Abstract
Molecular cloning and characterization of a novel cry gene, cry32Aa, of Bacillus thuringiensis subsp. yunnanensis was carried out. The Cry32Aa protein was predicted to have a molecular mass of 139.2 kDa and was found to have an unusual 42-amino-acid-long tail at the C terminus. The cry32Aa gene was localized on the 103-MDa plasmid of the organism. Bioassays showed no toxicity against several moths and mosquitoes. However, it exhibited weak toxicity against larvae of the diamondback moth, Plutella xylostella.
Bacillus thuringiensis is a rod-shaped, ubiquitous, gram-positive bacterium (1, 16). During sporulation, it produces large crystalline parasporal inclusions (Cry proteins), many of which are toxic to certain moths, flies, and beetles. However, ∼40% of the crystal forming strains exhibit no known insecticidal activity (16). Bacillus thuringiensis subsp. yunnanensis belongs to serotype 20ab of the ∼60 flagellar serotypes so far identified (M.-M. Lecadet and E. Frachon, XXVIIth Annu. Meet. Soc. Invertebr. Pathol., 1994). In B. thuringiensis subsp. yunnanensis, spore and crystal formation occurs in separate cells (12). The unique regulation of crystal production in B. thuringiensis subsp. yunnanensis maintained in each successive generation was found to be mediated by the Cry-encoding 103-MDa plasmid (18). A B. thuringiensis strain with similar properties was isolated from certain African soil samples and characterized by Zelazny et al. (24). The presence of the crystal inclusions only in asporogenous cells, the irregular shape of the crystals, and its weak toxicity for the diamondback moth (Plutella xylostella) are some of the identifying features of this strain.
In order to gain further molecular and biochemical understanding about this novel Cry protein, the aim of this study was to clone and characterize the cry gene of B. thuringiensis subsp. yunnanensis in Escherichia coli.
Construction of a partial genomic library.
The total DNA of B. thuringiensis subsp. yunnanensis was partially digested with MboI to obtain 9- to 23-kb fragments. The fragments were purified on a sucrose gradient and then cloned between the BamHI-digested λDASHII vector arms (Stratagene Co.). The library amplification and screening of recombinants in the provided XL1-Blue MRA (P2) cells were done according to the manufacturer’s instructions.
Cloning of the cry32Aa gene.
Two heterologous primers, K5UN2 (5′-AGGACCAGGATTTACAGGAGG-3′) and K3UN2 (5′-GCTGTGACAC GAAGGATATAGCCAC-3′), designed on two highly conserved regions of cry genes (6) were used to amplify a 2-kb amplicon from B. thuringiensis subsp. yunnanensis through PCR on a minicycler (MJ Research). The amplicon was cloned into pGEM-T (Stratagene Co.) vector to yield pCRS1. The amplicon was radiolabeled and subsequently used as a probe for screening the library of lambda clones. The positive lambda clones λSBK1 through λSBK6 were digested with various restriction enzymes (BglII, MscI, NcoI, PvuI, SalI, SmaI, and SphI), electrophoresed on a 1% agarose gel, and Southern blotted to screen for a suitable insert size. The 5.5-kb NcoI fragment from the clone λSBK6 showed up as a single band on Southern hybridization. This was subsequently cloned into pOK12 under the control of the lacZ promoter to yield pSDMK1 and transformed into E. coli strain DH5α for further characterization of the cry gene.
Localization of the gene.
The plasmid profile of B. thuringiensis subsp. yunnanensis was analyzed by vertical slot lysis essentially as described by Gonzalez et al. (5). The agarose gel showed the expected plasmid bands of 103, 91, 61, 52, 45, 4.7, and 3.2 MDa (18). The gel was blotted onto a Hybond N+ nylon membrane, and Southern hybridization was performed by standard procedures described in Bio-Rad’s Zeta probe membrane manual. An 862-bp SmaI-KpnI internal fragment of the coding region of cry32Aa gene from pSDMK1 was used as the probe. The Southern blot revealed that the gene is located on the 103-MDa plasmid of B. thuringiensis subsp. yunnanensis as predicted earlier by Srinivas et al. (18). The Bacillus thuringiensis subsp. kurstaki strain HD73 was used as a negative control which did not hybridize with the probe.
Analysis of the cry32Aa gene sequence.
Sequencing of the 5,589-bp insert in pSDMK1 was performed by Microsynth, Inc., Balgach, Switzerland.
