Abstract
We describe, for the first time, a detailed electroporation procedure for Lactobacillus delbrueckii. Three L. delbrueckii strains were successfully transformed. Under optimal conditions, the transformation efficiency was 104 transformants per μg of DNA. Using this procedure, we identified several plasmids able to replicate in L. delbrueckii and integrated an integrative vector based on phage integrative elements into the L. delbrueckii subsp. bulgaricus chromosome. These vectors provide a good basis for developing molecular tools for L. delbrueckii and open the field of genetic studies in L. delbrueckii.
Lactobacillus delbrueckii subsp. bulgaricus is widely used in the dairy industry as a starter for yogurt, fermented milk, and Italian cheese production. Recently, this organism has also been considered a putative probiotic microorganism (19, 22). Despite the importance of L. delbrueckii subsp . bulgaricus in the food industry, our understanding of the physiology and genetics of this bacterium is still limited. This fact can be explained in part by a lack of molecular tools, mainly due to the absence of a reliable transformation procedure.
Several reports have described introduction of DNA into L. delbrueckii strains by protoplast transformation (4), protoplast transfection (8), and electroporation 28, 36; T. Sasaki, Y. Ito, and Y. Sasaki, 4th Symp. Lactic Acid Bacteria, abstr. A8, 1993; T. Sasaki, 20 July 1993, Japanese Patent Office). However, the use of protoplast transformation and transfection methods has not been described since these reports, and there are major difficulties in using the briefly described electroporation procedure. Only one strain belonging to a public collection, ATCC 11842, can be transformed with the electroporation method, and it can be transformed only with very low reproducibility and efficiency.
The development of electroporation procedures for several Lactobacillus species, such as Lactobacillus helveticus (6, 23), Lactobacillus casei (11), Lactobacillus acidophilus (32), and Lactobacillus sakei (5), highlighted the conclusion that several parameters have to be tested in order to optimize electroporation efficiency for this group of organisms. Among these parameters are (i) the growth stage at which cells are harvested, which depends on the species or even the strain used (5, 6); (ii) the composition of the wash and electroporation buffers, which has been shown to play an important role in the transformation of several lactobacilli (2, 5, 32); (iii) the parameters of the electrical pulse (2); and (iv) the source of the DNA used to transform, since restriction-modification systems can severely inhibit transformation with foreign DNA (23).
Another difficulty in the development of a transformation procedure is the choice of the plasmid used. Indeed, the ability of a plasmid to replicate and to express its selectable marker in a given species is not predictable. The use of an endogenous plasmid carrying a native selectable marker often solves this problem. Unfortunately, it has been established that L. delbrueckii subsp. bulgaricus strains harbor very few plasmids. So far, three plasmids from L. delbrueckii subsp. bulgaricus and two plasmids from L. delbrueckii subsp. lactis have been isolated and at least partially sequenced. However, the replication mechanisms of these plasmids were not determined, and none of the plasmids carried an antibiotic resistance gene. The plasmids originating from L. delbrueckii subsp. bulgaricus are pBUL1 (Y. Ito, Y. Sasaki, and T. Sasaki, 4th Symp. Lactic Acid Bacteria, abstr. A9, 1993; Y. Ito, 3 March 1993, European Patent Office), pN42 (R. D. Pridmore, C. Blancpain, and B. Mollet, 4th Symp. Lactic Acid Bacteria, abstr. P10, 1993; B. Mollet, 15 March 1995, European Patent Office), and pLBB1 (GenBank accession number AF236060). Plasmid pWS58 (28) and, more recently, plasmid pLL1212 (GenBank accession number AF109691) were isolated from L. delbrueckii subsp. lactis. Only one plasmid, an erythromycin-resistant derivative of pBUL1 (designated pX3), was reintroduced into L. delbrueckii subsp. bulgaricus (Ito et al., 4th Symp. Lactic Acid Bacteria; Ito, European Patent Office). Other data have resulted from the use of heterologous plasmids; pIP501 (originating from Streptococcus agalactiae) was conjugated from Lactococcus lactis into L. delbrueckii subsp.bulgaricus IL-429 and IL-431 (30), pSY2 (originating from Lactococcus lactis) was electroporated into L. delbrueckki subsp. bulgaricus T-11 (36), and pGT633 (isolated from Lactobacillus reuteri [38]) and pCU1882 (a derivative of a Lactobacillus curvatus cryptic plasmid [29]) have been reported to transform L. delbrueckii. However, except for pSY2 (36), none of these plasmids was used for genetic experiments performed with L. delbrueckii. Because the plasmids mentioned above have different replication mechanisms, as well as different antibiotic resistance genes, no obvious criterion can be used to predict the ability of a plasmid to replicate in L. delbrueckii subsp. bulgaricus.
