Nitrosomonas spp. in a nitrifying activated sludge plant by quantitative FISH (with probe Nsm156), using CLSM and an epifluorescence microscope. In terms of both microcolony diameter and microcolony abundance, the values from epifluorescence microscopy were significantly lower than those obtained using CLSM (t test, P < 0.06). When these parameters were used to estimate the concentration of Nitrosomonas spp., the value obtained using epifluorescence microscopy was lower than that obtained using CLSM by about fourfold.
DISCUSSION
We believe that this is a valuable demonstration of the use of FISH to determine absolute numbers or biomass of AOB in activated sludge plants. Relating theoretically plausible and experimentally corroborated estimates of the productivity of the system clearly demonstrates the validity of the method (34). The method robustly estimates cell concentrations over 3 orders of magnitude in systems varying in scale over 7 orders of magnitude. Moreover, this has been achieved by counting manually, using relatively modest sample sizes at high magnification.
Belser's (7) critique of the use of cell-specific ammonia oxidation rates to corroborate quantification methods is well founded: it appears that these rates vary by approximately 3 orders of magnitude. This variation is not caused by over- or underestimation of AOB numbers in different plants, because (i) we are able to find more than 95% of the detectable AOB in a plant, and (ii) the variation in AOB biomass and cell numbers between plants is consistent with known variation in the ammonia consumed and plant characteristics. Thus, cell-specific ammonia oxidation rates emerge as a significant process variable. For example, the proportion of the biomass contributed by AOB in Wanlip (6%) is entirely consistent with theoretical estimates (7%), but the cell-specific ammonia oxidation rate observed here (0.03 femtomoles/cell/hour) is an order of magnitude lower than that previously observed in situ (0.22 femtomoles/cell/hour) (43). Thus, the estimate of cell numbers for this plant could have been dismissed as an overestimate. Conversely, we cannot confidently assume that data falling within the published range of cell-specific rates imply accuracy in quantification.
The description of the distribution of the cell numbers is a crucial part of the successful use of FISH to quantify AOB or indeed any other microbial community. Many authors have expressed dissatisfaction with the lack of precision obtained when counting even large numbers of cells by using FISH (26, 38). This has led to the belief that meaningful precision cannot be obtained with manual counting procedures, because of the difficulty of obtaining a sufficiently large sample size. In this and previous studies (12) we have determined the underlying distribution of the data, which has shown that log transformation of the data is required to permit the use of parametric statistical methods and modest sample sizes (without the use of image analysis). Calculation of arithmetic means from nontransformed data which are log-normally distributed (which is commonplace) not only will give large standard deviations but also will provide an erroneous mean value. For example, the data set describing the number of cells per microcolony (Fig. 5) was examined using the Box-Cox method and transformed using natural logarithms to give a back-transformed mean of 232 cells/microcolony and a standard deviation of 2. Using untransformed data, the arithmetic mean of the same data set was 300 cells/microcolony with a standard deviation of 241. Thus, by recognizing the underlying distribution, it is possible to reduce the variance in the data and make valid statistical comparisons using modest sample sizes in conjunction with powerful parametric statistics. A further advantage of examining the distribution of the AOB microcolony size is that we are able to establish the fraction of AOB that are detectable and demonstrate that those which occur as single cells or very small microcolonies represent a small proportion of the total AOB. We are thus able to disprove the tentative hypothesis of Rittmann et al. that discrepancies between modeled AOB biomass levels and those measured by FISH were due to single cells (34).
If quantitative methods are to improve, it is vital that we assess the nature and cause of our errors. A plot of theoretical versus experimental estimates provides a relatively plausible basis for such an assessment. Perfect agreement between a perfect theory and a perfect form of measurement would give us a slope of 1, an intercept of 0, and an r2 value of 100%.
In our comparison of measured AOB abundance and the abundance predicted from Rittman's model (34), the slope of the curve was statistically indistinguishable from 1, which also suggests good agreement between theory and measurement over a wide range of scales, ammonia concentrations, and sludge ages.
