Abstract
A novel Eubacterium cellulosolvens 5 gene encoding an endoglucanase (Cel5A) was cloned and expressed in Escherichia coli, and its enzymatic properties were characterized. The cel5A gene consists of a 3,444-bp open reading frame and encodes a 1,148-amino-acid protein with a molecular mass of 127,047 Da. Cel5A is a modular enzyme consisting of an N-terminal signal peptide, two glycosyl hydrolase family 5 catalytic modules, two novel carbohydrate-binding modules (CBMs), two linker sequences, and a C-terminal sequence with an unknown function. The amino acid sequences of the two catalytic modules and the two CBMs are 94% and 73% identical to each other, respectively. Two regions that consisted of one CBM and one catalytic module were tandemly connected via a linker sequence. The CBMs did not exhibit significant sequence similarity with any other CBMs. Analyses of the hydrolytic activity of the recombinant Cel5A (rCel5A) comprising the CBMs and the catalytic modules showed that the enzyme is an endoglucanase with activities with carboxymethyl cellulose, lichenan, acid-swollen cellulose, and oat spelt xylan. To investigate the functions of the CBMs and the catalytic modules, truncated derivatives of rCel5A were constructed and characterized. There were no differences in the hydrolytic activities with various polysaccharides or in the hydrolytic products obtained from cellooligosaccharides between the two catalytic modules. Both CBMs had the same substrate affinity with intact rCel5A. Removal of the CBMs from rCel5A reduced the catalytic activities with various polysaccharides remarkably. These observations show that CBMs play an important role in the catalytic function of the enzyme.
The rumen microbial ecosystem is composed of anaerobic microorganisms, such as bacteria, fungi, and protozoa. Some of these rumen microorganisms, the cellulolytic bacteria, are able to digest cellulosic material of plants and produce energy for the host animals. Many cellulolytic enzymes have been isolated from rumen microorganisms, and the genes encoding these enzymes have been cloned and sequenced (5, 9, 17). However, the precise mechanisms of lignocellulose degradation in the rumen are not yet fully understood. In order to clarify these mechanisms, it is necessary to study the microbial cellulolytic enzymes biochemically and genetically.
The anaerobic cellulolytic bacterium Eubacterium cellulosolvens is sporadically dominant in the rumen (18). It is known that E. cellulosolvens 5 adheres tightly to cellulose, so studies of this adhesion have been performed (13, 14, 23-26). Some cellulose-binding proteins (CBPs) have been found in culture supernatant and cell lysate of the organism (14, 26). A gene encoding cellulose-binding protein A (CBPA), which is one of these CBPs, has been cloned and characterized (23-25). Additionally, the presence of some proteins exhibiting carboxymethyl cellulase (CMCase) activity in culture supernatant and cell lysate of E. cellulosolvens 5 was revealed by zymogram analysis (26). In order to advance research on the mechanism of cellulose degradation by this bacterium, we tried to isolate a gene encoding CMCase from the genomic DNA library of E. cellulosolvens 5.
In this report, we describe cloning and nucleotide sequencing of the E. cellulosolvens 5 endoglucanase Cel5A gene (cel5A), the primary structure of Cel5A, and the enzymatic properties of Cel5A and derivatives of this protein expressed in Escherichia coli. We also describe the importance of novel carbohydrate-binding modules (CBMs) in cellulose hydrolysis by Cel5A.
MATERIALS AND METHODS
Bacterial strains and vectors.
The E. cellulosolvens 5 used in this study was kindly supplied by N.O. van Glyswyk (National Chemical Research Laboratory, Pretoria, South Africa). Recombinant ZAP Express (Stratagene) phages were grown on E. coli XL1-Blue MRF′. E. coli XLOLR (Stratagene) was used for in vivo excision of the pBK-CMV phagemid vector from the ZAP Express vector using the ExAssist/XLOLR system (Stratagene). XL1-Blue MRF′ and TOP10F′ were used as the hosts for pBluescript II SK(+) (Stratagene) and pCRT7-TOPO (Invitrogen) cloning, respectively. E. coli BL21(DE3) and M15(pREP4) (QIAGEN) were used as the hosts for pCRT7-TOPO and pQE-30 (QIAGEN) expression, respectively. The media and culture conditions used have been described in a previous report (23), except for pQE-30 expression. E. coli M15(pREP4) harboring pQE-30 plasmids was grown at 37°C or 18°C in Luria-Bertani (LB) broth or on LB agar supplemented with ampicillin (50 μg/ml) and kanamycin (25 μg/ml).
Construction of genomic DNA library, screening of CMCase-producing clones, and sequencing.
Ligation, transformation, and restriction enzyme analysis were performed by standard procedures (20). A genomic DNA library of E. cellulosolvens 5 was constructed in the ZAP Express vector as described previously (23). The genomic library was screened for CMCase-producing clones by using an overlay of 0.7% (wt/vol) top agar containing 0.2% (wt/vol) carboxymethyl cellulose (CMC). Plaques having CMCase activity were recognized by the formation of clear haloes on a red background after staining with 0.1% (wt/vol) Congo red and destaining with 1 M NaCl (27). Positive plaques were reisolated three times to ensure purity. The sequences of both strands were determined as described previously (23).
