Abstract
Bacteria presumably involved in oxygen- or nitrate-dependent sulfide oxidation in the biofilters of a recirculating marine aquaculture system were identified using a new application of reverse transcription-PCR denaturing gradient gel electrophoresis (DGGE) analysis termed differential-transcription (DT)-DGGE. Biofilter samples were incubated in various concentrations of sulfide or thiosulfate (0 to 5 mM) with either oxygen or nitrate as the sole electron acceptor. Before and after short-term incubations (10 to 20 h), total DNA and RNA were extracted, and a 550-bp fragment of the 16S rRNA genes was PCR amplified either directly or after reverse transcription. DGGE analysis of DNA showed no significant change of the original microbial consortia upon incubation. In contrast, DGGE of cDNA revealed several phylotypes whose relative band intensities markedly increased or decreased in response to certain incubation conditions, indicating enhanced or suppressed rRNA transcription and thus implying metabolic activity under these conditions. Specifically, species of the gammaproteobacterial genus Thiomicrospira and phylotypes related to symbiotic sulfide oxidizers could be linked to oxygen-dependent sulfide oxidation, while members of the Rhodobacteraceae (genera Roseobacter, Rhodobacter, and Rhodobium) were putatively active in anoxic, nitrate-dependent sulfide oxidation. For all these organisms, the physiology of their closest cultured relatives matches their DT-DGGE-inferred function. In addition, higher band intensities following exposure to 5 mM sulfide and nitrate were observed for Thauera-, Hydrogenophaga-, and Dethiosulfovibrio-like phylotypes. For these genera, nitrate-dependent sulfide oxidation has not been documented previously and therefore DT-DGGE might indicate a higher relative tolerance to high sulfide concentrations than that of other community members. We anticipate that DT-DGGE will be of general use in tracing functionally equivalent yet phylogenetically diverse microbial populations in nature.
Zero-discharge marine aquaculture (mariculture) systems provide a solution for the environmentally detrimental effects of conventional mariculture (16). These closed systems utilize biofilters to convert the main contaminants from the process water to harmless gases. Treatment encompasses oxidation of ammonia to nitrate in an aerobic trickling filter and anaerobic removal of organic matter and nitrate by denitrification in a parallel purification loop comprising a digestion/sedimentation basin (DB) and a fluidized-bed reactor (FBR) (16).
The high organic load entering the DB creates sharp redox potential gradients with extensive sulfate reduction within settled sludge. In these nitrate-depleted zones, sulfide concentrations can reach values exceeding 5,000 μM (11). Due to extensive sulfide oxidation, sulfide concentrations in the overlying water of the basin are generally more than 1 order of magnitude below these values (11). Remaining sulfide from the DB effluent is oxidized in the organic carbon-limited FBR (12). High concentrations of nitrate (1,000 to 5,000 μM) in the system suggest the occurrence of nitrate-dependent sulfide oxidation, particularly because oxygen is depleted at the surfaces of DB sludge and FBR flocs (50 to 100 μm). This argument is supported by previous studies that demonstrated the existence of sulfide oxidation in both oxic and anoxic zones of the FBR and DB biofilms (12; A. Gieseke, unpublished data).
Although several bacterial phylotypes with close affiliation to known sulfide oxidizers in these biofilms have been identified previously (11, 12), the application of 16S rRNA gene-based methods alone does not allow for final conclusions as to their functions in these biofilms, since sulfide-oxidizing bacteria are highly polyphyletic (15, 18, 21, 41, 44, 45, 49). Previously described methods geared toward linking the phylogeny and physiology of specific bacteria, such as RNA stable isotope probing (27) or combined microautoradiography and fluorescence in situ hybridization (MAR-FISH) (25, 32), require prior knowledge regarding the phylogenetic affiliation of the expected group of bacteria active in the specific process being studied and are extremely laborious. They are therefore not suitable for assessing the physiology of sulfide-oxidizing bacteria, which are highly polyphyletic and are characterized by extremely versatile (chemolithotrophic, mixotrophic, and/or facultatively heterotrophic) metabolic capacities (15, 19, 28, 37). Specifically, different organisms may be responsible for sulfide oxidation in different parts of the mariculture system, depending on the specific physiochemical conditions, i.e., with either oxygen or nitrate present.