The r2 values suggest that between 6% (if Hydburn is an outlier) and 11% (if Hydburn is not an outlier) of the observed variation between plants cannot be accounted for by our method. The former value implies only modest room for improvement, so the Hydburn data are worthy of further consideration, having far fewer AOB than theoretically predicted. This plant was close to failure shortly before the measurements were taken, and thus it is possible that the AOB community was not at equilibrium or not limited by ammonia, that other nitrification processes (e.g., heterotrophic nitrification) were at work, or that the organisms were so stressed that the yield was very low. Any of these possibilities would mean that the abundance of AOB was not described by the theory, which assumes an ammonia-limited system at equilibrium. Alternatively, the probe employed might not have detected all the AOB present in the Hydburn plant. The probe Nso1225 has a mismatch with the 16S rRNA of N. mobilis and so could underestimate the AOB community if this organism is abundant in the treatment plant. Organisms related to N. mobilis have been typically associated with wastewater treatment plants treating saline wastes (22, 37).
The value of the intercept on the y axis of the plot of measured versus predicted AOB biomass tells us whether the theoretical estimates systematically overestimated (intercept of <0) or underestimated (intercept of >0) the amount of measured AOB biomass. The method we present appears to systematically overestimate the amount of measured biomass (or vice versa). This systematic error must, in part at least, represent the assimilation of ammonia by the non-AOB biomass, an important sink of ammonia that is neglected in the original calculations by Rittmann et al. However, other simplifying assumptions include an assumed yield, measuring the removal of ammonia rather than total reduced nitrogen, the use of consensus data on the density of individual bacterial cells (15), and that the AOB were packed in microcolonies with the maximum possible efficiency (the validity of this last assumption is weakest in the largest microcolonies). We wish to draw particular attention to the assumed yield. The AOB biomass estimates vary in a linear manner with this value. Therefore, using a lower yield would also have brought the data closer to an intercept of 0. It is interesting that the yield employed is the maximum theoretical value (35). An authoritative review has suggested that AOB yields are nearly always close to this maximum in all autotrophic nitrifying organisms (32). However, the available empirical values represent data from a limited number of taxa under good laboratory conditions, and yield could vary with environmental conditions. AOB in “real life” might obtain slightly less than the theoretical maximum yield used in our theoretical estimates (especially if subject to stress). The advent of trustworthy molecular tools for the quantification of AOB should allow engineers to ascertain AOB yields and incorporate them into design and management strategies for wastewater treatment plants and to empirically relate them to environmental conditions.
Why did Rittmann et al., using the same approach, fail to find agreement between the theoretical and FISH-based estimates of AOB biomass (34)? This is probably because a conventional fluorescence microscope was used and the assumed value for the density of AOB cells was low (0.1 g/cm3). It appears that a CLSM, or a microscope of equivalent performance, is required for meaningful quantitative FISH in activated sludge and probably other complex three-dimensional environments.
The precision of our manual cell counts (coefficients of variation, 17 to 40%; mean, 27%) is as good, or better, than those previously reported (Schramm et al. [manual counting], 3 to 50% [38]; Wagner et al. [manual counting], 19% [44]; Daims et al. [quantification using image analysis], 20% [11]). These are minimum estimates of the errors, since some sources of error are unreported in those studies. Importantly, our method applies only to organisms that reliably form microcolonies. Thus, although the method of Daims et al. (11) is more complex than our own, their quantification strategy may be more widely applicable. Most of the random variation in our method appears to be attributable to variation in the measurement of the diameter of the microcolony. Thus, if desired, greater precision can be achieved by improving measurements of AOB microcolony diameter. Improving the precision of other elements in the method will yield only limited improvement in the precision of the method.
It would seem that, in general, cell counts will be preferable to biomass estimates, at least until the coefficient of variation in the estimation of the conversion factors can be reduced. The high r2 values for estimated and measured biomasses suggest that our precision is perhaps better than we suggest, probably because actual biovolume-to-carbon ratios are relatively constant even though the estimation of the exact value of such ratios is subject to error.