Cloning of a DNA fragment encoding the N-terminal end of Cel5A by targeted gene walking PCR.
Targeted gene walking PCR was performed as described previously (23). The first PCR was performed with a targeted sequence-specific primer, K-3 (Table 1), containing a known sequence in the cel5A gene, and a walking BK-reverse primer corresponding to the sequence present in the multiple-cloning site of the λZAP Express vector. After the first PCR, the PCR product was subjected to a second PCR with an internal detection primer, K-4 (Table 1), which contained a known sequence upstream from the K-3 sequence, and a walking T3 primer corresponding to the T3 promoter sequence that is located downstream from the BK-reverse sequence present in the multiple-cloning site of the λZAP Express vector. The first PCR amplification was performed for 32 cycles consisting of 94°C for 30 s, 52°C for 45 s, and 72°C for 2 min, using ExTaq (Takara). The second PCR was performed under the same conditions as the first PCR except that the annealing temperature was 56°C.
TABLE 1.
PCR primers used for screening of the N terminus of Cel5A and construction of rCel5A and its derivatives
| Primer | Oligonucleotide sequencea |
|---|---|
| K-3 | 5′-CCGTATGCTTTTGCGATCTG-3′ |
| K-4 | 5′-CTGCGGTTTAGAGCCATG-3′ |
| K-26 | 5′-ACTGAGGCTGCATCCGGGGA-3′ |
| K-36 | 5′-CAGGTCGATTACGGGATCAGGTTGCCGGA-3′ |
| K-39 | 5′-CCGTCGACCGGCAATACCCTTGAGA-3′ |
| K-40 | 5′-GCCGGATCCTGGGCAAACGGCGTGAAC-3′ |
| K-41 | 5′-TGTGTCGACTGCTCCGCCGTTTGCAGCA-3′ |
| K-42 | 5′-CTGTCGACTCGCCGGCAATTCCCTTGAGG-3′ |
| K-43 | 5′-CAGTCGACGGGATCAGGTTGCCGGAGCTTA-3′ |
| K-46 | 5′-CAGGATCCCAGGGCACTGAGGCTGCATC-3′ |
| K-47 | 5′-CAGGATCCTCAGGAGCTGATTCCGGCG-3′ |
The underlined sequences are the restriction sites of BamHI and SalI.
Plasmid construction.
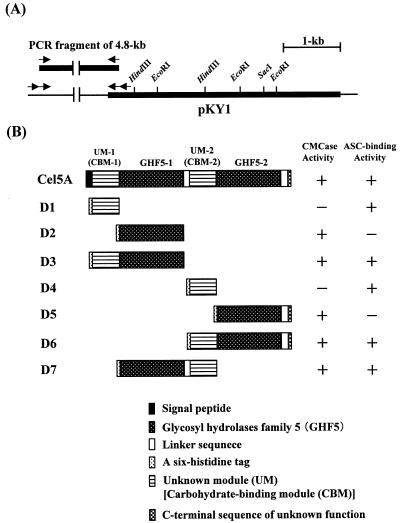
A DNA fragment encoding mature Cel5A was amplified by PCR from E. cellulosolvens 5 genomic DNA with LA Taq (Takara) and primers K-26 and K-36 (Table 1). The PCR product was cloned into the pCRT7-TOPO vector, which included the gene coding for six histidine residues (His6), as recommended by the supplier. The plasmid constructed was designated pKY12. The plasmids used to produce the truncated derivatives of Cel5A were constructed as follows. DNA fragments encoding derivatives were amplified by PCR from E. cellulosolvens 5 genomic DNA with ExTaq (Takara) and an appropriate combination of primers (Table 1) containing artificial BamHI or SalI restriction sites for cloning the PCR fragments into plasmid vectors. The resulting PCR fragments were cloned into pQE-30. The absence of mutations in the inserted fragments was confirmed by sequencing. The combinations of primers used were as follows: K-46 and K-39 to construct a plasmid yielding a polypeptide of CBM-1 (D1); K-40 and K-41 to construct a plasmid yielding a polypeptide of family 5 catalytic module GHF5-1 (D2); K-46 and K-41 to a construct plasmid yielding a polypeptide composed of CBM-1 and GHF5-1 (D3); K-47 and K-42 to construct a plasmid yielding a polypeptide of CBM-1 (D4); K-40 and K-43 to construct a plasmid yielding a polypeptide of GHF5-2 (D5); K-47 and K-43 to construct a plasmid yielding a polypeptide composed of CBM-2 and GHF5-2 (D6); and K-40 and K-42 to construct a plasmid yielding a polypeptide composed of CBM-2 and GHF5-1 (D7). Schematic diagrams of these proteins are shown in Fig. 1B.
FIG. 1.
Restriction map of pKY1 (A) and molecular architecture of Cel5A and its derivatives used in this study (B). The thin lines indicate vector plasmid DNA. The arrows indicate the primers used for targeted gene walking PCR. The different modules of Cel5A are indicated.