In order to circumvent this complexity, we have developed and applied differential-transcription denaturing gradient gel electrophoresis (DT-DGGE), a method that enables identification of specific microbial phylotypes that display enhanced activity in response to defined environmental conditions within a complex microbial community. The concept of this method is based on the observation that transcription of bacterial rRNA is often closely correlated with metabolic activity (1, 4, 10, 24, 34). Environmental samples are subjected to short-term incubations (10 to 20 h) in composite media aimed at increasing the metabolic activity of specific functional groups of the microbial community. Prior to and following incubation, both DNA and RNA are extracted, amplified by PCR (or reverse transcription-PCR [RT-PCR]) using general bacterial or group-specific 16S rRNA gene-targeting primers, and analyzed by DGGE. Substantial changes in band intensities of RT-PCR DGGE ribotypes following specific incubations (not detected in PCR DGGE profiles) are interpreted as changes in the relative metabolic activity of the corresponding microbial communities.
The objective of this study was to pinpoint specific bacterial phylotypes potentially responsible for oxygen- and nitrate-dependent sulfide oxidation in the biofilters of the recirculating mariculture system. In order to achieve this objective, DT-DGGE analyses were implemented using both DB sludge and FBR flocs incubated in marine media with and without oxygen or nitrate, and with different sulfide and thiosulfate concentrations.
MATERIALS AND METHODS
Description of the zero-discharge mariculture system.
The overall structure and performance of the intensive recirculating mariculture system studied here have been described in detail previously (16). Briefly, water from the upper part of a double-drain fish basin (2.3 m3) was pump-recirculated at a rate of 1 to 2 fish basin volumes h−1 over a trickling filter. Simultaneously, water from the bottom center of the fish basin flowed into the DB (working volume, 0.4 to 0.5 m3) at a rate of 0.1 to 0.25 fish basin volumes h−1. Water from the upper layers of the DB outlet was pumped (at rates of 5 to 7 liters min−1) into the cylindrical FBR through a vertical pipe that extended from the top center to approximately 3 cm above the base of the FBR. The FBR (working volume, 6.26 liters) was filled with sand (average diameter, 0.7 mm), which served as a carrier material for the biofilm. Upflow from the inlet pipe caused the flocs (biofilm-attached sand grains) to float within the column. Water from the FBR outlet at the top of the cylinder was drained back to the fish basin after passing through a settler for removal of particulate matter, which was funneled back into the DB. Artificial seawater (ASW; salinity, 20 ppt) used in the system was prepared by addition of sea salt (Red Sea Pharm Ltd., Eilat, Israel) to tap water. The system was supplemented daily with 100 to 200 g of feed (Matmoor Ltd., Ashdod, Israel) comprising 45% fat and 19% protein.
Chemical parameters in the DB and FBR.
Chemical parameters were measured at the inlet and outlet of both the DB and FBR biofilters, and sulfide and ammonia were also measured in the porewater within settled sludge of the DB. Oxygen and temperature were measured with a YSI temperature/oxygen probe (model 57; Yellow Spring Instruments, Yellow Springs, Ohio). Total ammonia (referred to as ammonia below) was analyzed spectrophotomically using the salicylate-hypochlorite method (6), and nitrate and nitrite were measured using a Quick Chem Ion Analyzer (Lachat Instruments, Milwaukee, Minn.) according to the protocol provided by the supplier. For sulfide analysis, 2.5-ml samples were fixed in 1 ml of 5% zinc-acetate solution, and total sulfide concentrations (referred to as sulfide below) were determined by the methylene blue method (9).