In this work, we adapted the previously described procedure for electrotransformation of L. delbrueckii subsp.bulgaricus (Sasaki et al., 4th Symp. Lactic Acid Bacteria, abstr. A8, 1993) to a strain, which resulted in about 104 transformants per μg of plasmid DNA in routine use. Using this protocol, we identified 9 replicative plasmids among 13 plasmids tested, and we showed that pMC1, a site-specific integrative plasmid (16), can integrate in the L. delbrueckii subsp. bulgaricus chromosome. In addition, two other L. delbrueckii strains were reproducibly transformed with this procedure.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The L. delbrueckii subsp. bulgaricus strains used in this work were CNRZ208 (which corresponds to type strain ATCC 11842), CNRZ397, CNRZ1057, and VI104 from our collection (Génétique Microbienne, Jouy-en-Josas, France). The L. delbrueckii subsp. lactis strains used were CNRZ700, CNRZ1003, CNRZ1059, and CNRZ327 (corresponding to ATCC 10697). All CNRZ strains were provided by the INRA National Culture Collection (URLGA, Jouy-en-Josas, France). L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis strains were distinguished on the basis of phenotypic criteria and randomly amplified polymorphic DNA analysis (P. Quéné and P. Tailliez, personal communication). Escherichia coli strains GM2929 [F−dam-13::Tn 9(Cmr)dcm-6 mcrB1 hsdR2(rk− mk+) mcrAara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galK2 galT22 supE44 hisG4 rpsL136(Strr) xyl-5 mtl-1], GM1674 { dam-3 dcm-6 Δ(lac-pro) tsx-78 glnV44 galK2 galT22 thi-1 F′ [lacIqΔ(lacZ)M15 pro +]}, and JJC128 [hsdR hsdM+araD139Δ(ara-leu)7696 galE15 galK16Δ(lac)X74 Strr] were used for plasmid propagation to check the effects of different restriction-modification mutations on transformation efficiency.
The plasmids used are listed in Table 1. Plasmids pX3, pJK650, pNZ12, pGB305Δ, and pG+host4 were isolated from Lactococcus lactis IL-1403 (12). Plasmid pLEM7 was isolated from L. reuteri LEM83 (17). Plasmids pE194 and pC194 were propagated in Bacillus subtilis SB202 (26), whereas plasmid pGT633 was isolated from B. subtilis BD170 (38). Plasmids pLEM415, pULP8, pCU1882, pAZ20, pJK300, and pMC1 were extracted from E. coli TG1 {hsdR supE thi Δ(lac-proAB) F′ [traD36 proAB lacIqΔ(lacZ)M15]} (20).
TABLE 1.
Efficiencies of L. delbrueckii subsp.bulgaricus VI104 transformation with various plasmids
| Plasmid (replicon, type)a | Size (kb) | Antibiotic markersb | Transformants/ μg | Refer- ence |
|---|---|---|---|---|
| pLEM415 (pLEM3, RC) | 6.3 | Em | 103−104 | 17 |
| pX3 (pBUL1, ND) | 9.0 | Emb | >103 | Itoc |
| pJK650 (pWS58, ND) | 8.96 | Ema | >103 | 28 |
| pULP8 (pLP1, RC) | 6.6 | Emc | >102 | 9 |
| pNZ12 (pSH71, RC) | 4.1 | Cmd | >102 | 15 |
| pG+ host4 (pWVO1, RC) | 3.8 | Ema | >102 | 33 |
| pGB305Δ (pIP501, θ) | 5.3 | Em | >102 | 31 |
| pGT633 (pGT633, ND) | 9.8 | Em | >10 | 38 |
| pCU1882 (pLC2, RC) | 6.7 | Cmd | >10 | 29 |
| pE194 (pE194, RC) | 3.7 | Em | 0 | 24 |
| pC194 (pC194, RC) | 2.9 | Cm | 0 | 25 |
| pJK300 (pWS97, ND) | 8.3 | Cmd | 0 | 40 |
| pAZ20 (pNCDO151, ND) | 6.8 | Cmd | 0 | 40 |
The replicon is the original plasmid containing the replication apparatus. The type is the type of replication mechanism used by the plasmid (RC, rolling circle; ND, not determined; θ, theta). pLEM415, pULP8, and pC194 belong to the pC194 family of rolling-circle plasmids. pNZ12, pG+host4, pCU1882, and pE194 belong to the pE194 family of rolling-circle plasmids.
Antibiotic resistance genes used to select for the plasmids. Em, native erythromycin resistance gene; Ema, gene originally identified in pE194; Emb, gene of pAMβ1; Emc, gene of pVA891 (9); Cm, native chloramphenicol resistance gene; Cmd, gene of pC194.
Ito, Y. Ito, European Patent Office.
Media and growth conditions.
L. delbrueckii subsp. bulgaricus was grown in MRS (Difco, Detroit, Mich.) (14) at 37 or 42°C. Plates were incubated under anaerobic conditions in jars containing GasPak (Oxoid, Basingstoke, England). E. coli and B. subtilis were grown on Luria-Bertani medium (Difco) (35) at 37°C. Lactococcus lactis was grown on M17 containing 0.5% glucose (39) at 30°C. Antibiotics were added at the following concentrations: erythromycin, 2.5 μg/ml for B. subtilis, 5 μg/ml for Lactococcus lactis, 7.5 μg/ml for L. delbrueckii subsp.bulgaricus, and 150 μg/ml for E. coli; chloramphenicol, 7.5 μg/ml for Lactococcus lactis and L. delbrueckii subsp. bulgaricus; and ampicillin, 75 μg/ml for E. coli.
