Abstract
The nitrogen-fixing, symbiotic bacterium Sinorhizobium meliloti reduces molecular dinitrogen to ammonia in a specific symbiotic context, supporting the nitrogen requirements of various forage legumes, including alfalfa. Determining the DNA sequence of the S. meliloti genome was an important step in plant-microbe interaction research, adding to the considerable information already available about this bacterium by suggesting possible functions for many of the >6,200 annotated open reading frames (ORFs). However, the predictive power of bioinformatic analysis is limited, and putting the role of these genes into a biological context will require more definitive functional approaches. We present here a strategy for genetic analysis of S. meliloti on a genomic scale and report the successful implementation of the first step of this strategy by constructing a set of plasmids representing 100% of the 6,317 annotated ORFs cloned into a mobilizable plasmid by using efficient PCR and recombination protocols. By using integrase recombination to insert these ORFs into other plasmids in vitro or in vivo (B. L. House et al., Appl. Environ. Microbiol. 70:2806-2815, 2004), this ORFeome can be used to generate various specialized genetic materials for functional analysis of S. meliloti, such as operon fusions, mutants, and protein expression plasmids. The strategy can be generalized to many other genome projects, and the S. meliloti clones should be useful for investigators wanting an accessible source of cloned genes encoding specific enzymes.
As the number of bacterial genomes with known DNA sequences increases, the important problem of assigning functional roles to predicted features of these genomes is becoming more obvious. Some of this assignment, such as predicting open reading frames (ORFs) or finding similarities between previously characterized sequences and those in the target organism, is currently done via various bioinformatic techniques. Unfortunately, many ORFs have no similarity to proteins with known function and, even when a match can be made, obtaining bioinformatic predictions specific enough to fit ORFs into a biological context can be difficult, especially in the situation where many proteins are predicted to have similar and related biochemical functions. These related enzymes might be involved in a variety of metabolic processes, depending on the specific substrates and products they actually interact with. For example, Sinorhizobium meliloti is predicted to contain about 22 sugar kinases but, since the current annotation does not strongly predict the substrates for most of these, understanding sugar metabolism in S. meliloti did not advance substantially based on the initial genomic analysis. The level of specificity needed to accomplish a functional level of understanding will emerge from studies that associate specific sugars with genes encoding predicted sugar kinases, that define gene expression conditions, and that determine phenotypes for mutants with defects in individual genes. Evaluating families of highly diverse transport and regulatory proteins will require similar efforts to put these ORFs into a biological context. One step in understanding the specific functions of ORFs would be to clone them in a way that facilitates further analysis.
S. meliloti is a gram-negative α-proteobacterium that is found both as a free-living soil microbe and in a nitrogen-fixing symbiotic relationship with forage legumes such as Medicago, Melilotus, and Trigonella spp. This type of symbiosis is ecologically important, providing a large fraction of the nitrogen available to natural ecosystems. It also provides an important nutrient input for many crop plants and can be used as a nitrogen source in sustainable agricultural systems. Establishing a symbiotic relationship depends on specialized development of the plant root and the bacterium during bacterial infection (9, 18). The root nodule, a plant organ formed as a result of the interaction, provides an environment within which the bacteria can differentiate into bacteroids and fix nitrogen to ammonia. Investigation of the rhizobium-legume symbiosis has provided unique opportunities to learn about plant signal transduction, plant development, and plant-microbe interactions (19, 23).
The genomic DNA sequences of the S. meliloti chromosome and two megaplasmids were annotated by a combination of computer programs and inspection by S. meliloti researchers and predicted to have approximately 6,200 ORFs (1, 4, 8, 10). At the time of annotation, only ca. 60% of the annotated ORFs had some predicted function (10). Analysis of these ORFs, combined with already existing genetic information, indicated that S. meliloti possesses a diverse set of genes that might be used in both its symbiotic and free-living niches. About 40% of the predicted ORFs belong to a gene family, indicating that there might be significant functional redundancy. Hundreds of ORFs were predicted to encode transport proteins, suggesting that S. meliloti is able to exchange a diverse set of compounds with its environment. More than 500 ORFs were predicted to encode regulatory proteins, providing evidence that S. meliloti possesses complex regulatory networks that allow it to adapt to different microenvironments.
Prior to the genomics era, most rhizobial genetics aimed to identify and characterize individual genes and to determine their roles in rhizobial physiology and in the symbiotic plant-microbe interaction. Understanding the role of individual S. meliloti genes is an enduring goal; however, now that the entire S. meliloti genome sequence is available, sequence information can be used to accelerate discovery and extend it to a global scale. One such approach is to use arrays of DNA sequences to measure transcription simultaneously across the genome (2, 21). Another is to analyze the complement of proteins produced under various conditions (5).
These two approaches for profiling gene expression are complemented by methods that allow functional manipulation of individual genes. We speculated that having a library of cloned ORFs would facilitate large-scale analysis of individual genes. However, manipulating thousands of genes simultaneously requires a significant commitment of resources. To create a functional genetics platform for S. meliloti, we sought a strategy that would enable a variety of genetic manipulations but minimize this commitment (14). We chose three objectives as essential for this kind of genomic strategy: (i) measuring levels of gene expression, either through hybridization assays or through reporter gene technology; (ii) generating mutants lacking gene function; and (iii) overproducing predicted proteins in order to alter the cell's physiology, to purify the proteins for further in vitro characterization, or to investigate interactions between proteins in the proteome.