Sampling and DT-DGGE experimental setup.
Two separate incubation experiments were conducted to detect changes in 16S rRNA gene transcription of bacterial communities in the DB and FBR under sulfidic conditions, the first in December 2003 (anoxic incubations) and the second in March 2004 (oxic and anoxic incubations). During both incubations, anoxic conditions were achieved as follows. Effluent water from the FBR (ASW) was filtered, aerated for 2 h to remove residual sulfide, purged with N2, and supplemented with NaNO3 and Na2HCO3 to give final NO3− and HCO3 concentrations of 5 mM and 1 mM, respectively. Aliquots of DB sludge from the middle portion of the basin and FBR flocs sampled from a sampling port in the middle of the FBR cylinder {0.75 and 2.5 g [wet weight], respectively} were washed in ASW and immediately transferred to 100-ml glass bottles under a constant flow of N2. Bottles were filled with ASW, amended with sulfide to final concentrations of 0, 100, 500, 1,000, or 5,000 μM or with thiosulfate (1,000 μM) under N2 flow, sealed with Teflon-coated butyl rubber stoppers, and cramped with aluminum seals. In the March 2004 experiment, oxic sulfidic conditions were attained as follows. DB sludge and FBR flocs {0.75 and 2.5 g [wet weight], respectively} were added to 250-ml cotton-capped glass beakers containing 100 ml of aerated ASW (without NO3−) supplemented with 1,000 μM S2O3. All incubations were done in the dark at 26°C for 10 and 20 h. Aliquots of DB sludge and FBR flocs (0.03 g and 0.3 g [wet weight], respectively) from the original biofilters and from the incubation experiments were transferred to 2.0-ml nucleic acid extraction tubes containing 6- to 8 5-mm-diameter glass beads (Marienfeld, Lauda-Koenigshofen, Germany) and 0.36 ± 0.02 g acid-washed glass beads (Sigma-Aldrich, St. Louis, MO), immediately frozen on a dry-ice-ethanol mixture, and stored at −80°C until nucleic acid extraction.
Nucleic acid extraction.
Crude extracts of DNA and RNA from DB and FBR samples were obtained via a preliminary extraction stage in a bead beater (Bio 101, Irvine, CA) at 5.5 S for 45 s. DNA samples were extracted in 900 μl extraction buffer (33), and RNA samples were extracted in 500 μl of Tissue and Cell Lysis Buffer (Epicenter, Madison, WI). DNA was further purified as described previously (33), and the purified DNA was stored at −20°C until further processing. RNA was purified from the DB and FBR extracts using the MasterPure RNA purification kit (Epicenter, Madison, WI). Following the preliminary bead-beating step (see above), samples supplemented with proteinase K to a final concentration of 0.5 μg ml−1 were incubated at 65°C for 15 min and centrifuged at 4°C for 15 min at 18,000 × g, after which RNA was purified according to the protocol provided by the manufacturer. The quality of extracted RNA was determined by agarose gel electrophoresis (2%) and staining with ethidium bromide. DNase treatment (performed according to the kit protocol) did not always suffice for total DNA removal, and samples contaminated with DNA were subjected to an additional DNase step. The absence of DNA was verified by PCR of RNA samples. Extracted RNA was stored at −80°C until further processing.
RT and PCR.