Expression and purification of recombinant Cel5A (rCel5A) and its derivatives.
To produce the recombinant proteins, E. coli recombinant cells harboring pYK12 or the plasmids constructed for expression of the derivatives of Cel5A were grown to the mid-log phase (absorbance at 600 nm, 0.6) and induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.1 or 0.5 mM. After additional incubation at 37°C for 4 h or at 18°C overnight, the cells were harvested and disrupted by sonication. Cell debris was removed by centrifugation. The cell extracts obtained in this way were used for purification of the recombinant proteins. The recombinant proteins were partially purified by using ProBond resin (Invitrogen) (23).
Enzyme assays.
The CMCase activities of the derivatives of Cel5A were detected by spotting the proteins on an agar plate containing 0.2% (wt/vol) CMC in 50 mM sodium citrate buffer (pH 5.5). The plate was incubated for 15 min at 37°C. The CMCase activities of the proteins were detected by Congo red staining as described above. The hydrolytic activities of the recombinant proteins were measured after 5 or 60 min of incubation at 37°C in 50 mM sodium citrate buffer (pH 5.0) in the presence of 1% (wt/vol) polysaccharides. The polysaccharides tested were CMC (Wako), acid-swollen cellulose (ASC) (28), Avicel (PH-101; Asahi Chemical Industry), oat spelt xylan (Sigma), lichenan (Sigma), laminarin (Sigma), chitin (Wako), and mannan (Sigma). The reducing sugars released from the substrates were determined with the 3,5-dinitrosalicylic acid reagent as described by Miller (12). One unit of activity was defined as the amount of enzyme that released 1 μmol of glucose equivalents per min from the substrate. The viscometric assay with recombinant Cel5A was performed using CMC as described previously (25).
SDS-PAGE and zymogram analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed using standard procedures, as described previously (26). Zymogram analysis was performed by the method of Toyoda et al. (26), with some modifications.
Analysis of hydrolysis products.
Cellooligosaccharides (cellobiose to cellohexaose; 1 mg each) were incubated with 0.1 U of the purified enzyme in 0.2 ml of 50 mM sodium citrate buffer (pH 5.5) at 37°C for 6 h. Thin-layer chromatography (TLC) of the hydrolysis products was performed on a silica gel plate (Sigma) developed with a solvent consisting of 1-propanol, acetic acid, and water (2:1:1, vol/vol/vol). Cellooligosaccharides were visualized by spraying the plate with an aniline-diphenylamine-phosphoric acid reagent and heating it at 120°C (30).
Polysaccharide binding assay.
Binding of Cel5A and its derivatives to the insoluble polysaccharides was determined as follows. The proteins were mixed with insoluble polysaccharides (5 mg) in 0.2 ml of 50 mM sodium phosphate buffer (pH 6.5) and were incubated on ice for 1 h with occasional stirring. After centrifugation, the pellets were washed four times with 50 mM sodium phosphate buffer (pH 6.5). Then the polysaccharides with bound proteins were eluted with 100 μl of 5% SDS for 30 min at 37°C. The eluted proteins were collected by centrifugation and subjected to SDS-PAGE. The polysaccharides tested were ASC, Avicel, oat spelt xylan, lichenan, chitin, agarose (Agarose S; Nippon Gene), Sephadex G-25 (Amersham Biosciences), and corn starch (Wako).
The affinities of the proteins for soluble polysaccharides, including methylcellulose (Wako), birchwood xylan (Sigma), laminarin, and soluble starch (Wako), were examined by native affinity gel electrophoresis as described by Arai et al. (4). The separating gel contained 7.5% acrylamide. The polysaccharides were incorporated into the gel at a concentration of 0.1% prior to polymerization. A control gel without polysaccharides was prepared and run simultaneously.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB179780.
RESULTS
Cloning of the gene encoding Cel5A.
An E. cellulosolvens 5 genomic DNA library was screened for clones (plaques) hydrolyzing CMC. Eight positive plaques were selected from approximately 45,000 plaques. The DNA fragments of these eight clones were subcloned into the pBK-CMV phagemid by in vivo excision. The restriction fragment patterns of all positive clones were identical, and one of the plasmids constructed was designated pKY1 (Fig. 1A). pKY1 expressed an approximately 120-kDa protein having CMCase activity (data not shown). Since sequencing analysis revealed that pKY1 did not carry the entire genetic region encoding Cel5A, gene walking PCR was employed to determine the complete sequence of the cel5A gene. The size of the fragment amplified by a nested PCR of a genomic DNA library of E. cellulosolvens 5 was 4.8 kb. Sequence analysis of the PCR fragment revealed that the DNA fragment contained the nucleotide sequence encoding the N-terminal end of Cel5A (Fig. 1A).
Nucleotide sequence of the cel5A gene.
