Abstract
We compared fecal samples with samples collected with rectoanal mucosa swabs (RAMS) to determine the prevalence of Escherichia coli O157 in feedlot cattle (n = 747). Escherichia coli O157 was detected in 9.5% of samples collected with RAMS and 4.7% of samples tested by fecal culture. Pulsed-field gel electrophoresis analysis of isolates suggested that the strains colonizing the rectoanal junction were the same as those from the feces. Mucosal swab sampling was more sensitive than fecal sampling for determining the prevalence of E. coli O157 in feedlot cattle.
Escherichia coli O157 is an important food-borne pathogen (15), and cattle are considered the major reservoir (2, 5). Early studies indicated the prevalence of E. coli O157 in cattle feces to be less than 2% of the cattle population (8, 10). However, molecular and immunomagnetic separation techniques have estimated that number to be closer to 20 to 30% in the summer months (4, 7, 9, 17, 22). Two major patterns of E. coli O157 shedding in the feces of cattle have been suggested: transient, in which animals shed the organism only briefly after picking it up from the environment, and colonized, in which the organism is shed for an extended period of time after presumptive colonization in the gastrointestinal tract (18). Recent findings suggest that E. coli O157 specifically colonizes the lymphoid follicle-dense mucosal epithelium at the terminal rectum, located approximately 2 to 5 cm proximal to the rectoanal junction (16). Based on this observation, Rice et al. (18) developed a rectoanal mucosal swab (RAMS) technique to sample the mucosal surface of the rectoanal junction. The objectives of this study were to compare the two collection methods, feces sampling and the RAMS technique, for detecting the prevalence of E. coli O157 in feedlot cattle and to use genetic subtyping by pulsed-field gel electrophoresis (PFGE) to compare isolates obtained by the two collection methods.
Crossbred beef steers (n = 747) from two different feedlots were used in this study. Diets consisted of approximately 94% concentrate (dry-rolled or steam-flaked corn) and 6% forage (alfalfa hay). The collection of RAMS samples was performed according to Rice et al. (18). The swabs were placed into culture tubes containing 3 ml of gram-negative broth (BD, Franklin Lakes, N.J.) with 0.05 mg/liter of cefixime, 10 mg/liter of cefsulodin, and 8 mg/liter of vancomycin (GNccv) and held on ice until transported to the laboratory. Fecal samples (at least 10 g) were taken immediately following the RAMS procedure by rectal palpation and stored on ice in individually sealed bags until transported to the laboratory. The RAMS sample was vortexed for 1 min, and 1 ml of the broth was transferred to 9 ml of GNccv. Approximately 1 g of subsample was placed in 9 ml of GNccv by using a sterile transfer stick. The procedures for enrichment, immunomagnetic separation, selective isolation, and identification of E. coli O157 from RAMS and fecal samples were according to Sargeant et al. (22). PFGE analysis was performed according to the Centers for Disease Control and Prevention PulseNet protocol (http://www.cdc.gov/ncidod/dbmd/pulsenet/pulsenet.htm). Isolates were grouped into subtypes and types based on fingerprint pattern similarities. Subtypes and types were defined as isolates having fingerprint patterns of 100% and >95% Dice similarities, respectively. The prevalences of E. coli O157 between methods were compared using chi-square analysis, with significance determined at a P of ≤0.05.
There was no difference in prevalence of E. coli O157 in cattle located at the two sites (P > 0.10); therefore, data were pooled across collection sites. The RAMS technique detected a higher prevalence (P < 0.01) of E. coli O157 in cattle than fecal culture (9.5% versus 4.7%) (Table 1). Of the 747 cattle tested, 82 (11.0%) were identified as positive by at least one detection method. Of those animals that tested positive for E. coli O157 (n = 82), 87% (71 of 82) were detected by the RAMS method and only 43% (35 of 82) were detected by the fecal culture method. Among RAMS-positive cattle (n = 71), 24 animals (33.8%) were positive by fecal culture. Among cattle whose feces were positive (n = 35), 24 (68.6%) were positive by the RAMS method. Only a small number of cattle (11 of 82 [13.4%]) were positive by fecal culture alone. However, 36 of 82 positive cattle (43.9%) were positive by the RAMS technique and negative by fecal culture.