We designed a single PCR primer set to amplify each ORF in the S. meliloti genome, generating DNA fragments that can be cloned into a plasmid by using an integrative recombination protocol (3, 12). This manipulation results in a comprehensive set of plasmid clones from which the ORFs could be recombined into other plasmids specifically useful for various kinds of functional analysis. We describe here the completion of the first phase of this project with the cloning of 100% of the predicted S. meliloti ORFs, 13 of which are represented by a significant portion of the 5′ end of the ORF. These methods should be applicable to other genomes where considerable DNA sequence information is available.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli was cultured at 37°C in Luria-Bertani (LB) broth or on LB agar (22) with the appropriate antibiotics or incubated at 30°C when preparing electroporation competent cells. Routine procedures were used to prepare electrocompetent cells (22) with an efficiency of 107 CFU/μg of DNA or higher. Cells were incubated for 1.5 h without agitation at 37°C in SOC broth (22) when recovered after electroporation. Kanamycin and chloramphenicol (Sigma Chemical Co., St. Louis, MO) were added as required to media at concentrations of 75 and 50 μg/ml, respectively.
Genomic and plasmid DNA isolation.
S. meliloti genomic DNA was purified by using a DNeasy tissue kit according to the manufacturer's recommendations (QIAGEN, Inc., Valencia, CA). Briefly, 1.5 ml of bacterial cells from a 48-h culture of S. meliloti 1021 was harvested by centrifugation and then resuspended in lysis buffer. The genomic DNA was then bound to a silica gel membrane, washed with provided buffers to remove salts, and eluted from the membrane by using 100 μl of sterile distilled water. Plasmid DNA was purified by using QIAGEN′s QIAprep 96 Turbo Miniprep kit or QIAprep Spin Miniprep kit. Bacterial cells were harvested by centrifugation from a 24-h culture of E. coli cells and lysed by using a modified alkaline lysis procedure, and the DNA was bound to a silica gel membrane. DNA was washed and then eluted with 80 or 50 μl of sterile water, respectively.
Primer design and construction.
S. meliloti 1021 DNA sequence information was obtained from the database maintained at CNRS (Toulouse, France) by Jerome Gouzy, Daniel Kahn, and Jacques Batut (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/). Primary forward primers were constructed by adding the sequence 5′-GGAGGCTCTTCA-3′ to the 5′ end of the first 20 nucleotides of each ORF in the S. meliloti genome. If the start codon predicted in the sequence was not AUG, the DNA sequence was changed to ATG in order to specify an AUG start codon. The primary reverse primers were constructed by taking the reverse complement of the last 20 nucleotides of the respective ORF, removing the 3 nucleotides encompassing the stop codon and adding the sequence 5′-AGCTGGGTTCTA-3′ to the 5′ end of the sequence. This manipulation changes the stop codon to UAG, which is potentially suppressible. Secondary primers were constructed by modifying the secondary primer sequences suggested in the nested primer scheme used in GATEWAY Cloning (Invitrogen, Carlsbad, CA), adding a putative S. meliloti ribosome-binding sequence (GGAGGC) upstream of the start codon. A Bsp1407I restriction site (isoschizomer SspBI) was included in both the forward and the reverse secondary primers to obtain a secondary forward primer with the sequence 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGGAGGCTCTTCAATG -3′ and a secondary reverse primer with the sequence 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTTCTA-3′. The forward sequence contains a SapI restriction endonuclease recognition site. SapI cuts +1/+4, and the site was positioned next to the ATG so that SapI digestion would leave a consistent three-base single-stranded extension in order to allow control sequences at the 5′ end of the ORF to be replaced by using restriction enzyme methods. When needed, longer primary primers were constructed that contained the first 26 nucleotides and last 26 nucleotides of each ORF. Gene-specific primers (Invitrogen) were synthesized at a 10-nmol scale, and secondary primers were synthesized in larger quantity. All primers were used without further purification.
Nested PCR.
A nested PCR protocol was used to amplify DNA corresponding to the putative ORFs in the S. meliloti strain 1021 genome. In the primary PCR, DNA regions were amplified by using a touchdown protocol from 50 ng of S. meliloti genomic DNA with a reaction mixture that contained 0.5 U of KOD Hot Start polymerase (Novagen, Inc., Madison, WI), 0.3 mM deoxynucleoside triphosphates (dNTPs), 1.0 mM MgSO4, 5.8% glycerol, 5% dimethyl sulfoxide (DMSO), 1× polymerase buffer, and 0.05 μM concentrations of each forward and reverse primary primer in a final volume of 25 μl contained in strip cap tubes (MJ Research, Inc., San Francisco, CA). The parameters for the primary PCR were 94°C for 2 min for one cycle to activate the polymerase, followed by one cycle of 94°C for 1 min, 67°C for 1 min, and 68°C for 6 min. The annealing temperature, which starts at 67°C, was successively decreased by 1°C for each of the next seven cycles. When the annealing temperature reached 60°C, PCR was continued for 12 cycles of 94°C for 1 min, 60°C for 1 min, and 68°C for 6 min. After the 20th cycle, the temperature was lowered to 23°C. Secondary primers were added in 5 μl to a final concentration of 0.83 μM, and the secondary PCR was run for 25 cycles in an MJ Research PTC-200 DNA Engine (MJ Research, Inc., San Francisco, CA) using the parameters of 94°C for 1 min, 55°C for 1 min, and 68°C for 6 min. The expected sizes of all PCR products were confirmed by using agarose gel electrophoresis. PCR products were stored at −80°C.