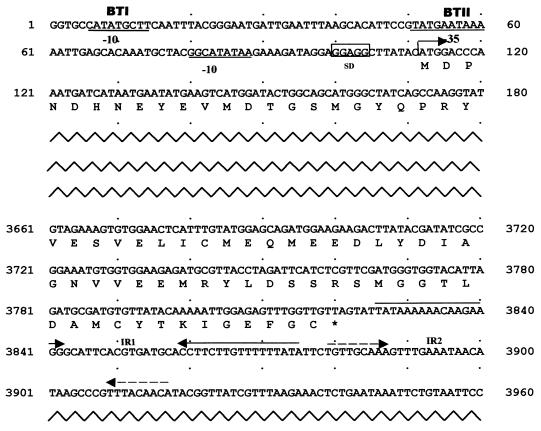
A complete open reading frame composed of 3,711 nucleotides encoding 1,236 amino acid residues was detected within the sequenced region by using the Wisconsin Package (version 9.1; Genetics Computer Group, Madison, Wis.). The protein was predicted to have a molecular mass of 139.2 kDa. A BLAST analysis of the protein revealed close homology to the Cry4 class of proteins and showed nearly 51 and 48% similarity to Cry4Ba and Cry4Aa, respectively. A CLUSTALW comparison (19) of the new Cry protein sequence with several other known Cry protein sequences helped to identify the characteristic eight conserved blocks predicted by Schnepf et al. (16) and also an unusual 42-amino-acid tail at the C terminus. Of the two transcriptional start sites (BtI and BtII) defining two overlapping sequentially activated promoters (22) in cry genes, we were able to partially characterize a putative BtI region and a complete BtII region in the new cry gene represented schematically in Fig. 1. A palindromic sequence (IR1) located downstream of the gene which can form a mRNA hairpin loop with ΔG of −23.04 kcal/mol and a shorter one (IR2) could act as putative factor-independent transcriptional terminators (4, 16, 21). The usual translational termination codon (after the ELICMNE motif that is present at the C terminus of the protoxin in most Cry proteins; 1188ELICMEQ1194 in Cry32Aa) was found to be replaced by a methionine residue (1195M), resulting in a 42-amino-acid tail at the carboxy terminus. The fact that this tail was the only significant difference we could find between this protein and the other closely related Cry proteins lends credence to the view that it could somehow be related to the nontoxicity of this protein, which requires further study. The evolutionary divergence of the Cry protein was estimated with phylogenetic tree algorithms as described by Crickmore et al. (3). The protein was identified as a new Cry toxin named Cry32Aa, which was confirmed by the B. thuringiensis Pesticidal Crystal Protein Nomenclature Committee (3). The phylogram can be viewed at http://www.biols.susx.ac.uk/home/Neil_Crickmore/Bt/.
FIG. 1.
Positions of the regulatory regions of cry32Aa. The promoter region (−10 and −35 sequences) is underlined. The ribosomal binding site is boxed. The deduced amino acid sequence is below the coding region. The terminator region (IR1 and IR2) is indicated with inverted arrows. The initiation codon, ATG (at position 112), and the termination codon, TAG (at position 3822), are indicated with an arrow and an asterisk, respectively.
The protein sequence reveals putative proteolytic cleavage sites near the N terminus (R58) and at the C terminus of the toxin (the K613 residue) which are known to be more frequently associated with proteolytic cleavage (2, 11, 16). Hence, the toxin is estimated to have a length of 561 amino acids (position 58 to 618) after proteolytic cleavage, with a predicted molecular mass of 63.5 kDa. A three-dimensional model of the protein was constructed by the unpublished method of Shindyalov and Bourne at the San Diego Supercomputer Center, University of California, San Diego, through the Web-based tool PREDICTPROTEIN. A hydropathic profiling of the protein with the Kyte and Doolittle algorithm (7) did not reveal significant differences from closely related Cry proteins, but analysis of the model with INSIGHTII software (Biosym Technologies, Inc., San Diego, Calif.) revealed that loops 1 and 2 of domain II, which are involved in receptor binding (13, 16), are rich in hydrophilic residues (336DSQDAE341 and 419RTDANN424, respectively) instead of the usual hydrophobic residues, which is detrimental to strong receptor binding (14, 16, 17, 23) and may also contribute to the lack of toxicity.
Expression studies on Cry32Aa.