TABLE 1.
Detection of E. coli O157 in feedlot cattle by RAMS and fecal culture
| Item (total no. tested) | No. of positive samples (%)
|
|||
|---|---|---|---|---|
| RAMSa | Feces | RAMS or fecesc | RAMS and fecesd | |
| No. of animals (747) | 71 (9.5) | 35 (4.7)b | 82 (11.0) | 24 (3.2) |
| Positive animalse (82) | 71 (87.0) | 35 (42.7) | 82 (100) | 24 (29.3) |
| RAMS positive (71) | 71 (100) | 24 (33.8) | ||
| Feces positive (35) | 24 (68.6) | 35 (100) | ||
RAMSs were obtained by inserting a sterile foam-tipped applicator approximately 2 to 5 cm into the anus of each steer, using a rapid in and out motion, to swab the entire mucosal surface of the rectoanal junction.
Significantly different from the value for RAMS at a P of <0.01.
Animals that tested positive by either sampling method.
Animals that tested positive by both sampling methods.
Animals that tested positive by either sampling method.
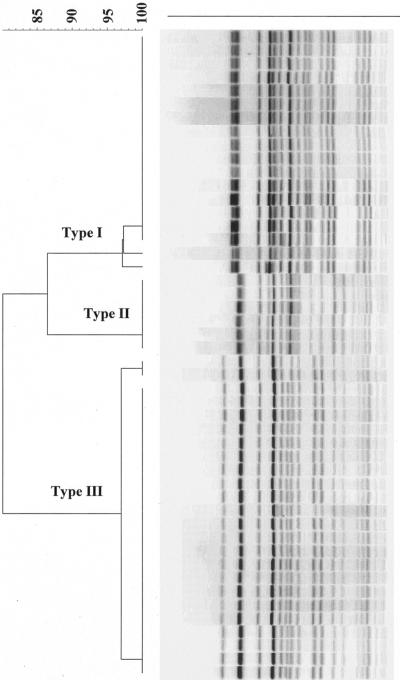
Genomic fingerprints of isolates were analyzed by PFGE, and only isolates that were obtained by both sampling methods from animals (n = 24) were used. As shown by XbaI restriction endonuclease digestion patterns, only three genetic types (>95% Dice similarity) were present in the RAMS and fecal isolates (n = 48) of 24 cattle. Types I (18 isolates) and II (6 isolates) showed 86% similarity, whereas type III (24 isolates) was the most distant strain, with only 80% similarity (Fig. 1). Type I was derived from cattle at one location, and types II and III were derived from cattle at the second location. Of the 24 pairs of isolates evaluated, 20 had 100% similarity and four had >95% similarity in PFGE banding patterns, suggesting that strains colonizing the rectoanal junction were the same as those isolated from feces.
FIG. 1.
Dendrogram generated to show the relationship of 48 isolates (24 from RAMSs and 24 from fecal cultures) of E. coli O157 from 24 cattle. The bands were analyzed by Dice similarity coefficient and the unweighted pair group method for clustering, with a position tolerance setting of 1.5% for optimization and a position tolerance of 1.5% for band comparison. The scale at the top of the dendrogram indicates the levels of similarity between isolates.