Modified GATEWAY cloning technology protocol.
All PCR products were precipitated by the addition of 87.5 μl of TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]) and 50 μl of polyethylene glycol solution (30% PEG 8000, 30 mM MgCl2) to 27 μl of the PCR. The tubes were inverted several times to mix the phases thoroughly and were then placed at −20°C. The tubes were inverted every 10 min for approximately 1 h in order to prevent freezing. The samples were then centrifuged for 20 min at 11,500 rpm (8,800 × g) in a modified Eppendorf centrifuge rotor that held the tubes horizontally. The supernatant was carefully removed, and the pellet was resuspended in 20 μl of sterile distilled H2O. The BP clonase reaction was completed by using a protocol modified from the procedures recommended by GATEWAY cloning technology (Invitrogen). A 2-μl aliquot of each PCR product was added to 150 ng of entry vector pMK2010 and 1 μl of BP clonase enzyme mix in a final volume of 10 μl that contained 1× BP reaction buffer. The reaction was incubated at room temperature for 2 h. Then, 1 μl of proteinase K (20 mg/ml in H2O) was mixed well into each sample, and the reaction was incubated at 37°C for 30 min. A 3-μl aliquot was removed from the modified BP clonase reaction, mixed with 50 μl of electrocompetent cells, and transferred to a precooled (on ice) electroporation cuvette with a 0.1-cm gap. This was placed into a BTX TransPorator Plus (Harvard Apparatus, Inc., Holliston, MA) and shocked at a voltage of 1.5 kV. SOC was added to the cuvette at a volume of 950 μl, and the cell suspension was transferred to a 1.5-ml Eppendorf tube and incubated at 37°C without agitation for 1.5 h. After incubation, the cell suspension was mixed and a 100-μl aliquot of the cell suspension was plated onto LBkan75 agar plates. The plates were then incubated at 37°C for 18 to 24 h in order to obtain individual colonies. Electroporation cuvettes were cleaned with a cuvette washer, soaked in 70% ethanol for 5 min, allowed to dry, UV irradiated for 2 min, and stored at −20°C. Electroporation cuvettes were recycled 10 times. The remainder of the BP clonase reaction was stored at −80°C and can be used to recover additional clones if needed.
Confirmation protocol.
Single colonies from each transformation were chosen, and broth cultures were grown in 1.5 ml of LBkan75 with shaking overnight at 37°C. These cultures were arrayed in a 96-well deep-well microplate provided with the QIAprep 96 Turbo miniprep kit (QIAGEN, Inc., Valencia, CA). Plasmid DNA was isolated by using the QIAprep 96 Turbo miniprep kit as described above. To determine whether the size of the insert corresponded to the size expected for each putative ORF, this plasmid DNA was used as a template in a PCR with the attL1 (5′-TCGCGTTAACGCTAGCATGGATCTC-3′) and attL2 (5′-GTAACATCAGAGATTTTGAGACAC-3′) primers. For this confirmatory PCR, DNA regions were amplified from ∼50 ng of plasmid DNA with 0.25 U of KOD Hot Start DNA polymerase (Novagen) with 0.3 mM dNTPs, 1.0 mM MgSO4, 5.8% glycerol, 5% DMSO, 1× polymerase buffer, and 2 μM concentrations of both attL1 and attL2 primers in a final volume of 12.5 μl. The parameters for the confirmation PCR were 94°C for 2 min for one cycle to activate the polymerase, followed by 94°C for 1 min, 67°C for 1 min, and 68°C for 6 min for one cycle. The annealing temperature was successively decreased by 1°C for each of the next seven cycles. When the annealing temperature reached 60°C, treatment continued at 94°C for 1 min, 60°C for 1 min, and 68°C for 6 min for 32 cycles. When ORFs of greater than 2 kb were to be screened, the time at 68°C was increased to 8 min.
Colony screening.
Additional colonies were screened, as necessary, by using a cell lysis PCR from several individual colonies from each transformation. In this PCR, a toothpick was used to lightly touch each colony and transfer a small amount of cells into a 0.2-ml reaction tube. The DNA regions were amplified by using 0.25 U of Taq polymerase (Promega, Inc., Madison, WI) with 0.3 mM dNTPs, 1.0 mM MgCl2, 5.8% glycerol, 5% DMSO, × Taq DNA polymerase buffer (10 mM Tris-HCl [pH 9.0 at 25°C], 50 mM KCl, and 0.1% Triton X-100), and 2 μM concentrations of attL1 and attL2 primers in a final volume of 12.5 μl. The parameters for this PCR were 95°C for 3 min for one cycle to aid in cell lysis and DNA denaturation, followed by PCR using the confirmation protocol described above. Our confirmation strategy differs from that used by Dricot et al. (7) for the Brucella melitensis ORFeome, in which they did PCR amplifications on pools of an average of 50 transformants. Their strategy identifies situations in which the desired clone is present within the pool, but there may also be a number of incorrect plasmids present. We considered it important to have a more definitive identification of the plasmid in a purified colony before moving forward, although for some purposes a less rigorous confirmation may be adequate.