The nucleotide sequence of the gene encoding Cel5A was determined from both the sequence of the inserted DNA in pKY1 and the sequence of the 4.8-kb PCR product obtained by the gene walking PCR. The open reading frame of the cel5A gene consisted of 3,444 nucleotides encoding a 1,148-amino-acid protein with a deduced molecular mass of 127,047 Da. The putative ATG translational start codon was preceded at a spacing of 6 bp by a potential ribosome-binding site (GGGGA). Two possible promoter sequences, TTGCGT and TTGCAG for the −35 region and TATAAT and TAAAAA for the −10 region, with 17-bp spacing, were located upstream of the open reading frame. A presumptive transcription terminator that consisted of a 12-bp palindrome was found downstream of the TAA termination codon.
Structural features of Cel5A.
The deduced primary structure of Cel5A is shown in Fig. 1B. The 36-amino-acid sequence of the N terminus of the deduced polypeptide had features similar to features of the signal sequences of prokaryotes. A comparison of the amino acid sequence of Cel5A with the amino acid sequences of other proteins using the BLAST program revealed that the mature Cel5A consisted of four modules, two linker sequences, and a C-terminal sequence with an unknown function. In particular, Cel5A comprised mainly two catalytic modules belonging to glycosyl hydrolase family 5 and two modules with unknown functions. The two modules with unknown functions had carbohydrate-binding ability, as described below. The C-terminal sequence with an unknown function was composed of 17 amino acid residues. The two family 5 catalytic modules of Cel5A were designated GHF5-1 and GHF5-2. The amino acid sequences of GHF5-1 and GHF5-2 were 94% identical. An amino acid sequence alignment of the catalytic modules of Cel5A and family 5 glycosyl hydrolases is shown in Fig. 2. The amino acid sequence of the GHF5-1 module of Cel5A exhibited high degrees of similarity to the amino acid sequences of endoglucanase VII from Ruminococcus albus (43% sequence identity for 355 amino acid residues) (16), CelE from Orpinomyces sp. strain PC-2 (39% sequence identity for 359 amino acid residues) (7), CelB29 from Orpinomyces joyonii (39% sequence identity for 359 amino acid residues) (19), and CelA from Clostridium longisporum (37% sequence identity for 353 amino acid residues) (15). All of these enzymes had a family 5 catalytic module. Two modules with unknown functions (UM-1 and UM-2) were linked to the N termini of GHF5-1 and GHF5-2, respectively (Fig. 1B). The level amino acid sequence identity between UM-1 and UM-2 was 73%. Furthermore, two linker sequences rich in proline, threonine, and serine linked to the C termini of GHF5-1 and GHF5-2.
FIG. 2.
Alignment of the amino acid sequences of the catalytic modules of E. cellulosolvens 5 (e.c.) Cel5A, C. longisporum (c.l.) CelA (GenBank accession no. L02868), O. joyonii (o.p.) CelB29 (GenBank accession no. AF015248), Orpinomyces sp. strain PC-2 (o.sp.) CelE (EMBL accession no. U97153), and R. albus (r.a.) endoglucanase VII (GenBank accession no. AB028321). Amino acids that are conserved in at least four of the six sequences are shaded. Dashes indicate gaps left to improve the alignment. The numbers indicate the amino acid residues at the beginning of each line; all sequences are numbered from Met-1 of the peptide.
Expression of Cel5A and its derivatives in E. coli.
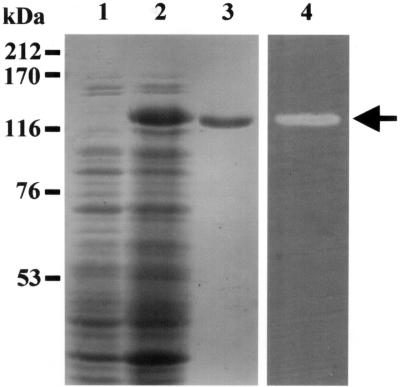
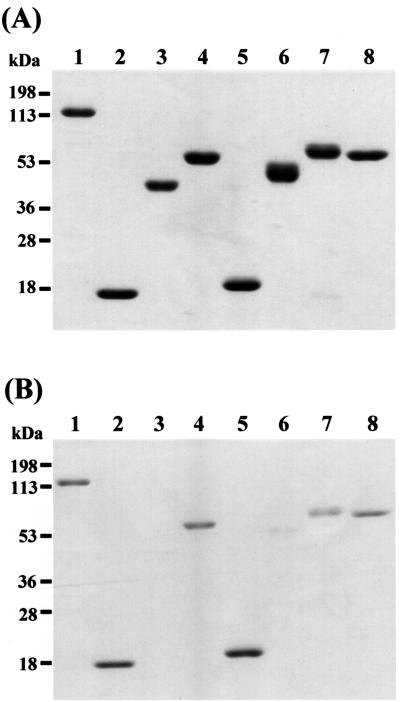
SDS-PAGE analysis showed that the molecular mass of His6-tagged Cel5A (rCel5A) expressed in E. coli harboring plasmid pKY12 was approximately 127 kDa (Fig. 3, lanes 1 to 3). Zymogram analysis showed that recombinant Cel5A exhibited CMCase activity (Fig. 3, lane 4). The proteins of truncated forms of Cel5A, which were constructed as described in Materials and Methods, were D1, D2, D3, D4, D5, D6, and D7 (Fig. 1B). These proteins were partially purified from whole-cell lysates of E. coli recombinants by Ni+ affinity chromatography. Each of the purified proteins produced a single band on SDS-PAGE gels, and their molecular sizes were in good agreement with those deduced from the nucleotide sequences (Fig. 4A).