The effect of glycine as a cell wall-weakening agent was tested by using the following concentrations: 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 1, 1.5, 2.0, 2.5, and 3.0%.
Transformation procedures.
The initial electrotransformation procedure was described by Sasaki et al. (Sasaki et al., 4th Symp. Lactic Acid Bacteria; Sasaki, Japanese Patent Office) and is outlined in Table 2. The optimized transformation procedure was as follows. Serial dilutions of a fresh bacterial culture were inoculated into 100 ml of MRS and incubated at 42°C. After overnight growth, a culture at the beginning of the stationary phase (optical density at 600 nm [OD600], 1.7) was harvested by centrifugation. The bacteria were washed once with 100 ml of cold electroporation buffer (EB) (0.4 M sucrose [Merck, Darmstadt, Germany], 1 mM MgCl2 [Merck], 5 mM KH2PO4 [Merck]; pH 6) and twice with 30 ml of cold EB. The cells were then resuspended in EB to an OD600 of about 50. The cell suspension was incubated at 45°C for 20 min and then kept on ice for 10 min. For electroporation, 80 μl of the cell suspension was mixed with 0.3 to 2μg of plasmid DNA. The sample was subjected to a 1-kV, 800-Ω, 25-μF electric pulse in a 0.2-cm cuvette by using a Gene Pulser and a Pulse Controller apparatus (Bio-Rad, Richmond, Calif.). Two milliliters of milk medium (0.2 M sucrose, 5% skim milk [Regilait, Saint-Martin-Belle-Roche, France], 0.1% yeast extract [Difco], 1% Casamino Acids [Difco], 25 mM MgCl2) was immediately added, and the cells were incubated for 3 h at 37°C before they were plated on MRS supplemented with the appropriate antibiotic. The plates were incubated at 37°C for 2 to 3 days under anaerobic conditions.
TABLE 2.
Outline of the electroporation procedures
| Step | Initial procedurea | Modified procedure |
|---|---|---|
| Cell culture | MRS overnight, 2 h in MRS (pH 5.5) | MRS-0.1% Gly, onset of stationary phase |
| Wash buffer | Tris (20 mM, pH 7.0) | EBa (pH 6.0)b |
| Thermal shock | EBb (pH 7.0), 30 min, 45°Cc | EBa (pH 6.0), 20 min, 45°C |
| Electrical pulse | 1.5 kV, 200 Ω, 25μF | 1 kV, 800 Ω, 25 μF |
| Expression | Milk mediumd | Milk mediume |
| Plating, selection | Skim milk agar + antibiotic | MRS agar + antibiotic |
Described by Sasaki et al. (4th Symp. Lactic Acid Bacteria) and Sasaki (Japanese Patent Office).
EBa contained 0.4 M sucrose, 1 mM MgCl2, and 5 mM KH2PO4 (pH 6.0).
EBb contained 0.3 M raffinose, 1 mM MgCl2, and 1 mM KH2PO4 (pH 7.0).
The milk medium contained 0.15 M raffinose, 5% skim milk, 0.1% yeast extract, 1% Casamino Acids, and 25 mM MgCl2.
The milk medium contained 0.2 M sucrose, 5% skim milk, 0.1% yeast extract, 1% Casamino Acids, and 25 mM MgCl2.
For each parameter tested at least three independent transformation assays were performed. The transformation efficiencies were expressed as the number of transformants per microgram of plasmid DNA, and means ± the standard errors were determined.
DNA techniques.
For DNA extraction, L. delbrueckii subsp. bulgaricus cells were first protoplasted by 45 min of incubation at 37°C in TES (50 mM Tris [pH 7], 20 mM EDTA, 0.2 mM sucrose) containing lysozyme (10 mg/ml) and mutanolysin (70 U/ml). Plasmid extraction was then performed by the alkaline lysis procedure (7), followed by cesium chloride density gradient centrifugation (35). After lysozyme and mutanolysin treatment, total DNA was extracted as previously described (18). Plasmids were extracted from E. coli, B. subtilis, L. reuteri, and Lactococcus lactis by the alkaline lysis procedure. Restriction enzyme digestion and gel electrophoresis were carried out as recommended by the suppliers or by standard methods (35). DNA hybridization was performed as described by Southern (37). DNA probes were labeled with [α -32P]dCTP (ICN Biomedicals, Costa Mesa, Calif.) by using a random priming kit (Boehringer, Mannheim, Germany).