DNA sequencing.
DNA sequencing reactions used fluorescence-based dideoxy terminators and Ampli-Taq polymerase (Perkin-Elmer Life and Analytical Sciences, Inc., Boston, MA). Sequences were determined by using an Applied Biosystems model 373A DNA sequencer (Perkin-Elmer Applied Biosystems, Inc., Norwalk, CT) in the Washington State University Laboratory for Biotechnology and Bioanalysis. All PCR and sequencing reactions were performed by using a MJ Research PTC-200 DNA engine.
RESULTS
Development of a functional genomics platform.
To accomplish the goals listed in the introduction, our strategy uses one set of primers to PCR amplify and clone each ORF and makes it relatively easy to then move each ORF into a new plasmid context for different types of analysis. This strategy should be able to be adapted to analyze any organism of interest. The goal of expressing proteins dictated that we clone predicted ORFs and was the starting point for primer design. Our cloning strategy used bacteriophage lambda-mediated integrase recombination (Int) to construct a set of plasmids that contain single ORFs and then uses the Int and excisionase (Xis) reactions to recombine the ORFs into “destination” plasmids for specialized purposes (12). Cloning with Int has a significant virtue because the recognition sequence for Int is rare enough that the ORF is unlikely to contain a target sequence. By including origin of transfer (oriT) sequences on the plasmids used for the initial cloning as well as the destination plasmids, the required recombination reactions can be done by using conjugation to bring the plasmids together in vivo, with a considerable savings in cost (13).
PCR-generated DNA fragments corresponding to the ORFs were cloned into pMK2010, an “entry” plasmid derived by inserting the oriT from plasmid RP4 into the GATEWAY plasmid pDONR201 (13). pMK2010 contains two versions of the lambda attP sites flanking the F plasmid ccdB gene, whose protein product is toxic to many E. coli strains. Efficient replacement of ccdB by Int-mediated recombination provides the selection for cloning each ORF. An ORF cloned in pMK2010 can then be transferred to destination plasmids via Int/Xis-mediated recombination in vitro or in vivo by using a pentaparental mating scheme (13).
Determining the ORFs to clone.
In addition to the 6,204 ORFs identified by the original annotation, 113 ORFs were added to the set as the result of reevaluating the S. meliloti genome sequence using sequence information that had accumulated since the original annotation. The quality of the initial annotation is likely to be very good, since only 1% of the proteins identified in a proteomic study (6) were not among those originally predicted from the sequence. The genome sequence of the closely related species, Agrobacterium tumefaciens, was especially useful in reannotation since it reinforced the earlier assignment of several “hypothetical proteins” by showing they were “conserved hypothetical proteins” (11, 25). We also included seven additional ORFs that correspond to proteins actually expressed in the S. meliloti proteome (6). We had already included one of the new ORFs as a result of our reannotation. We also included a few genes (e.g., nolR and expR) for which it is known that the S. meliloti 1021 genome contains mutant versions of genes active in other, closely related S. meliloti strains.
Primer design.
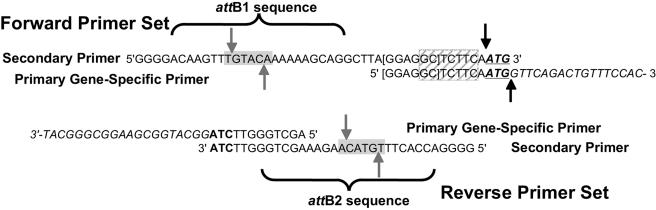
Since considerable effort would be needed to clone >6,300 PCR products and since subsequent manipulations would be limited by the exact choice of what was cloned, we designed primers including several features to facilitate later manipulation (Fig. 1). With minor modifications, these features can also be incorporated into schemes for analyzing other genomes. GGAGGC, a sequence predicted to be a strong ribosome-binding site (RBS), was inserted upstream of the predicted start codon for the target ORF. Optimal sequence and spacing of this site were determined by examining proteins likely to be strongly expressed in S. meliloti (15) and the sequence of the S. meliloti 16S rRNA. Starting translation at this RBS would allow the predicted protein to be expressed from the cloned DNA without any N-terminal or C-terminal extension. The RBS shown in Fig. 1 contains no stop codons in the reading frame of the protein, which allows the protein to be tagged at the N terminus when translation begins at a start codon upstream of the attB1 sequence (M. W. Mortimer, J. J. Bovitz, and M. L. Kahn, unpublished data). The DNA sequence corresponding to all start codons was standardized to be ATG, which required changing about 1,000 predicted start codons and, between the RBS of the primer and the start codon, we included the sequence TCTTC. Together with the GC at the end of the RBS, these sequences generate a SapI recognition sequence (GCTCTTC) between the start codon and the ribosome binding site such that, by design, there is a consistent 3′-TAC-5′ overhang after cleavage +1/+4 relative to the SapI recognition site. SapI digestion would permit complete replacement of the 5′ transcription and translation signals by restriction enzyme/ligase methods, should this be desired. TAG was used as the stop codon to enable the possibility of C-terminal tagging of the protein by readthrough of this codon in amber suppressor strains. To reduce the cost of the PCR primers, a nested primer strategy was used in which the gene-specific primers had a 20-base overlap with the gene and a 12-base overlap with the secondary primer pair that contained the attB sequences (Fig. 1). Finally, Bsp1407I restriction sites were included in both secondary forward and reverse primers to enable excision of the insert.