FIG. 3.
Recombinant Cel5A expressed in E. coli and its CMCase activity. Proteins in gels containing 0.1% CMC were stained with Coomassie brilliant blue (lanes 1 to 3), and CMCase in gels containing 0.1% CMC was detected by Congo red staining (lane 4). Whole-cell lysates of both E. coli BL21(DE3) (control) (lane 1) and E. coli BL21(DE3) harboring plasmid pKY12 (lane 2) were included. The rCel5A expressed was partially purified with ProBond resin (lanes 3 and 4). The arrow indicates the position of the approximately 127-kDa band. The locations of molecular mass markers are indicated on the left.
FIG. 4.
Expression of Cel5A and its derivatives in E. coli (A) and binding of the proteins to acid-swollen cellulose (B). Lane 1, Cel5A; lane 2, D1; lane 3, D2; lane 4, D3; lane 5, D4; lane 6, D5; lane 7, D6; lane 8, D7. The locations of molecular mass makers are indicated on the left.
Binding of rCel5A and its derivatives to insoluble and soluble polysaccharides.
rCel5A bound to ASC, lichenan, and oat spelt xylan. On the other hand, rCel5A had no affinity for Avicel, agarose, Sephadex G-25, and chitin (data not shown). Since rCel5A could bind to ASC, the abilities of the partially purified derivatives of Cel5A to bind to ASC were examined. An ability to bind to ASC was detected for D1, D3, D4, D6, and D7. These proteins commonly included one of the unknown modules (UM-1 or UM-2) of Cel5A. On the other hand, D2 and D5, the derivatives that lacked the unknown modules, did not bind to ASC (Fig. 4B). Furthermore, in order to characterize the unknown modules of Cel5A, the abilities of the derivatives largely consisting of UM-1 or UM-2 of Cel5A (D1 and D4) to bind to various insoluble polysaccharides were examined. D1 and D4 could bind to lichenan and oat spelt xylan in addition to ASC. On the other hand, these proteins did not bind to Avicel, agarose, Sephadex G-25, or chitin (data not shown). From these results, UM-1 and UM-2 were identified as CBMs of Cel5A.
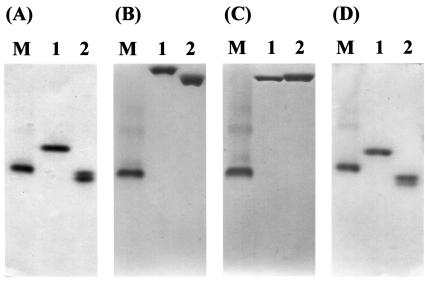
The affinities of the CBMs (D1 and D4) for a series of soluble polysaccharides were also qualitatively evaluated by native affinity gel electrophoresis (Fig. 5). The migration of these proteins was significantly retarded by inclusion of methylcellulose and lichenan in gels but was not affected by CMC, laminarin, or soluble starch, whereas the CBMs showed very low affinity for the soluble fraction of oat spelt xylan (data for oat spelt xylan, laminarin, and soluble starch are not shown).
FIG. 5.
Affinity analysis of derivatives D1 (lane 1) and D2 (lane 2) of Cel5A with soluble polysaccharides by native affinity gel electrophoresis. Purified protein samples were separated in nondenaturing polyacrylamide gels containing 0.1% (wt/vol) soluble polysaccharides, including methylcellulose (B), lichenan (C), and CMC (D). A gel without polysaccharide was used as a noninteracting control for migration (A). Lane M contained bovine serum albumin as a control protein.
Enzymatic properties of rCel5A and its derivatives.
rCel5A and its derivatives were spotted on an agarose plate containing CMC. CMCase activity was detected in rCel5A and its derivatives which contained the family 5 catalytic module (D2, D3, D5, D6, and D7). On the other hand, the D1 and D4 derivatives, which consisted mostly of the CBM, could not hydrolyze CMC (Fig. 1B). Then the hydrolytic specific activities of rCel5A and the D2, D3, D5, D6, and D7 derivatives with various polysaccharides were examined (Table 2). Partially purified rCel5A exhibited relatively high specific activities with CMC (4,800 IU/μmol) and lichenan (2,900 IU/μmol) and low activities with ASC (48 IU/μmol) and oat spelt xylan (170 IU/μmol). In addition, the activities of the D2, D3, D5, D6, and D7 derivatives with CMC and lichenan were also high, and the activities with ASC and oat spelt xylan were low. On the other hand, rCel5A and its derivatives exhibited no hydrolytic activity with Avicel, laminarin, chitin, and mannan.
TABLE 2.
Activities of rCel5A and its derivatives with various substratesa
| Substrateb | Sp act (IU/μmol) of:
|
|||||
|---|---|---|---|---|---|---|
| rCel5A | D2 | D3 | D5 | D6 | D7 | |
| CMC | 4,800 | 1,500 | 2,800 | 2,200 | 3,400 | 2,700 |
| ASC | 48 | 12 | 26 | 13 | 26 | 29 |
| Oat spelt xylan | 170 | 46 | 170 | 64 | 220 | 140 |
| Lichenan | 2,900 | 400 | 980 | 800 | 1,800 | 1,800 |
The measurements were done at least in triplicate.