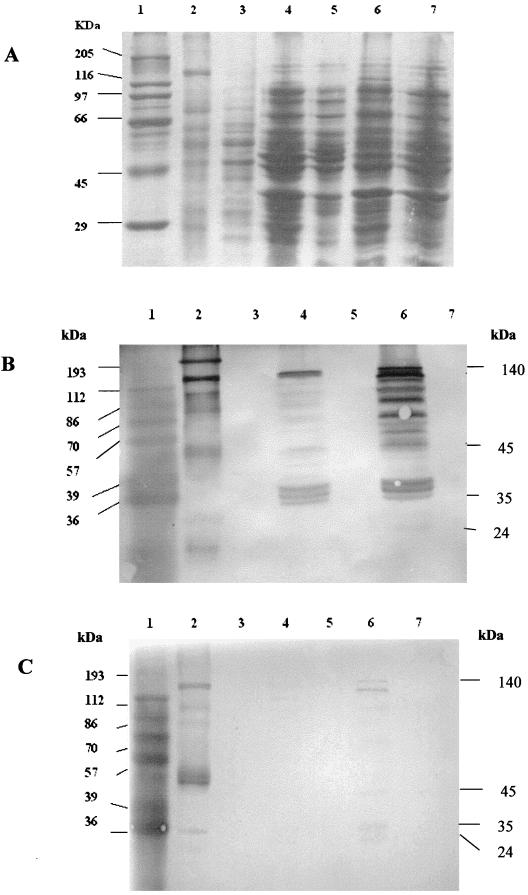
The orientation of the 5.5-kb NcoI fragment in pSDMK1 was reversed and put under the control of the T7 promoter of pOK12 to yield pPSVS. The transformant E. coli strain BL21(DE3) was grown in Luria-Bertani broth (15) at 37°C with shaking and induced with 0.4 mM IPTG (isopropyl-β-d-galactopyranoside) at an optical density at 600 nm of 0.5 to induce Cry32Aa expression. The expression of the Cry32Aa protein in the transformant BL21(DE3)(pPSVS) was higher than that in DH5α(pSDMK1), where the gene is under the control of the lacZ promoter. Total protein extracts from both transformed and untransformed E. coli strains were prepared by spinning down the 8-h cultures, suspending the pellet in a suitable quantity of water, and boiling it for 5 min. It was again spun briefly, the supernatant was collected, and the protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 1 mM) was added to it. Crude protein extracts from B. thuringiensis subsp. yunnanensis as a positive control and B. thuringiensis subsp. kurstaki HD73 as a negative control were prepared as described by Murty et al. (10). The crude preparation was solubilized for 3 h at 37°C in 50 mM NaHCO3 supplemented with 10 mM dithiothreitol and 1 mM PMSF for protease inhibition. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8) was performed with the E. coli extracts on a 10% polyacrylamide gel (Fig. 2A). The gel was immunoblotted (20) with rabbit antiserum raised against the 140- and 45-kDa proteins of B. thuringiensis subsp. yunnanensis to check the expression of the Cry32Aa protein. The immunoblots showed a major 140-kDa reacting band and several smaller cross-reacting bands in the transformant E. coli and the positive B. thuringiensis subsp. yunnanensis lanes for both the antisera used (Fig. 2B and C), even though PMSF was added in all protein preparations. The extensive degradation of Cry32Aa observed in E. coli may be due to proteolytic cleavage of the protein in the heterologous host.
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (A) and Western blot analysis (B and C) of Cry32Aa protein extracts from strains DH5α and BL21(DE3). Immunoblot analysis was performed with the antisera to the 140-kDa (B) and 45-kDa (C) proteins. Lanes: 1, protein marker; 2, solubilized crystal of B. thuringiensis subsp. yunnanensis; 3, B. thuringiensis subsp. kurstaki strain HD 73; 4, E. coli DH5α(pSDMK1); 5, E. coli DH5α; 6, E. coli BL21(DE3)(pPSVS) induced with 0.4 mM IPTG; 7, E. coli BL21(DE3).
Bioassays.
Due to the close homology of Cry32Aa to Cry4 and Cry1 classes of proteins, it was thought prudent to check its toxicity against mosquitoes (Diptera) and moths (Lepidoptera). The protein preparation from B. thuringiensis subsp. yunnanensis was quantified by the method of Lowry et al. (9).
Bioassays against moths (Lepidoptera) were carried out with B. thuringiensis subsp. kurstaki HD73 as a positive control (1 μg/cm2). The solubilization buffer and water were used as negative controls. Bioassays were performed by coating suitable leaves cut to the desired size (young mulberry leaves for silkworm Bombyx mori and cotton leaves for the tobacco cutworm Helicoverpa armigera) with 50 μg of the preparations per cm2. Bioassays with P. xylostella were done with B. thuringiensis subsp. yunnanensis toxins as crude, solubilized, and trypsin-digested preparations. For each treatment, five larvae were used and the assays included four replicates. The surfaces of detached mustard leaves were coated with the protein preparations (10 and 25 μg/cm2), and 12 such leaves were placed in a petri dish lined with moist filter paper. The mortality of the larvae was scored after 24 and 72 h. The toxin showed weak toxicity only for P. xylostella (40 to 55% mortality at 10 μg/cm2 and 90% at 25 μg/cm2 after 72 h). The positive control strain (HD73) showed very strong toxicity (100% after 24 h) for all moths. Weak toxicity of B. thuringiensis B109 for P. xylostella larvae has been reported by Zelazny et al. (24).