FIG. 1.
Diagram of the nested PCRs. The gene-specific forward primer (shown here for gene SMa1985) included the first 20 bases of the ORF (in italics) with an ATG start codon (in boldface, underlined). The ribosome-binding sequence (brackets) added to the ORF and sequences between the ribosome-binding sequence and the start codon generate a SapI restriction site (hatched gray box) that generates a consistent overhang around the start codon (black solid arrows). The gene-specific reverse primer included the last 20 bases of the ORF (in italics) and an ATC sequence (in boldface) that will generate TAG at the end of the ORF to insert an amber suppressible stop codon. The secondary forward and reverse primers included the attB1 and attB2 sequences, respectively, as indicated by the brackets below the sequence and a Bsp1407I restriction site (gray-shaded box, gray solid arrows).
PCR amplification of the ORFeome.
A major concern at the beginning of the project was that we would be unable to obtain consistent PCR success with a high-fidelity DNA polymerase since, because the primer sequences were to be determined by the DNA sequences at the ends of the ORFs, there was no opportunity to choose primer sequences that would optimize the PCR. We therefore screened several DNA polymerases and protocols for their ability to work with nonoptimized primer sets and with the relatively high (62%) G+C template (data not shown). A touchdown PCR protocol using a hot-start version of the high-fidelity KOD DNA polymerase (Novagen) was superior to all other protocols we tried with any of several DNA polymerases (data not shown). However, when KOD DNA polymerase was used with the secondary primer sequences recommended by Invitrogen, a more prominent band of short PCR products was observed, and it appeared that more of our clones contained short inserts than was seen with Taq DNA polymerase. Minor changes to the secondary primer sequences as indicated in Fig. 1 reduced the proportion of short PCR products and appeared to reduce the proportion of short inserts in the clones. We did not systematically optimize this change.
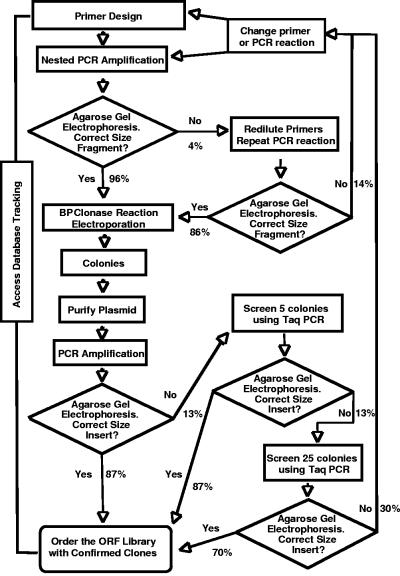
Using the nested PCR described in Materials and Methods, the first attempt at amplification produced DNA fragments of the size expected for the putative ORFs in 96% of the PCRs (Fig. 2). Primer stocks corresponding to the ∼4% of ORFs where the PCR had failed were rediluted and the standard nested PCR was repeated. A total of 86% of this second group of PCRs were successful. Approximately 50 ORFs needed additional attention in order to get the PCR fragments to amplify. In some cases success was achieved after six additional bases were added to the primary PCR primers or, especially for long ORFs, the elongation time used in the PCR protocol was increased to 8 min. It is possible that differences between the template and published sequence were causing some of our difficulties, since the ∼260,000 bases of sequence in the primers is large compared to the target sequence quality of less than 1/10,000 errors per base (1, 4, 8, 10). However, despite the fact that the primers were not optimized the final tally indicates that only one of the 6,317 ORFs, SMb21548, did not amplify as a complete ORF. This ORF is unusually long (6,522 bp), is unusually G+C rich (68%), and has many internally repeated sequences.
FIG. 2.
Flow chart of the ORFeome construction. The various steps and the transition probabilities are indicated.
Cloning the ORFeome.