Hydrolytic activities were not detected with Avicel, chitin, laminarin, and mannan.
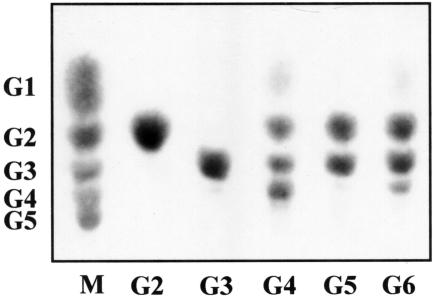
To investigate the activities of rCel5A and the D2 and D5 derivatives with various cellooligosaccharides, the hydrolysis products obtained from the substrates were qualitatively analyzed by TLC (Fig. 6). rCel5A produced mainly cellobiose and cellotriose along with glucose as a minor product from cellotetraose and cellohexaose and produced only cellobiose and cellotriose from cellopentaose. As shown in Fig. 6, the hydrolysis products obtained from these cellooligosaccharides after hydrolysis by D2 and D5 were identical to the products obtained after hydrolysis by rCel5A (data for D5 are not shown). rCel5A, D2, and D5 did not hydrolyze cellobiose and cellotriose. These proteins were less active with cellotetraose than with cellopentaose and cellohexaose.
FIG. 6.
TLC analysis of products from hydrolysis of cellooligosaccharides by the derivatives of Cel5A. Each cellooligosaccharide (G2 to G6; 1 μg) was incubated with the D2 derivative (0.1 U) for 6 h, and the hydrolysates were analyzed by TLC. Lane M contained authentic oligosaccharides. G1, glucose; G2, cellobiose; G3, cellotriose; G4, cellotetraose; G5, cellopentaose; G6, cellohexaose.
To investigate the properties of rCel5A as a cellulolytic enzyme, a viscometric analysis of CMC hydrolysis was performed. The viscosity of a CMC solution was reduced rapidly by rCel5A during the first 5 min of a reaction. After this, the reduction in viscosity was small (data not shown). This behavior of rCel5A is typical for endoglucanase activity.
DISCUSSION
Most glycoside hydrolases are modular enzymes that consist of two or more discrete modules, such as catalytic modules and CBMs. Catalytic modules, which are involved in the hydrolysis of polysaccharides, are now classified into 96 families on the basis of amino acid sequence similarities (10; http://afmb.cors-mrs.fr/∼cazy/CAZY/index.html). On the other hand, CBMs are classified into 49 families on the basis of amino acid sequence similarities (http://afmb.cors-mrs.fr/∼cazy/CAZY/index.html). It is thought that CBMs promote close proximity of enzymes to polysaccharides and assist the more rapid degradation of polysaccharides with their catalytic modules (2, 6).
E. cellulosolvens 5 Cel5A is a modular enzyme consisting of an N-terminal signal peptide, two catalytic modules belonging to glycosyl hydrolase family 5, two novel CBMs, two linker sequences, and a C-terminal sequence with an unknown function (Fig. 1B). The two catalytic modules were classified into glycosyl hydrolase family 5 on the basis of amino acid sequence similarity (Fig. 2). It has been reported previously that the amino acid sequence of the CBM of E. cellulosolvens 5 CBPA did not exhibit significant homology with the amino acid sequences of other CBMs (23); the amino acid sequences of CBMs of E. cellulosolvens 5 Cel5A also did not exhibit significant homology to the amino acid sequences of other CBMs as determined by a BLAST search. These findings indicate that CBMs of both Cel5A and CBPA from E. cellulosolvens 5 are members of new types of CBM families.
To compare the enzymatic properties of the two catalytic modules (GHF5-1 and GHF5-2) of E. cellulosolvens 5 Cel5A, truncated derivatives (D2, D3, D4, D5, D5, and D6) of Cel5A were constructed by PCR, and their hydrolytic activities with various substrates and activities with cellooligosaccharides were examined. All of the derivatives that commonly contained one catalytic module of Cel5A hydrolyzed CMC, ASC, oat spelt xylan, and lichenan. rCel5A showed the same substrate specificity with these derivatives (Table 2). In addition, all of the derivatives produced cellobiose and cellotriose and a minor amount of glucose from cellotetraose and cellohexaose and produced only cellobiose and cellotriose from cellopentaose. rCel5A showed the same activity with cellooligosaccharides with these derivatives (Fig. 6). These results indicate that there are no differences in substrate specificity and activity with cellooligosaccharides between GHF5-1 and GHF5-2.