Toxicity for dipteran insects (mosquitoes like Culex quinquefasciatus, Culex tritaeniorhynchus, Aedes aegypti, and Anopheles stephensi) was assayed by using 50 μg of Cry proteins from B. thuringiensis subsp. yunnanensis per ml. B. thuringiensis subsp. israelensis was used as a positive control. The bioassays did not reveal any mortality for any mosquito species.
Nucleotide sequence accession number.
The sequence of the 5,589-bp insert in pSDMK1 has been deposited in EMBL/DDBBJ/GenBank with the accession number AY008143.
Acknowledgments
P.B. and R. J. thank the Council for Scientific and Industrial Research for senior research fellowships. P.S. is grateful to the Department of Biotechnology, Government of India, for a studentship.
We thank G. Srinivas for his help during the initial stages of this work.
The first two authors contributed equally to this work.
REFERENCES
- 1.Beegle, C. C., and T. Yamamoto. 1992. History of Bacillus thuringiensis Berliner research and development. Can. Entomol. 124:587–616. [Google Scholar]
- 2.Bietlot, H. P., P. R. Carey, M. Pozsgay, and H. Kaplan. 1989. Isolation of carboxyl-terminal peptides from proteins by diagonal electrophoresis: application to the entomocidal toxin from Bacillus thuringiensis. Anal. Biochem. 181:212–215. [DOI] [PubMed] [Google Scholar]
- 3.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Vane Rie, D. Lereclus, J. Baum and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glatron, M. F., and G. Rapoport. 1972. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half life of its corresponding messenger RNA. Biochimie 54:1291–1301. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez, J. M., Jr., H. T. Dulmage, and B. C. Carlton. 1981. Correlation between specific plasmids and δ-endotoxin production in Bacillus thuringiensis. Plasmid 5:351–365. [DOI] [PubMed] [Google Scholar]
- 6.Kuo, W., and K. Chak. 1996. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl. Environ. Microbiol. 62:1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriology T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 9.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 10.Murty, M. G., G. Srinivas, R. S. Bora, and V. Sekar. 1994. A simple method for separation of the protein crystals from Bacillus thuringiensis using carboxymethyl cellulose column chromatography. J. Microbiol. Methods 19:103–110. [Google Scholar]
- 11.Nagamatsu, Y., Y. Itai, C. Hatanaka, G. Funatsu, and K. Hayashi. 1984. A toxic fragment from the entomocidal crystal protein of Bacillus thuringiensis. Agric. Biol. Chem. 48:611–619. [Google Scholar]
- 12.Ohba, M., and K. Aizawai. 1986. Crystals of Bacillus thuringiensis subsp. yunnanensis are produced only in asporogenous cells. J. Invertebr. Pathol. 60:3847–3853. [Google Scholar]
- 13.Rajamohan, F., J. A. Cotrill, F. Gould, and D. H. Dean. 1996. Role of domain II, loop 2 residues of Bacillus thuringiensis Cry1Ab δ-endotoxin in reversible and irreversible binding to Manduca sexta and Heliothis virescens. J. Biol. Chem. 271:2390–2396. [DOI] [PubMed] [Google Scholar]
- 14.Rajamohan, F., O. Alzate, J. A. Cotrill, A. Curtiss, and D. H. Dean. 1996. Protein engineering of Bacillus thuringiensis δ-endotoxin: mutations at domain II of Cry1Ab enhance receptor affinity and toxicity towards gypsy moth larvae. Proc. Natl. Acad. Sci. USA 93:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, G. P., and D. J. Ellar. 1994. Mutagenesis of two surface-exposed loops of the Bacillus thuringiensis Cry1C δ-endotoxin affects insecticidal specificity. Biochem. J. 302:611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivas, G., S. J. Vennison, S. N. Sudha, P. Balasubramanian, and V. Sekar. 1997. Unique regulation of crystal protein production in Bacillus thuringiensis subsp. yunnanensis is mediated by the Cry protein-encoding 103-megadalton plasmid. Appl. Environ. Microbiol. 63:2792–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gets to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong, H. C., and S. Chang. 1986. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc. Natl. Acad. Sci. USA 83:3233–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong, H. C., H. E. Schnepf, and H. R. Whiteley. 1983. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J. Biol. Chem. 258:1960–1967. [PubMed] [Google Scholar]
- 23.Wu, S.-J., and D. H. Dean. 1996. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA δ-endotoxin. J. Mol. Biol. 255:628–640. [DOI] [PubMed] [Google Scholar]
- 24.Zelazny, B., D. Stephan, and J. Hamacher. 1994. Irregular crystal formation in some isolates of Bacillus thuringiensis. J. Invertebr. Pathol. 63:229–234. [Google Scholar]