RT was performed using the Improm-II reverse transcriptase kit (Promega, Madison, WI) according to the standard reverse transcription protocol provided by the manufacturer, using 1 μl (∼80 ng) of the RNA template. RT extension was carried out by adding 20 pmol of the general bacterial primer 907R (30) using a T-gradient thermocycler (Biometra, Goettingen, Germany) with annealing at 25°C for 5 min, extension at 42°C for 60 min, and heat inactivation of the reverse transcriptase at 70°C for 15 min. PCR amplifications for DGGE were performed with the previously described bacterial primer pair 341F (with a GC clamp at its 5′ end) and 907R (30). Each 50-μl reaction mixture contained the following components: 1.5 U Taq DNA polymerase (Red Taq; Sigma, St. Louis, MO), Taq buffer containing a final magnesium concentration of 2.5 mM, deoxynucleoside triphosphates (20 nmol each), 12.5 μg bovine serum albumin, 25 pmol of each primer, and 1.2 μl DNA (or cDNA) template. The PCR program was carried out with an initial denaturation step of 95°C for 30 s, followed by 33 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s. Cycling was completed with a final elongation step at 72°C for 2 min. The presence and size of PCR fragments was determined by agarose gel electrophoresis (2%) and staining with ethidium bromide.
DGGE analysis.
DGGE of PCR- and RT-PCR-amplified 16S rRNA gene fragments was performed using a D-gene system (Bio-Rad, Hercules, CA), as previously described (30). In this analysis, 1-mm-thick 6% (wt/vol) polyacrylamide gels with a 20 to 60% denaturing gradient were run for 20 h in 1× Tris-acetate-EDTA buffer at a constant voltage of 90 V. Following electrophoresis, the gels were stained with GelStar nucleic acid stain (BioWhittaker Molecular Applications, Inc., Rockland, ME) and photographed on a UV transillumination table (302 nm) with a Kodak KDS digital camera (Kodak Co., New Haven, CT). Prevalent DGGE bands were carefully excised on a Dark Reader Transilluminator (Clare Chemical Research, Inc., Dolores, CO) and purified as described previously (30). Purified bands were reamplified and verified by a second DGGE analysis. Band areas of RT-DGGE profiles were measured using LabImage software, version 2.7.1 (Kapelan GmbH, Halle, Germany). Excised bands that showed substantial differences in RT-PCR DGGE analyses were evaluated by measuring their intensity relative to that of selected reference bands (from the corresponding lane) that showed relatively little change in all of the PCR and RT-PCR DGGE patterns. In addition, these bands were evaluated in terms of relative band intensity (RBI) units, the fraction of a defined band's intensity from a specific lane relative to the total intensity of that lane.
Cloning of PCR products and sequencing.
The pGEM-T Easy vector system (Promega, Madison, WI) was used to clone PCR products amplified from excised DGGE bands. Ligation and transformation reactions were performed according to the protocol described by the manufacturer. Clones were screened by colony PCR amplification and verified by DGGE (as described above). Plasmids from selected colonies were purified with the Concert Rapid Miniprep plasmid purification system (Genomed GmbH, Löhne, Germany) according to the protocol provided by the manufacturer.
Clones were sequenced using the Applied Biosystems PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with Ampli Taq DNA polymerase and the T7 primer as suggested by the manufacturer. The sequencing products were analyzed with an Applied Biosystems 377 DNA sequencer.
Phylogenetic analysis.
DT-DGGE band sequences (530 bp) were incorporated into a prealigned database of 16S rRNA gene sequences using the aligning tool implemented in the ARB phylogenetic program package (26). Parsimony was used to add the DT-DGGE band sequences to neighbor joining, maximum likelihood, and parsimony trees constructed with >1,000-bp sequences using the Felsenstein correction method. A consensus tree was then constructed based on the merged topologies of the three trees (26).
Nucleotide sequence accession numbers.
Twenty-four 16S rRNA gene sequences determined in this study were deposited in GenBank under accession numbers AY905666 to AY905689.
RESULTS
Chemical parameters in DB and FBR.