Moreover, to investigate the effect of the repetition of a catalytic module of Cel5A on enzymatic activities, the hydrolytic activities of rCel5A and its derivatives with various substrates were determined. The specific activity of rCel5A with ASC or lichenan was almost identical to the sum of the specific activities of D3 containing GHF5-1 and CBM-1 and D6 containing GHF5-2 and CBM-2. On the other hand, the specific activity of rCel5A with oat spelt xylan was about one-half the sum of the D3 and D6 specific activities (Table 2). This suggests that only one of the two catalytic modules of rCel5A is involved in the hydrolysis of xylan. One possible explanation for this result is that β-1,4-linked hexoses present a planar three-dimensional structure, with 180° rotation between monomers, whereas xylan has a threefold helical structure, with approximately 120° rotation between every second monomer (1, 11). Therefore, it is possible that the substrate-binding groove of rCel5A has a completely complementary shape with a flat conformation of β-1,4-linked hexoses but has an incomplete complementary shape with a threefold helix conformation of xylan. Furthermore, the results described above indicate that the duplication of a catalytic module in Cel5A does not result in synergistic activity between the catalytic modules. In general, it is known that aerobic microorganisms produce higher concentrations of the enzymes than anaerobic microorganisms produce (21). Therefore, E. cellulosolvens and other rumen microorganisms may have overcome the disadvantage mentioned above by using a strategy such as reiteration of the catalytic module in an enzyme and insertion of a different kind of catalytic module into an enzyme.
It is known that the removal of CBMs from cellulolytic enzymes reduces their catalytic activities with various substrates (2, 4). In this study, the hydrolytic specific activities of the derivatives containing a catalytic module and a CBM with insoluble polysaccharides (ASC, oat spelt xylan, and lichenan) and soluble polysaccharide (CMC) were evidently higher than those of the derivatives devoid of CBMs (Table 2). These results indicate that for Cel5A and its derivatives the hydrolytic activities with insoluble and soluble polysaccharides are remarkably reduced by removal of CBMs and that the CBMs of Cel5A play an important role in assisting hydrolysis of polysaccharides by the catalytic module. Additionally, CBMs of Cel5A showed no affinity for CMC (Fig. 5), whereas CBMs adjacent to catalytic modules were involved in reinforcing the hydrolytic activity of catalytic modules (Table 2). A likely assumption is that the carboxymethyl side chains of CMC interfere with the association with the substrate-binding sites of Cel5A and that CBMs adjacent to catalytic modules accommodate a flat CMC structure on the substrate-binding surface of Cel5A and facilitate an association between active sites and ligands.
Cel5A from E. cellulosolvens 5 contained two homologous family 5 catalytic modules. Other cellulases comprising multiple family 5 catalytic modules have been found mainly in rumen fungi (3, 8, 29). For example, Neocallimastix patriciarum CelD consists of three family 5 catalytic modules that exhibit a very high degree of amino acid identity (3, 29), and Piromyces equi Cel5A comprises four family 5 catalytic modules that are more than 99% identical to each other on the nucleic acid level (8). Recently, genes encoding cellulases with multiple family 5 catalytic modules have been found in the rumen protozoa Polyplastron multivesiculatum and Epidinium caudatum (22). Additionally, the region consisting of a CBM and a catalytic module is tandemly reiterated in the primary structure of Cel5A. The two catalytic modules (GHF5-1 and GHF5-2) of Cel5A exhibited 94% sequence identity. Based on these findings, it is assumed that Cel5A arose through duplication of an ancestral gene that originally encoded a single region consisting of one CBM and one catalytic module.
In this study, the cel5A gene from E. cellulosolvens 5 was expressed as a His6-tagged Cel5A protein in E. coli, and the properties of the recombinant Cel5A were examined. However, further work is needed to compare the characteristics of the recombinant Cel5A with those of native Cel5A from E. cellulosolvens 5.
Acknowledgments
We are grateful to S. Karita (Mie University, Tsu, Japan) for helpful discussions. We thank M. Yoshimatsu for technical assistance.
This work was supported in part by Grant-in-Aid for Encouragement of Young Scientists 14760171 from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Abou Hachem, M., E. Nordberg Karlsson, E. Bartonek-Roxa, S. Raghothama, P. J. Simpson, H. J. Gilbert, M. P. Williamson, and O. Holst. 2000. Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem. J. 345:53-60. [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, M. K., H. Hayashi, S. Karita, M. Goto, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65:41-47. [DOI] [PubMed] [Google Scholar]
- 3.Alyward, J. H., K. S. Gobius, G.-P. Xue, G. D. Simpson, and B. P. Dalrymple. 1999. The Neocallimastix patriciarum cellulase, CelD, contains three almost identical catalytic domains with high specific activities on avicel. Enzyme. Microb. Technol. 24:609-614. [Google Scholar]
- 4.Arai, T., R. Araki, A. Tanaka, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barros, M. E., and J. A. Thomson. 1987. Cloning and expression in Escherichia coli of a cellulase gene from Ruminococcus flavefaciens. J. Bacteriol. 169:1760-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrard, G., A. Koivula, H. Soderlund, and P. Beguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., X. L. Li, D. L. Blum, and L. G. Ljungdahl. 1998. Two genes of the anaerobic fungus Orpinomyces sp. strain PC-2 encoding cellulases with endoglucanase activities may have arisen by gene duplication. FEMS Microbiol. Lett. 159:63-68. [DOI] [PubMed] [Google Scholar]
- 8.Eberhardt, R. Y., H. J. Gilbert, and G. P. Hazlewood. 2000. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology 146:1999-2008. [DOI] [PubMed] [Google Scholar]
- 9.Forsberg, C. W., J. Gong, L. M. J. Malburg, H. Zue, A. Iyo, K.-J. Cheng, and P. J. Krell. 1993. Cellulases and hemicellulases of Fibrobacter succinogenes and their role in fiber digestion, p. 125-136. In K. Shimada, K. Ohmiya, Y. Kobayashi, S. Hoshino, K. Sakka, and S. Karita (ed.), Genetics, biochemistry and ecology of lignocellulose degradation. Uni Publishers, Tokyo, Japan.