Quantification is a strategically important aspect of microbial ecology. However, there is a world of difference between a number and the correct number. Important insights and practical applications will be gained if we can improve accuracy and precision in our methods.
One such insight might be that cell-specific ammonia oxidation rates vary by 2 to 3 orders of magnitude. Thus, AOB in some plants may be working 1,000 times harder than AOB in other plants. It could be significant that the highest cell-specific ammonia oxidation rate was seen in a treatment plant that was close to failure (Hydburn). It is also possible that the very low cell-specific ammonia oxidation rate in another plant means that this plant could be run more cheaply. We hypothesize that there is a threshold cell-specific ammonia oxidation rate below which stable performance may be expected. Operating a plant significantly below this threshold could incur needless extra aeration costs (typically the largest or second largest recurring expense in a treatment plant), and operating above this threshold may increase the risk of failure. Thus, the ability to count AOB (and indeed other functional groups) could help those operating biological treatment plants to more rationally balance costs and the risk of failure.
Acknowledgments
We thank Trevor Booth for guidance with the CLSM, Andrew Metcalfe for statistical advice, Severn-Trent Water PLC and Northwest Water PLC for providing access to the plants and operational data, and two anonymous reviewers and J. Prosser for their helpful comments.
S.J.B. and G.C. thank Shell/NERC and Cumhuriyet University, respectively, for financial support.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1995. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 4.Ballinger, S. J. 2000. PhD thesis. University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom.
- 5.Ballinger, S. J., T. Curtis, A. R. Godley, and I. M. Head. 2002. The effect of C/N ratio on ammonia oxidising bacteria community structure in a laboratory nitrification denitrification reactor. Water Sci. Technol. 46:543-550. [PubMed] [Google Scholar]
- 6.Ballinger, S. J., I. M. Head, T. P. Curtis, and A. R. Godley. 1998. Molecular microbial ecology of nitrification in an activated sludge process treating refinery wastewater. Water Sci. Technol. 37:105-108. [Google Scholar]
- 7.Belser, L. W. 1979. Population ecology of nitrifying bacteria. Annu. Rev. Microbiol. 33:309-333. [DOI] [PubMed] [Google Scholar]
- 8.Biesterfeld, S., L. Figueroa, M. Hernandez, and P. Russell. 2001. Quantification of nitrifying bacterial populations in a full-scale nitrifying trickling filter using fluorescent in situ hybridization. Water Environ. Res. 73:329-338. [DOI] [PubMed] [Google Scholar]
- 9.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daims, H., N. B. Ramsing, K. H. Schleifer, and M. Wagner. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davenport, R. J., T. P. Curtis, M. Goodfellow, F. M. Stainsby, and M. Bingley. 2000. Quantitative use of fluorescent in situ hybridization to examine relationships between mycolic acid-containing actinomycetes and foaming in activated sludge plants. Appl. Environ. Microbiol. 66:1158-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dionisi, H. M., A. C. Layton, G. Harms, I. R. Gregory, K. G. Robinson, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebie, Y., H. Miura, N. Noda, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2002. Detection and quantification of expression of amoA by competitive reverse transcription-PCR. Water Sci. Technol. 46:281-288. [PubMed] [Google Scholar]
- 15.Fry, J. C. 1990. Direct methods and biomass estimation. Methods Microbiol. 22:41-85. [Google Scholar]
- 16.Furumai, H., and B. E. Rittmann. 1994. Evaluation of multiple-species biofilm and floc processes using a simplified aggregate model. Water Sci. Technol. 29:439-446. [Google Scholar]
- 17.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 19.Hermansson, A., and P. E. Lindgren. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikuta, H., N. Noda, Y. Ebie, A. Hirata, S. Tsuneda, M. Matsumura, and Y. Inamori. 2000. The rapid quantification and detection of nitrifying bacteria by using monoclonal antibody method. Water Sci. Technol. 42:1-7. [Google Scholar]