After carrying out a modified BP clonase reaction with the PCR product and electroporating the circularized plasmids carrying a PCR fragment insert into E. coli strain DH5α, kanamycin-resistant transformants were selected (Fig. 2). A single colony was chosen from each electroporation, and plasmid DNA was isolated from a small broth culture of this colony. PCR with KOD DNA polymerase followed by agarose gel electrophoresis was used to determine the size of the cloned insert. Around 88% of the samples contained a PCR fragment of the expected size. If a fragment of the expected size was not observed, more colonies were screened by using colony PCR with Taq DNA polymerase as described in Materials and Methods. When five additional colonies were examined from electroporations that had been unsuccessful in the “one colony screen,” at least one colony carrying the expected size fragment was identified in 86% of the constructions. We selected 25 more colonies from the unsuccessful “five colony screens,” and at least one of these contained a correct insert ca. 70% of the time. If a fragment of the expected size was still not found in the 25 colony screens, we repeated recombination and electroporation steps and rescreened them. If this failed again, new PCR products were generated before reattempting to clone. At least part of each of the 6,317 putative Sinorhizobium meliloti ORFs has been cloned. Complete sequences of 13 predicted ORFs have not yet been cloned, and these (SMb21298, SMb21548, SMb20514, SMc00852, SMc01316, SMc01710, SMc02086, SMc02273, SMc03761, SMc04028, SMc04382, TGc2812, and TGc1901) are represented in the ORFeome by clones containing substantial portions of the 5′ end of the ORF. Although these are not suitable for protein production, they can be used for some operations that require recombination of ORF constructs into the chromosome.
Confirmation of the clones in the ORFeome.
The size of DNA corresponding to each of the ORFs was examined by PCR and agarose gel electrophoresis at several steps during the cloning process. DNA sequencing of 409 distinct random clones for which the correct size had been confirmed by PCR was used to confirm that the correct DNA sequences were present and that the junctions were as predicted in the plasmid design. This success rate indicated that we would obtain little, if any, information about errors in the clones by further sequencing so they have not been confirmed base by base. From the predicted error frequency of the KOD DNA polymerase (24), we anticipate that fewer than 0.5% of ORFs (most likely ca. 0.1%) will have an error due to PCR. This number was comparable to the target error rate for the original sequence.
Database management of the project workflow.
It was evident at the start of this project that the ability to move each ORF forward at its own pace would be essential. This required us to organize the workflow to deal with considerable asynchrony in the rates of progress. Tracking each ORF—from deciding on and ordering primer sequences to confirming the cloned ORF—required close attention. Although we judged that this would be extremely difficult to do by hand, appropriate software for managing this project did not seem to be available. A Microsoft Access database with tables linked to Microsoft Excel spreadsheets was developed (S. N. Yurgel, B. K. Schroeder, and M. L. Kahn, unpublished data). This approach used the familiar Excel format for data entry and some data display and custom Visual Basic and Perl programs for data manipulation, database updating, and error checking. Each manipulation in the cloning process was tracked, and the database was updated when each step was completed. So, for example, a query would select ORF sequences from the database, a routine would then alter the primer start and stop codons as described above, and these data could be used to generate an order form that could be sent to Invitrogen. When the ordered primers were received, another routine compared the file received from Invitrogen with the order and marked the primers according to whether they had been delivered or back-ordered. The database could then be asked to prepare worksheets for PCRs consisting only of ORFs for which complete primer sets were available. In a similar way, worksheets could be generated for each successive step in the cloning process, and summary information related to overall progress of the project could be prepared.
During peak efficiency for the project, PCR amplification of ∼470 ORFs were started in a week. Two to three weeks were required to move from PCR to confirmed plasmid. When problems arose, the step that had failed could be repeated by rescheduling the ORF into the workflow, either as a normal procedure or with the annotation that it had been problematic and might need special care.
Database and clone access.
Information about the entry plasmid clones is available through a Web site at www.bioinformatics.wsu.edu/kahn. This now operates as a working Web site that contains descriptions of the plasmids constructed in the work reported here and information about other projects related to the mobilization of ORFs into destination vectors. The Web site also contains information about relevant protocols. Plasmids corresponding to a given gene are designated pEGENEID. For example, the entry plasmid clone carrying the dctA gene, SMb20611, is pESMb20611, and this can be accessed through the gene name or identification number. In addition to providing plasmids directly, we are now arranging for plasmids to be distributed through Addgene, Inc. (http://www.addgene.org/).
DISCUSSION
We have developed a generalizable strategy that has allowed us to construct an ORFeome of S. meliloti strain 1021 that contains 6,317 plasmids and represents 100% of the ORFs predicted to be encoded in the genome. Several recent projects (7, 16, 17, 20) have used Invitrogen's GATEWAY system for recombinational cloning as a starting point for dissecting the genomics of an organism. We chose this method for the high-throughput cloning of the S. meliloti ORFeome because of its efficiency and relative flexibility. However, aspects of our cloning strategy are unique. First, a nested PCR strategy was used in which a universal set of secondary primers was used to add attB sequences to PCR fragments generated by primers specific for each gene. Using this nested primer scheme significantly reduced the cost of the PCR primers, since each custom primer was only 32 bases long, including 20 bases of homology with the ORF and 12 bases of overlap with the secondary primer. Second, because the pMK2010 entry vector and the various destination vectors we are using (13, 14) contain a mobilization origin of transfer (oriT), recombination between the ORFeome plasmids and destination plasmids can be carried out in vivo via a pentaparental mating (13). Although the ORFeome plasmids can be recombined in vitro, the use of in vivo recombination can significantly reduce the effort and expense of transferring the ORFs into a new context. Third, the design of the gene specific primers should enable versatile manipulation of later constructs. For example, the placement of a strong RBS 5′ to the start codon, as well as the change of all of the start codons to AUG, should promote the expression of protein when the ORF is transferred to a destination vector that contains a strong promoter. Sequences upstream of the ORF contain a continuous ORF that allows additional peptide sequences to be attached to the amino terminus in some destination vehicles (Mortimer et al., unpublished). In addition, substituting a UAG stop codon for the normal stop codon for each ORF should allow additions to the C terminus of the protein in strains that carry an appropriate tRNA suppressor. A SapI restriction endonuclease recognition site was also positioned near the start codon to facilitate the insertion of alternate sequences immediately 5′ to the ORF, and Bsp1407I sites can be used to excise the entire insert.