The DNA corresponding to the 16S rRNA was PCR amplified with primers W001 (5′-AGAGTTTGATCCTGGCTC-3′) and W025 (5′ -CACGTCCTTCATCGGCT-3′) (21). Fluorescent sequencing was performed by using the recommendations of Perkin-Elmer Biosystems (Foster City, Calif.).
RESULTS AND DISCUSSION
Electrotransformation of L. delbrueckii subsp. bulgaricus.
Because pX3 could be reintroduced into L. delbrueckii subsp. bulgaricus by electrotransformation (Ito et al., 4th Symp. Lactic Acid Bacteria), we first used this plasmid and the previously described electroporation protocol (Table 2) (Sasakiet al., 4th Symp. Lactic Acid Bacteria) to transform a few L. delbrueckii subsp. bulgaricus strains, including ATCC 11842 and VI104. Transformants were obtained sporadically with ATCC 11842, whereas transformation was reproducible with VI104, although the number of transformants was variable. For example, in three independent experiments, the number of VI104 transformants varied from 55 to less than 10 transformants per μg of DNA. The presence of free plasmid forms in transformants was confirmed. Several plasmids were then tested to determine their ability to transform VI104. One of these plasmids, pLEM7 (originating from L. reuteri) (17), gave the highest transformation efficiency (160 transformants per μg of DNA), and analysis of the transformants confirmed the presence of free forms of pLEM7.
These findings indicated that (i) we were able to reproducibly transform VI104 (although with variable efficiency) and (ii) the heterologous pLEM7 plasmid seemed to transform slightly more efficiently than pX3. Therefore, strain VI104 and plasmid pLEM7 were used to optimize L. delbrueckii subsp. bulgaricus electroporation.
Optimization of the electroporation procedure.
As mentioned above, several parameters can influence electroporation efficiency; we tested most of them as described below.
(i) Growth stage and medium effects.
In the initial procedure, 10 ml of cells from an overnight culture (OD600,>2) was centrifuged, and the cells were suspended in 100 ml of MRS at pH 5.5. The suspension was incubated for 2 h at 42°C prior to the washing steps (Table 2). We checked whether this 2-h incubation step could be eliminated. Serial dilutions of a bacterial culture were grown overnight at 42°C. An overnight culture whose pH was 5.5 (OD600, 0.8) was harvested and prepared for electroporation with or without the incubation step. Cells that were incubated at pH 5.5 gave an average of ∼50 transformants per μg of DNA, compared to ∼300 transformants per μg of DNA for cells that were not incubated at pH 5.5. In further experiments, cells were harvested at different OD600s from the early exponential phase to the early stationary phase and electroporated with or without the pH 5.5 incubation step. Our results showed that the transformation efficiency was always higher when the cells were not incubated at pH 5.5 prior to electroporation (data not shown). These experiments also revealed that the transformation efficiency was as high as 750 transformants per μg of DNA when cells harvested at the beginning of the stationary phase (OD600, 1.7) were used, whereas cells harvested in the exponential phase (OD600, 0.2 to 1.0) or in the stationary phase (OD600, >2.0) yielded lower transformation efficiencies (<200 transformants per μg of pLEM7).
Glycine was added at several concentrations to the growth medium used for overnight culture in order to determine whether it weakened the cells. None of the glycine concentrations tested significantly improved the transformation efficiency, but we noticed a less-than-twofold improvement when we used VI104 grown in MRS containing 0.1% glycine.
In summary, using cells grown in MRS containing 0.1% glycine until the OD600 was 1.7 and omitting the pH 5.5 incubation step shortened the procedure and improved the transformation efficiency at least 10-fold. These conditions were used for the subsequent experiments.
(ii) Source and amount of plasmid DNA.
To facilitate plasmid preparation, instead of pLEM7 we used a derivative of pLEM7, pLEM415, an E. coli-L. reuteri shuttle vector (17). In order to identify the best E. coli strain to propagate the plasmid before its introduction into VI104, we tested three restriction-modification mutants of E. coli (see Materials and Methods). The average transformation efficiencies with pLEM415 prepared from L. delbrueckii subsp.bulgaricus and the average transformation efficiencies with pLEM415 prepared from E. coli were of the same order of magnitude (7.4 × 102 ± 4.5 × 102 transformants per μg). However, with DNA originating from GM1674 (dam dcm), the average transformation efficiency was 1 × 103 ± 0.4 × 103 transformants per μg. Such a slight difference cannot be attributed to restriction of DNA entering L. delbrueckii subsp. bulgaricus; it could have been due to differences in the monomer/multimer ratio in the plasmid DNA preparations. Using different amounts of DNA, we observed that the transformation efficiency reached a plateau between 0.3 and 2 μg of DNA; 1 to 1.5 μg of pLEM415 was used in the experiments described below.
(iii) Wash solution and EB.