- 21.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 22.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Knowles, G., A. L. Downing, and M. J. Barrett. 1965. Determination of kinetic constants for nitrifying bacteria with the aid of an electronic computer. J. Gen. Microbiol. 38:263-273. [DOI] [PubMed]
- 23.Konuma, S., H. Satoh, T. Mino, and T. Matsuo. 2001. Comparison of enumeration methods for ammonia-oxidizing bacteria. Water Sci. Technol. 43:107-114. [PubMed] [Google Scholar]
- 24.Laanbroek, H. J., and S. Gerards. 1993. Competition for limiting amounts of oxygen between Nitrosomonas europaea and Nitrobacter winogradskyi grown in mixed continuous culture. Arch. Microbiol. 159:453-459. [Google Scholar]
- 25.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria—problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 26.Manz, W., M. Wagner, R. Amann, and K. Schleifer. 1994. In-situ characterisation of the microbial consortia active in 2 waste-water treatment plants. Water Res. 28:1715-1723. [Google Scholar]
- 27.Matulewich, V. A., P. A. Strom, and M. S. Finstein. 1975. Length of incubation for enumerating nitrifying bacteria present in various environments. Appl. Microbiol. 29:265-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalfe, A. V. 1994. Statistics in engineering. Chapman and Hall, London, United Kingdom.
- 29.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. (Erratum, 63: 815, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pentz, M., and M. Shott. 1988. Handling experimental data. Open University Press, Milton Keynes, United Kingdom.
- 31.Powell, S. J., and J. I. Prosser. 1985. The effect of nitrapyrin and chloropicolinic acid on ammonium oxidation by Nitrosomonas europaea. FEMS Microbiol. Lett. 28:51-54. [Google Scholar]
- 32.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 33.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rittmann, B. E., C. S. Laspidou, J. Flax, D. A. Stahl, V. Urbain, H. Harduin, J. J. van der Waarde, B. Geurkink, M. J. C. Henssen, H. Brouwer, A. Klapwijk, and M. Wetterauw. 1999. Molecular and modeling analyses of the structure and function of nitrifying activated sludge. Water Sci. Technol. 39:51-59. [Google Scholar]
- 35.Rittmann, B. E., and P. L. McCarty. 2001. Environmental biotechnology: principles and applications. McGraw-Hill, Boston, Mass.
- 36.Rogers, C. A. 1958. The packing of equal spheres. Proc. London Math. Soc. 1958:609-620. [Google Scholar]
- 37.Rowan, A. K., J. R. Snape, D. Fearnside, M. R. Barer, T. P. Curtis, and I. M. Head. 2003. Composition and diversity of ammonia-oxidising bacterial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol. Ecol. 43:195-206. [DOI] [PubMed] [Google Scholar]
- 38.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skinner, F. A., and N. Walker. 1961. Growth of Nitrosomonas europaea in batch and continuous culture. Arch. Mikrobiol. 38:339-349. [DOI] [PubMed] [Google Scholar]
- 40.Sokal, R. R., and F. J. Rohlf. 1995. Biometry, the principles and practice of statistics in biological research. W. H. Freeman and Company, New York, N.Y.
- 41.Strom, P. A., V. A. Matulewich, and M. S. Finstein. 1976. Concentrations of nitrifying bacteria in sewages, effluents and a recieving stream and resistance of these organisms to chlorination. Appl. Environ. Microbiol. 31:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utåker, J. B., L. Bakken, Q. Q. Jiang, and I. Nes. 1995. Phylogenetic analysis of seven new isolates of ammonia-oxidising bacteria based on 16S rRNA gene sequences. Syst. Appl. Microbiol. 18:549-559. [Google Scholar]
- 43.Wagner, M., G. Rath, R. Amann, H. P. Koops, and K. H. Schleifer. 1995. In-situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 44.Wagner, M., G. Rath, H. P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237-244. [Google Scholar]
- 45.Watson, S. W., and M. Mandel. 1971. Comparison of the morphology and deoxyribonucleic acid composition of 27 strains of nitrifying bacteria. J. Bacteriol. 107:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]