We were able to recover a larger proportion of the ORFs than similar cloning efforts in the smaller genomes of Treponema pallidum (17) and Brucella melitensis (7), where the ORFs were cloned from genomic DNA. LaBaer et al. (16) cloned 100% of the ORFs from the 5.5-Mb genome of Pseudomonas aeruginosa PAO1 into pDONR201, the parent of pMK2010, using PCR with Taq polymerase and a GATEWAY in vitro reaction. Although the high proportion of ORFs that could be cloned indicated that few S. meliloti ORFs carried in pMK2010 are toxic to the E. coli host, our inability to recover 13 ORFs as full-length clones might indicate either toxicity or some problem with the integrase cloning procedure.
One key to the construction of the ORFeome was the greater than expected proportion of successful PCRs. Rüberg et al. (21) used optimized primers and Taq DNA polymerase to amplify fragments internal to the S. meliloti 1021 genomic ORFs that were 80 to 350 bp long. These authors were able to amplify only 6,046 of 6,207 ORFs and then constructed 161 70mer oligonucleotides to complete their array. The higher processivity of the high-fidelity KOD DNA polymerase probably contributed to our success with PCR, as did the touchdown protocol. PCR depends on numerous components, including the reaction buffer, Mg2+ concentration, nucleotide concentration, and primer length and base composition. Attempts were made to standardize the reaction conditions as much as possible, and additives such as DMSO and glycerol (26) were found to be useful in increasing the rate of success. Despite the fact that the gene-specific primer sets could not be systematically optimized to match Tm or eliminate the formation of primer dimers, only one of 6,317 ORFs was not amplified. When problems in amplification did occur, using longer primers was almost always successful. This suggested that the initial failure was due to problems with the primers.
With regard to the cloning itself, there tended to be a higher efficiency in cloning the shorter ORFs and, were we to begin a new project, we would initiate the longer ORFs early in the project in order to integrate repetitions of unsuccessful attempts more efficiently into the general workflow. No systematic bias was seen in cloning various classes of protein with one exception. At the point where >90% of the ORFs had been cloned, only 1 of 10 acrB-related ORFs had been recovered. Since AcrB is a membrane protein and forms complexes with AcrA proteins, we obtained a strain lacking AcrA-related proteins and successfully used this strain for transformation. However, these transformants were easily transformed into DH5α, and it may be that the apparent pattern in the earlier lack of success was coincidental.
Cloning the ORFeome is the first step in developing a functional genomics platform for S. meliloti. To be most effective for manipulations in S. meliloti itself, a set of useful destination vectors needs to be developed. A reporter-type destination vector has been constructed (M. W. Mortimer and M. L. Kahn, unpublished data) that allows the cloned ORF to be placed upstream of GUS and green fluorescent protein reporter genes. This destination vector uses an R6Kγ origin of replication, which is not active in the absence of a specific replicator protein and thus should be useful as a suicide plasmid in many different bacteria. This resulting plasmid can be introduced into S. meliloti via a triparental mating and, after homologous recombination with the copy of the ORF contained in the genome, expression of these reporter genes will be under control of the native promoter for the ORF, enabling expression of each ORF to be measured under various free-living and symbiotic conditions. It also contains yeast flipase recognition target sequences and can be used as one end of a deletion mutation constructed by site-specific recombination with deletion destination plasmids previously described (13). We are in the process of transferring the ORFeome into our reporter-type destination plasmid for introduction into S. meliloti, as well as designing new destination plasmids to further our genomic analysis of S. meliloti. These constructs should be an excellent addition to the genetic tools available for investigating bacterial gene expression patterns used when the bacteria are growing under both symbiotic and free-living conditions.
It is our hope that the availability of this set of ORF clones will enable better functional characterization of S. meliloti, both by researchers interested in the bacterium itself and those interested in the properties of the enzymes it contains. Although the procedures described here are similar to those used in previous large-scale cloning efforts, they have been refined to include differences in primer design, the use of a high-fidelity polymerase and a touchdown PCR protocol that may be useful in similar efforts. Validating procedures on the scale of this and similar efforts (7, 16, 17) is difficult since even rare problems can lead to a significant number of failures when procedures are repeated thousands of times. Thus, it may be appropriate to consider each successive effort to construct an ORFeome as a kind of large experiment that produces a useful set of plasmids and experience that may be useful in the next attempt to construct genetic materials on a genomic scale.