Addition of sucrose (0.2 to 0.4 M), an osmotic stabilizer, to the original wash solution (Tris buffer, pH 7.0) improved the transformation efficiency. Since sucrose is more soluble and cheaper than raffinose, which was used in the original EB (designated EBb) (Table 2), we replaced raffinose with sucrose in the EB, which resulted in a buffer designated EBa (Table 2). Washing the cells with EBa reproducibly increased the transformation efficiency twofold. Although MgCl2 has been reported to improve electroporation by eliminating extracellular polysaccharides (1, 5), no increase in the transformation efficiency was observed with various concentrations of MgCl2. Finally, competent cells were washed three times with EBa before thermal treatment and electroporation.
(iv) Thermal treatment.
Induction of autolysis of L. delbrueckii subsp. bulgaricus at 45°C in EBb (pH 7.0) is used in the electroporation procedure to weaken the cell wall (T. Sasaki, Y. Asami, and N. Taketomo, 2nd Symp. Lactic Acid Bacteria, abstr. D18, 1987; Sasaki et al., 4th Symp. Lactic Acid Bacteria). The optimal parameters for autolysin induction and activity are different for different bacterial strains (10). In order to determine the optimal parameters for VI104, we tested EBa at several pH values (pH 7.0, 6.5, 6.0, 5.5, and 5.0) combined with various incubation times at 45°C. These experiments showed that the highest transformation efficiencies obtained with EBa at pH 5.5 and 5.0 were 25- and 190-fold lower, respectively, than the transformation efficiency obtained with EBa at pH 6.0. Next, we found that for any incubation time (from 7 to 75 min), the pH 6.0 buffer led to better transformation efficiencies than the pH 6.5 or 7.0 EBa (Fig. 1). The highest transformation efficiency (8 × 103± 1.3 × 103 transformants per μg) was obtained after 20 min of incubation in EBa at pH 6.0. In further experiments, the cell wall was weakened by induction of autolysis by using similar conditions.
FIG. 1.
Optimization of thermal treatment: effects of various incubation times at 45°C in pH 6.0 (black bars), pH 6.5 (grey bars), and pH 7.0 (white bars) EB on the transformation efficiency of VI104. The results are representative of the results obtained in five independent experiments. Cells were prepared by the modified procedure (Table 2). Electroporation was performed with pLEM415 DNA at 1.5 kV, 25μF, and 200 Ω. T, transformants.
(v) Electrical parameters.
Finally, cells were electroporated by using various electrical settings (voltage and resistance) at a capacitance of 25 μF. With the different combinations, the transformation efficiencies varied from 0.4 to 1.4 times the initial value obtained with 1.5 kV and 200 Ω. The maximum transformation efficiency was reproducibly obtained when cells were subjected to 1 kV and 800 Ω at 25 μF.
In conclusion, the modifications of the electrotransformation procedure (Table 2) reliably increased the transformation efficiency of VI104 from a maximum of about 102 transformants perμg to several thousand transformants per μg (5 × 103 to 104 transformants per μg). The new procedure described here is reproducible for a batch of prepared cells, but a two- to fourfold variation of the transformation efficiency was observed when different electrocompetent cell batches prepared simultaneously were used.
Transformation with various plasmids. (i) Replicative plasmids.
Using the optimized transformation procedure, we tested several plasmids (Table 1) to determine their abilities to replicate and to be maintained in L. delbrueckii subsp.bulgaricus VI104. Based on the transformation efficiencies, three groups of plasmids were distinguished. These groups correspond to plasmids yielding (i) at least 102 transformants per μg, (ii) less than 102 transformants perμg, and (iii) no transformants (Table 1). The presence of free plasmids in the transformants was confirmed by DNA extraction and comparison with the initial DNA.
In addition to pLEM415, which was used in all our previous tests, six plasmids gave at least 102 transformants per μg (Table 1). They were pX3 and pJK650, both of which originated from L. delbrueckii, and the heterologous plasmids pNZ12, pG+host4, pGB305Δ, and pULP8. The copy numbers of these plasmids were variable in L. delbrueckii subsp. bulgaricus (Fig. 2). pJK650 and pX3 were easily detected by ethidium bromide staining of total DNA extracts, whereas pNZ12, pG+host4, and pGB305Δ had low copy numbers and were barely detectable. pULP8 was detected only by hybridization, which also revealed the presence of high-molecular-weight multimers (13) in the transformants.
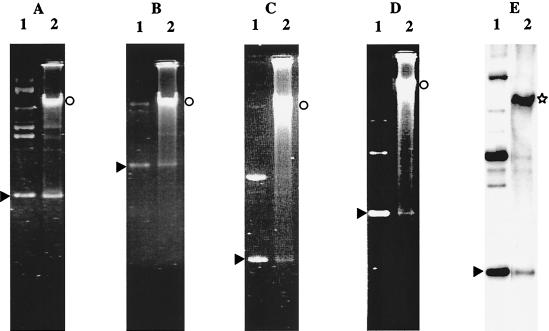
FIG. 2.