- 10.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leggio, L. L., J. L. Nours, C. Ryttersgaard, S. Kaneko, Z. Fujimoto, A. Kuno, S. Diertavitian, and S. Larsen. 2004. Structure and specificity of polysaccharides belonging to clan GH-A, p. 296-300. In Y. Onishi (ed.), Biotechnology of lignocellulose degradation and biomass utilization. Uni Publishers, Tokyo, Japan.
- 12.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 13.Minato, H., and T. Suto. 1978. Technique for fractionation of bacteria in rumen microbial ecosystem. II. Attachment of bacteria isolated from bovine rumen to cellulose powder in vitro and elution of bacteria attached therefrom. J. Gen. Appl. Microbiol. 24:1-16. [Google Scholar]
- 14.Mitsumori, M., and H. Minato. 1995. Distribution of cellulose-binding proteins among the representative strains of rumen bacteria. J. Gen. Appl. Microbiol. 41:297-306. [Google Scholar]
- 15.Mittendorf, V., and J. A. Thomson. 1995. Transcriptional induction and expression of the endoglucanase celA gene from a ruminal Clostridium sp. (“C. longisporum”). J. Bacteriol. 177:4805-4808. [DOI] [PMC free article] [PubMed]
- 16.Ohara, H., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci. Biotechnol. Biochem. 64:254-260. [DOI] [PubMed] [Google Scholar]
- 17.Ohara, H., J. Noguchi, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Sequence of egV and properties of EgV, a Ruminococcus albus endoglucanase containing a dockerin domain. Biosci. Biotechnol. Biochem. 64:80-88. [DOI] [PubMed] [Google Scholar]
- 18.Prins, R. A., F. Van Vugt, R. E. Hungate, and C. J. Van Vorstenbosch. 1972. A comparison of strains of Eubacterium cellulosolvens from the rumen. Antonie Leeuwenhoek 38:153-161. [DOI] [PubMed] [Google Scholar]
- 19.Qiu, X., B. Selinger, L. Yanke, and K. Cheng. 2000. Isolation and analysis of two cellulase cDNAs from Orpinomyces joyonii. Gene 245:119-126. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka, A., K. Tajima, M. Mitsumori, and H. Kajikawa. 2004. Properties of the fibrolytic enzyme gene from rumen ciliate protozoa, p. 296-300. In K. Ohmiya, K. Sakka, S. Karita, T. Kimura, M. Sakka, and Y. Onishi (ed.), Biotechnology of lignocellulose degradation and biomass utilization. Uni Publishers, Tokyo.
- 23.Toyoda, A., and H. Minato. 2002. Cloning, nucleotide sequence and expression of the gene encoding the cellulose-binding protein A (CBPA) of Eubacterium cellulosolvens 5. FEMS Microbiol. Lett. 207:141-146. [DOI] [PubMed] [Google Scholar]
- 24.Toyoda, A., and H. Minato. 2002. Identification of the cellulose-binding and the cell wall-binding domains of Eubacterium cellulosolvens 5 cellulose-binding protein A (CBPA). FEMS Microbiol. Lett. 214:113-118. [DOI] [PubMed] [Google Scholar]
- 25.Toyoda, A., K. Takano, and H. Minato. 2003. A possible role of cellulose-binding protein A (CBPA) in the adhesion of Eubacterium cellulosolvens 5 to cellulose. J. Gen. Appl. Microbiol. 49:245-250. [DOI] [PubMed] [Google Scholar]
- 26.Toyoda, A., K. Yoda, Y. Nakamura, and H. Minato. 2001. Presence of several cellulose-binding proteins in culture supernatant and cell lysate of Eubacterium cellulosolvens 5. J. Gen. Appl. Microbiol. 47:321-328. [DOI] [PubMed] [Google Scholar]
- 27.Wood, P. J., J. D. Erfle, and R. M. Teather. 1988. Use of complex formation between Congo Red and polysaccharides in detection and assay of polysaccharide hydrolases. Methods Enzymol. 160:59-74. [Google Scholar]
- 28.Wood, T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19-25. [Google Scholar]
- 29.Xue, G. P., K. S. Gobius, and C. G. Orpin. 1992. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarum encoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J. Gen. Microbiol. 138:2397-2403. [DOI] [PubMed] [Google Scholar]
- 30.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2002. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology 148:247-255. [DOI] [PubMed] [Google Scholar]