Acknowledgments
We acknowledge helpful discussions with Robert Fisher, Melanie Barnett, and Sharon Long concerning the cloning strategy. No robots were used in the cloning of the S. meliloti ORFeome, but a very capable group of Washington State University students provided significant and very valuable labor. These students included Sharon Ankrah, Cynthia Wilson, Chi Meng Fong, Cyndi White, Katherine Santiago, Jhoanna Berrocal, Jamie Pearsall, Rebecca Thompson, Elizabeth Daghighi, Hillery Krebs, David Kahn, Jody Thomas, Rebecca Flanagan, Malia Haddock, Austin Harvey, Michelle Mace, Tishra Mattox, Amanda Grimm, Harvey Doty, Joseph Powers, and Rebecca Eastman.
Early phases of this project were supported by a grant from the U.S. Department of Energy Biosciences Program, but most of the “heavy lifting” depended on a grant from the Microbial Genetics Program at the U.S. National Science Foundation.
REFERENCES
- 1.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K.-C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasch, M. A., J. L. Hartley, and M. Vidal. 2004. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 14:2001-2009. [DOI] [PubMed] [Google Scholar]
- 4.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 2001 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 6.Djordjevic, M. A., H. C. Chen, S. Natera, G. Van Noorden, C. Menzel, S. Taylor, C. Renard, O. Geiger, G. F. Weiller, et al. 2003. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant-Microbe Interact. 16:508-524. [DOI] [PubMed] [Google Scholar]
- 7.Dricot, A., J. F. Rual, P. Lamesch, N. Bertin, D. Dupuy, T. Hao, C. Lambert, R. Hallez, J. M. Delroisse, J. Vandenhaute, I. Lopez-Goni, I. Moriyon, J. M. Garcia-Lobo, F. J. Sangari, A. P. Macmillan, S. J. Cutler, A. M. Whatmore, S. Bozak, R. Sequerra, L. Doucette-Stamm, M. Vidal, D. E. Hill, J. J. Letesson, and X. De Bolle. 2004. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 14:2201-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 11.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 12.Hartley, J. L., G. F. Temple, and M. A. Brasch. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn, M. L., B. K. Schroeder, B. L. House, M. M. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Warren, R. F. Fisher, M. J. Barnett, C. Toman, and S. R. Long. 2004. Foraging for meaning: postgenome approaches to Sinorhizobium meliloti, p. 416-422. In B. Lugtenberg, I. Tikhonovich, and N. Provorov (ed.), Biology of molecular plant-microbe interactions, vol. 4. IS-MPMI Press, St. Paul, MN. [Google Scholar]
- 15.Karlin, S., M. J. Barnett, A. M. Campbell, R. F. Fisher, and J. Mrazek. 2003. Predicting gene expression levels from codon biases in alpha-proteobacterial genomes. Proc. Natl. Acad. Sci. USA 100:7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaBaer, J., Q. Qiu, A. Anumanthan, W. Mar, D. Zuo, T. V. Murthy, H. Taycher, A. Halleck, E. Hainsworth, S. Lory, and L. Brizuela. 2004. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 14:2190-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKevitt, M., K. Patel, D. Smajs, M. Marsh, M. McLoughlin, S. J. Norris, G. M. Weinstock, and T. Palzkill. 2003. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 13:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patriarca, E. J., R. Tate, S. Ferraioli, and M. Iaccarino. 2004. Organogenesis of legume root nodules. Int. Rev. Cytol. 234:201-262. [DOI] [PubMed] [Google Scholar]
- 19.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reboul, J., P. Vaglio, J. F. Rual, P. Lamesch, M. Martinez, C. M. Armstrong, S. Li, L. Jacotot, N. Bertin, R. Janky, T. Moore, J. R. Hudson, Jr., J. L. Hartley, M. A. Brasch, J. Vandenhaute, S. Boulton, G. A. Endress, S. Jenna, E. Chevet, V. Papasotiropoulos, P. P. Tolias, J. Ptacek, M. Snyder, R. Huang, M. R. Chance, H. Lee, L. Doucette-Stamm, D. E. Hill, and M. Vidal. 2003. Caenorhabditis elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34:35-41. [DOI] [PubMed] [Google Scholar]
- 21.Rüberg, S., Z. X. Tian, E. Krol, B. Linke, F. Meyer, Y. Wang, A. Puhler, S. Weidner, and A. Becker. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255-268. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 23.Schultze, M., and A. Kondorosi. 1998. Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32:33-57. [DOI] [PubMed] [Google Scholar]
- 24.Takagi, M., M. Nishioka, H. Kakihara, M. Kitabayashi, H. Inoue, B. Kawakami, M. Oka, and T. Imanaka. 1997. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 63:4504-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, L. Chen, G. E. Wood, Y. Chen, L. Woo, J. P. Kitajima, V. K. Okura, N. F. Almeida, Jr., Y. Zhou, D. Bovee Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, D. Guenthner, T. Kutyavin, R. Levy, M. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, D. Gordon, J. A. Eisen, I. Paulsen, P. Karp, P. Romero, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Zhao, M. Dolan, S. V. Tingey, J. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 26.Varadaraj, K., and D. M. Skinner. 1994. Denaturants or cosolvents improve the specificity of PCR amplification of a G+C-rich DNA using genetically engineered DNA polymerases. Gene 140:1-5. [DOI] [PubMed] [Google Scholar]