Analysis of total DNA of L. delbrueckii subsp. bulgaricus VI104 transformants by agarose gel electrophoresis (A to D) and by hybridization with pULP8 as the probe (E). Total DNA extracted from one transformant (lanes 2) was compared with the following plasmids used for transformation (lanes 1): pLEM415 (A), pJK650 (B), pNZ12 (C), pG+host4 (D), and pULP8 (E). The arrowheads, the circles, and the star indicate the positions of the covalently closed plasmid monomers, the high-molecular-weight multimers, and the chromosomal DNA, respectively.
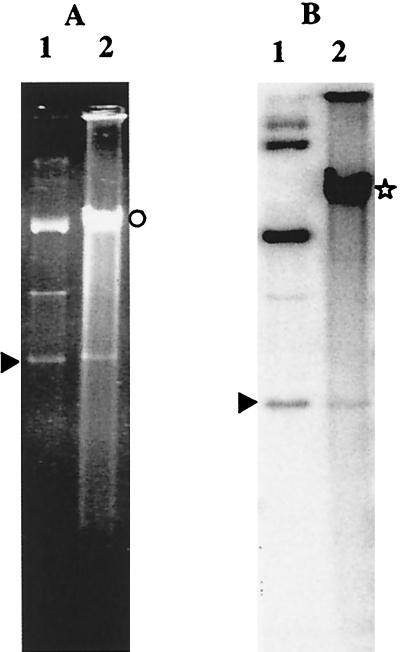
Plasmids pGT633 and pCU1882 yielded between 10 and 102 transformants per μg (Table 1). Extraction of the total DNA of these transformants indicated that both plasmids were present at low copy numbers (Fig. 3). pGT633 could be detected on ethidium bromide-stained gels, whereas hybridization was needed for detection of monomers and high-molecular-weight multimers of pCU1882.
FIG. 3.
Analysis of total DNA of L. delbrueckii subsp. bulgaricus VI104 transformants by agarose gel electrophoresis (A) and by hybridization with pCU1882 as the probe (B). Total DNA extracted from one transformant (lanes 2) was compared with the following plasmids used for transformation (lanes 1): pGT633 (A) and pCU1882 (B). The arrowheads, the circle, and the star indicate the positions of the covalently closed plasmid monomers, the high-molecular-weight multimers, and the chromosomal DNA, respectively.
Attempts to transform VI104 with pC194, pE194, pAZ20, and pJK300 failed. This result suggests that these plasmids cannot replicate or establish themselves efficiently in L. delbrueckii subsp.bulgaricus.
(ii) Site-specific integrative vector.
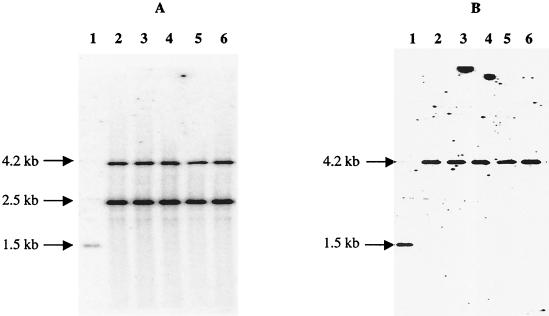
Plasmid pMC1 contains the attP site and the integrase (int) gene of the L. delbrueckii mv4 bacteriophage (16). Since no transformation procedure was available for L. delbrueckii at the time, Auvray et al. showed that pMC1 was able to integrate into the tRNASer gene of a wide range of lactic acid bacteria, such as Lactobacillus plantarum, L. casei, Lactococcus lactis, Enterococcus faecalis, and Streptococcus pneumoniae (3). As expected, pMC1 transformed VI104, although with a lower transformation efficiency (between 30 and 200 transformants perμg depending on the DNA preparation) than that of replicating plasmids, such as pX3 (9 × 103 transformants per μg). The values given above allowed us to estimate that the pMC1 integration efficiency was between 3 × 10−3 and 2 × 10−2 integrants per transformant of VI104. We verified by using Southern analysis that all transformants resulted from site-specific integration of pMC1 at the tRNASer locus of VI104 (Fig. 4). This plasmid provides an efficient way to generate stable DNA integration in L. delbrueckii subsp. bulgaricus.
FIG. 4.
Analysis of total DNA of L. delbrueckii subsp. bulgaricus VI104 pMC1 integrants. Total DNA from VI104 (lanes 1) and from five integrants (lanes 2 to 6) were digested with EcoRI and hybridized. (A) The pMC1 probe revealed a 1.5-kb EcoRI fragment on the chromosomal DNA of VI104 (lane 1) and two fragments (4.2 and 2.5 kb) corresponding to the two EcoRI junction fragments resulting from pMC1 integration (lanes 2 to 6). (B) The tRNASer gene probe revealed a 1.5-kb EcoRI fragment on the chromosomal DNA of VI104 (lane 1) and a 4.2-kb fragment, as expected after pMC1 integration.
The plasmids used carry different selectable markers, such as the chloramphenicol resistance gene of pC194 and the erythromycin resistance genes of pAMβ1, pIP501, pVA891, pE194, pGT633, and pLEM3. Selection of transformants indicated that these markers can be used in L. delbrueckii subsp. bulgaricus, although we should mention that the pAMβ1, pVA891, and pE194 erythromycin resistance genes seem to be expressed better than the other genes.