Our data involving 747 cattle suggest that the RAMS technique is a more sensitive method than fecal culture for detecting the prevalence of E. coli O157 in cattle. Our finding is in agreement with the previous report by Rice et al. (18); however, the cultural procedure employed in our study was different from the one used by Rice et al. (18). In their study, among naturally infected cattle, enriched RAMS and fecal cultures were equally sensitive, but direct plating of RAMS culture detected a significantly higher prevalence than either direct or enriched fecal cultures (18). Because all our samples were enriched before being plated, the prevalence data were entirely qualitative. In addition, we used a sample size of 1 g for enrichment compared to 10 g used by Rice et al. (18). Generally, use of 10-g samples has been shown to increase the sensitivity of the isolation method by severalfold (11, 21). However, a number of studies have used a 1-g sample size for the detection of fecal shedding of E. coli O157 (12, 17, 20, 24). Additionally, our observation has relevance for epidemiological investigations that involve thousands of fecal samples when, for logistical reasons, a 1-g sample size is more practical than 10-g samples (10, 17, 22). It is not likely that the difference that we observed between the RAMS technique and fecal culture was because of a 1-g sample size. Others (18, 23) have observed a similar difference even with a 10-g sample size. The higher sensitivity with the RAMS technique was possibly because the swabbing allowed direct sampling of the rectal mucosa, the site of E. coli O157 colonization (16). Also, the RAMS sample with minimal fecal contamination probably had fewer competing organisms, thereby allowing target bacteria to become enriched in the selective medium (18).
Cattle that were RAMS positive but negative by fecal culture (n = 47) indicate that animals can harbor the organism and not shed it in detectable numbers in the feces. These cattle potentially represent an on-farm reservoir not previously recognized (18). Cattle that were feces positive but RAMS negative may represent cattle that have been recently exposed to E. coli O157 but whose rectoanal junction has not yet been colonized. Rice et al. (18) observed that within the first 2 weeks after experimental exposure from culture-positive calves the enrichment fecal cultures were more sensitive than enrichment RAMS cultures at detecting E. coli O157. In contrast, in the late period (>40 days), the enrichment RAMS cultures were more sensitive, indicating that perhaps the environmental exposure to the bacteria shows up in the feces relatively quickly whether the rectoanal junction has been colonized or not. Later in the exposure period, the RAMS technique appeared to become more sensitive at identifying animals that may no longer be shedding environmental contaminants but rather were shedding due to colonization of the animals. The idea that rectoanal mucosa is a site of colonization is further supported by Sheng et al. (23), who reported that rectal swab inoculation of E. coli O157 resulted in a consistent, long-term colonization in cattle and was superior to traditional oral inoculation.
Naylor et al. (16) reported that counts of E. coli O157 cells from the surfaces of fecal samples were 1,000-fold higher than the counts in the core samples in experimentally inoculated calves believed to be colonized by the organism. Therefore, it was theorized that the source of E. coli O157 in the feces was the rectoanal junction during the passage of the feces through this region. Recently, Low et al. (14) have reported that high levels of fecal shedding of E. coli O157 were associated with mucosal carriage at the terminal rectum. During the sampling of animals, it is impossible to sample by both techniques without the risk of cross-contamination from the feces to the RAMS and vice versa. This cross-contamination at either location has the potential to overwhelm the sample and increases the difficulty of showing the strains from the two locations to be different.
Pulsed-field gel electrophoresis has been used widely as a molecular subtyping tool in outbreak investigations and surveillance in human infections (3, 19) and also to determine genetic relatedness between isolates from cattle (1, 6, 13). Of the 82 cattle that were positive for E. coli O157, only 24 animals tested positive by both sampling methods. These isolates were compared by PFGE to determine genetic similarities of the two isolates from the same animal. Only three genetically related clusters were observed among the 48 isolates. All 24 pairs of isolates showed >95% similarity between RAMS and fecal isolates from the same animal, suggesting that the clones in the feces were the same as those that colonized the mucosal region of the rectoanal junction. This provides support for the theory proposed by Naylor et al. (16) that the feces become coated with the organism colonizing the mucosal region when they come into contact with the mucosal region. However, it is possible that fecal contamination occurs when the mucosal region is swabbed, thus explaining genetic similarities between the two isolates. Although precautions were taken to minimize fecal contamination, it was evident that in many instances there was visible fecal staining of the swabs. Nevertheless, the RAMS technique of sampling was superior to the use of conventional fecal samples for detecting the prevalence of E. coli O157:H7 in cattle.
Acknowledgments
This work was supported by a special grant from the USDA Cooperative State Research, Education, and Extension Service (2004-34359-13008).