Transformation of other L. delbrueckii strains.
Using the conditions optimized for VI104, we tested three L. delbrueckii subsp. bulgaricus strains (ATCC 11842, CNRZ397, and CNRZ1057) and four L. delbrueckii subsp.lactis strains (CNRZ327, CNRZ700, CNRZ1003, and CNRZ1059) for transformation with pLEM415 and pJK650. Only two strains gave transformants. L. delbrueckii subsp. bulgaricus ATCC 11842 was transformed only by pJK650, at a frequency of 2 × 103 ± 1 × 103 transformants per μg of DNA. CNRZ327, an L. delbrueckii subsp. lactis strain, was transformed at a frequency of about 50 ± 10 transformants per μg of DNA with both plasmids. Replicative forms of the plasmids were always detected in the transformants (data not shown).
The negative transformation results obtained with the other five strains tested might have been due to a nonoptimal electroporation protocol, although the possibility of restriction of incoming DNA cannot be eliminated. We noticed that most of the L. delbrueckii subsp. bulgaricus strains were phage resistant (T. Smokvina, personal communication), which could have been due to restriction. Similarly, the low transformation efficiency obtained with CNRZ327 when pJK650 or pLEM415 was used might have been due to a nonoptimal electroporation procedure or to restriction of the incoming DNA.
However, our results also suggest that transformation of L. delbrueckii might be strain dependent. First, L. delbrueckii subsp. bulgaricus strains ATCC 11842 and VI104 gave comparable transformation efficiencies with pJK650 but not with pLEM415. To evaluate the genetic similarity between these two strains, their 16S DNAs were sequenced. The sequences were found to be identical and corresponded to the sequence of L. delbrueckii subsp. bulgaricus. Although this result suggested that VI104 and ATCC 11842 are closely related, VI104 exhibited a broader plasmid range than ATCC 11842. Second, L. delbrueckii subsp.bulgaricus T-11 (36) was reproducibly transformed with pX3 and pJK650 at an efficiency of 104 transformants per μg, but no transformants were obtained with plasmids pNZ12 and pG+host5 (data not shown); in contrast, these four plasmids transformed VI104. This kind of observation is not limited to L. delbrueckii. Indeed, transformation efficiency is frequently strain dependent in various species of lactic acid bacteria, including L. sakei (5), L. casei, and L. plantarum (34). It has been proposed that in L. sakei (5) and Propionibacterium strains (27), plasmid replication may be strain specific. Similarly, in L. delbrueckii, differences in restriction-modification systems or in expression of host factors involved in plasmid replication could modify the transformation abilities of strains.
Conclusion.
Although there have been several reports of transformation of L. delbrueckii, the transformability of this species has been questioned many times on the basis of numerous unsuccessful attempts. For the first time, we describe a detailed and reproducible electroporation procedure which can be used for at least three L. delbrueckii strains, ATCC 11842, VI104, and CNRZ327. Using various strains, we also confirmed that the transformation abilities of strains vary. We suspect that the autolysis induction step is critical and has to be optimized for each strain in order to successfully electroporate L. delbrueckii.
Several plasmids which replicate efficiently in L. delbrueckii subsp. bulgaricus were also identified. Our data show that plasmids of different replication types are active in L. delbrueckii. Besides the L. delbrueckii replicons, whose replication mechanism was not determined (pBUL1 and pWS58), heterologous replicons of the theta type (pIP501) and the rolling-circle type (pWVO1Ts, pSH71, pLEM3, pLC2, and maybe pLP1) transform L. delbrueckii VI104 or CNRZ327. However, it may still be necessary to distinguish between plasmids originating from L. delbrueckii and heterologous plasmids. Plasmid pJK650 (from L. delbrueckii) transforms ATCC 11842 and CNRZ327, whereas the pLEM derivative (from L. reuteri) does not transform ATCC 11842 although it transforms VI104 and CNRZ327. The basis for this difference between endogenous and heterologous plasmids is not understood yet.
The replicative plasmids, the integrative vector pMC1, and the antibiotic resistance genes all provide a solid basis for development of genetic and molecular studies of L. delbrueckii.
Acknowledgments
We thank S. Kulakauskas for the gift of primers W001 and W025 and E. coli strains, P. Bourgeois, W. de Vos, M. Fons, G. W. Tannock, J. Klein, P. Ritzenthaler, P. Tailliez, and P. Quéné for the gift of strains and plasmids, T. Smokvina and members of M. C. Chopin’s group for helpful discussions, and M. van de Guchte for reading the manuscript. R. Dervyn and S. François are gratefully acknowledged for their technical participation in this work. We also thank L. Benbadis, C. Fremaux, and A. Sepulchre for their interest in this study.
Part of this work was supported by Rhodia Food (Texel) and Danone (Vitapole).
REFERENCES
- 1.Aukrust, T. W., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253–261. [Google Scholar]
- 2.Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201–208. [DOI] [PubMed] [Google Scholar]
- 3.Auvray, F., M. Coddeville, P. Ritzenthaler, and L. Dupont. 1997. Plasmid integration in a wide range of bacteria mediated by the integrase of Lactobacillus delbrueckii bacteriophage mv4. J. Bacteriol. 179:1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batt, C. A. 1986. Genetic engineering of Lactobacillus. Food Technol. 40:95–98. [Google Scholar]
- 5.Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273–1279. [DOI] [PubMed] [Google Scholar]
- 6.Bhowmik, T., and J. L. Steele. 1993. Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ 32. J. Gen. Microbiol. 139:1433–1439. [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boizet, B., J. L. Flickinger, and B. M. Chassy. 1988. Transfection of Lactobacillus bulgaricus protoplasts by bacteriophage DNA. Appl. Environ. Microbiol. 54:3014–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bringel, F., L. Frey, and J. C. Hubert. 1989. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid 22:193–202. [DOI] [PubMed] [Google Scholar]
- 10.Chapot-Chartier, M.-P. 1996. Les autolysines des bactéries lactiques. Lait 76:91–109. [Google Scholar]
- 11.Chassy, B. M., and J. L. Flickinger. 1987. Transformation of Lactobacillus casei by electroporation. FEMS Microbiol. Lett. 44:173–177. [Google Scholar]
- 12.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263. [DOI] [PubMed] [Google Scholar]
- 13.Dabert, P., S. D. Ehrlich, and A. Gruss. 1992. High-molecular-weight linear multimer formation by single-stranded DNA plasmids in Escherichia coli. J. Bacteriol. 174:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130–135. [Google Scholar]
- 15.De Vos, W. M. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281–295. [Google Scholar]
- 16.Dupont, L., B. Boizet-Bonhoure, M. Coddeville, F. Auvray, and P. Ritzenthaler. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fons, M., T. Hege, M. Ladire, P. Raibaud, R. Ducluzeau, and E. Maguin. 1997. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid 37:199–203. [DOI] [PubMed] [Google Scholar]
- 18.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365–378. [PubMed] [Google Scholar]
- 20.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge University, Cambridge, United Kingdom.
- 21.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldin, B. R. 1998. Health benefits of probiotics. Br. J. Nutr. 80:S203–S207. [PubMed] [Google Scholar]
- 23.Hashiba, H., R. Takiguchi, S. Ishii, and K. Aoyama. 1990. Transformation of Lactobacillus helveticus subsp.jugurti with plasmid pLHR by electroporation. Agric. Biol. Chem. 54:1537–1541. [PubMed] [Google Scholar]
- 24.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jannière, L., B. Niaudet, E. Pierre, and S. D. Ehrlich. 1985. Stable gene amplification in the chromosome of Bacillus subtilis. Gene 40:47–55. [DOI] [PubMed] [Google Scholar]
- 27.Jore, J. P., N. van Luijk, R. G. Luiten, M. J. van der Werf, and P. H. Pouwels. 2001. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, J. R., C. Ulrich, U. Wegmann, E. Meyer-Barton, R. Plapp, and B. Henrich. 1995. Molecular tools for the genetic modification of dairy lactobacilli. Syst. Appl. Microbiol. 18:493–503. [Google Scholar]
- 29.Klein, J. R., C. Ulrich, and R. Plapp. 1993. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid 30:14–29. [DOI] [PubMed] [Google Scholar]
- 30.Langella, P., and A. Chopin. 1989. Conjugal transfer of plasmid pIP501 from Lactococcus lactis to Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus helveticus. FEMS Microbiol. Lett. 51:149–152. [DOI] [PubMed] [Google Scholar]
- 31.Le Chatelier, E., S. D. Ehrlich, and L. Jannière. 1993. Biochemical and genetic analysis of the unidirectional theta replication of the S. agalactiae plasmid pIP501. Plasmid 29:50–56. [DOI] [PubMed] [Google Scholar]
- 32.Luchansky, J. B., P. M. Muriana, and T. R. Klaenhammer. 1988. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus,Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol. Microbiol. 2:637–646. [DOI] [PubMed] [Google Scholar]
- 33.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posno, M., R. J. Leer, N. van Luijk, M. J. F. van Giezen, P. T. H. M. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Satoh, E., Y. Ito, Y. Sasaki, and T. Sasaki. 1997. Application of the extracellular alpha-amylase gene from Streptococcus bovis 148 to construction of a secretion vector for yogurt starter strains. Appl. Environ. Microbiol. 63:4593–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517. [DOI] [PubMed] [Google Scholar]
- 38.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100–63. Plasmid 31:60–71. [DOI] [PubMed] [Google Scholar]
- 39.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zink, A., J. R. Klein, and R. Plapp. 1991. Transformation of Lactobacillus delbrueckii ssp.lactis by electroporation and cloning of origins of replication by use of a positive selection vector. FEMS Microbiol. Lett. 78:207–212. [Google Scholar]