We thank Amy Hanson and Neil Wallace for their assistance in the laboratory.
REFERENCES
- 1.Avery, S. M., E. Liebana, M. L. Hutchinson, and S. Buncic. 2004. Pulsed-field gel electrophoresis of related Escherichia coli O157 isolates associated with beef cattle and comparison with unrelated isolates from animals, meats and humans. Int. J. Food Microbiol. 92:161-169. [DOI] [PubMed] [Google Scholar]
- 2.Bach, S. J., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2003. Transmission and control of Escherichia coli O157:H7. A review. Can. J. Anim. Sci. 82:475-490. [Google Scholar]
- 3.Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, R. Baron, and J. Kobayashi. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 272:1349-1353. [PubMed] [Google Scholar]
- 4.Buchko, S. J., R. A. Holley, W. O. Olson, V. P. J. Gannon, and M. Veira. 2000. The effect of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J. Food Prot. 63:1467-1474. [DOI] [PubMed] [Google Scholar]
- 5.Callaway, T. R., R. C. Anderson, T. S. Edrington, R. O. Elder, K. J. Genoese, K. M. Bischoff, T. L. Pools, Y. S. Jang, R. B. Harvey, and D. J. Nisbet. 2003. Preslaughter intervention strategies to reduce food-borne pathogens in food animals. J. Anim. Sci. 81(Suppl. 2):E17-E23. [Google Scholar]
- 6.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Lagreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M.-S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gansheroff, L. J., and A. D. O'Brien. 2000. Escherichia coli O157 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated. Proc. Natl. Acad. Sci. USA 97:2959-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, D. D., D. H. Rice, L. A. Thomas, D. A. Dargatz, and T. E. Besser. 1997. Epidemiology of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 11.Lahti, E., O. Ruoho, L. Rantala, M. L. Hänninen, and T. Honkanen-Buzalski. 2003. Longitudinal study of Escherichia coli O157 in a cattle finishing unit. Appl. Environ. Microbiol. 69:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laven, R. A., A. Ashmore, and C. S. Stewart. 2003. Escherichia coli in the rumen and colon of slaughter cattle, with particular reference to E. coli O157. Vet. J. 165:78-83. [DOI] [PubMed] [Google Scholar]
- 13.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low, J. C., I. J. McKendrick, C. McKechnie, D. Fenlon, S. W. Naylor, C. Currie, D. G. E. Smith, L. Allison, and D. L. Gally. 2005. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 71:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice, D. H., H. Q. Sheng, S. A. Wynia, and C. J. Hovde. 2003. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 41:4924-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rios, M., V. Prado, M. Trucksis, C. Arellano, C. Borie, M. Alexandre, A. Fica, and M. M. Levine. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson, S. E., E. J. Wright, C. A. Hart, M. Bennett, and N. P. French. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045-1053. [DOI] [PubMed] [Google Scholar]
- 21.Sanderson, M. W., J. M. Gay, D. D. Hancock, C. C. Gay, L. K. Fox, and T. E. Besser. 1995. Sensitivity of bacteriologic culture for detection of Escherichia coli O157:H7 in bovine feces. J. Clin. Microbiol. 33:2616-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sargeant, J. M., M. W. Sanderson, R. A. Smith, and D. D. Griffin. 2003. Escherichia coli O157 in feedlot cattle feces and water in four major feeder-cattle states in the USA. Prev. Vet. Med. 61:127-135. [DOI] [PubMed] [Google Scholar]
- 23.Sheng, H., M. A. Davis, H. J. Knecht, and C. J. Hovde. 2004. Rectal administration of Escherichia coli O157:H7: novel model for colonization of ruminants. Appl. Environ. Microbiol. 70:4588-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Baale, M. J., J. M. Sargeant, D. P. Gnad, B. M. DeBey, K. F. Lechtenberg, and T. G. Nagaraja. 2004. Effect of forage or grain diets with or without monensin on ruminal persistence and fecal Escherichia coli O157:H7 in cattle. Appl. Environ Microbiol. 70:5336-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]