Chemical parameters were periodically measured at selected points of the biofiltration components of the mariculture system prior to and during the two DT-DGGE sampling periods (November 2003 and March 2004). The system temperature averaged 22 ± 3°C, and oxygen concentrations averaged 188, 86, and 39 μM at the DB inlet, DB outlet (same as FBR inlet), and FBR outlet, respectively. Nitrate concentrations averaged 1,570, 1,120, and 845 μM at the DB inlet, DB outlet (FBR inlet), and FBR outlet, respectively, indicating nitrate reduction in both the DB and FBR. Average ammonia concentrations measured at the DB inlet, DB outlet, and FBR outlet were 25, 80, and 45 μM, respectively, and ammonia concentrations within the sludge at the bottom DB averaged 7,000 μM. Nitrite values averaged 15, 25, and 10 μM at the DB inlet, DB outlet, and FBR outlet, respectively. Sulfide concentrations at the DB outlet (43 μM) were substantially lower than those measured within the sludge at the bottom of the DB (3,000 μM), indicating sulfide oxidation in certain zones of the DB. Average inlet and outlet sulfide concentrations in the FBR were 43 and 17 μM, respectively, giving an estimated average daily sulfide removal rate of 0.9 mol of total sulfide day−1. The maximal standard deviation between the chemical parameters measured was 20%.
DT-DGGE.
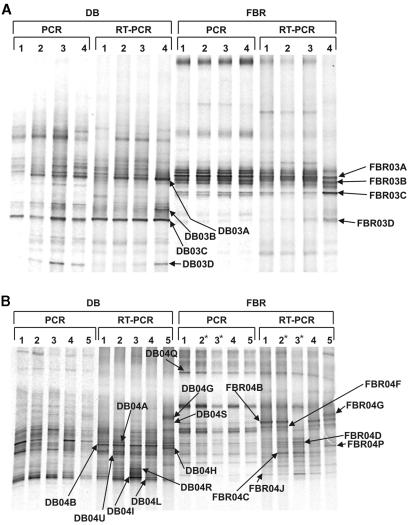
DGGE ribotypes obtained from DNA extracted from the DB and the FBR from both the December 2003 and the March 2004 experiments showed relatively high similarity between different incubation treatments in comparison to RT-PCR DGGE profiles of RNA extracts, where several bands showed increased or decreased intensity relative to that for the original, untreated biofilter sample (Fig. 1A and B). Duplicate samples showed high similarity between RT-PCR DGGE ribotypes, and therefore only one replicate of each is shown in the analyses (Fig. 1A and B). Prevalent bands from the RT-PCR DGGE analyses were excised and sequenced, and their phylogenetic affiliations were determined (Fig. 2). RT-PCR DGGE profiles for anoxic thiosulfate-amended incubations were almost identical to those for anoxic incubations amended with low sulfide concentrations, confirming that changes observed in 16S rRNA gene transcription in the oxic incubations stemmed from the aerobic conditions and not from the effect of thiosulfate as opposed to sulfide amendment.
FIG. 1.
(A) DT-DGGE analysis of DB and FBR samples following anoxic, nitrate-amended incubations (December 2003). Samples were incubated for 10 h in degassed nitrate-amended marine medium. PCR (DNA) and RT-PCR (RNA) treatments were supplemented with the following sulfide concentrations: lanes 1, original (nonincubated) biofilter sample; lanes 2, 0 μM H2S; lanes 3, 500 μM H2S; lanes 4, 5,000 μM H2S. (B) DT-DGGE analysis of DB and FBR samples following anoxic, nitrate-amended, and oxic incubations (March 2004). Samples were incubated for 20 h in degassed nitrate-amended marine medium (anoxic) or aerated medium (oxic). PCR (DNA) and RT-PCR (RNA) treatments were supplemented with the following sulfide concentrations: lanes 1, original (nonincubated) biofilter sample; lanes 2, 1,000 μM H2S; lanes 3, 5,000 μM H2S; lanes 2*, 100 μM H2S; lanes 3*, 1,000 μM H2S; lanes 4, 1,000 μM S2O3 (anoxic); lanes 5, 1,000 μM S2O3 (oxic). Asterisks indicate sulfide concentration discrepancies for FBR treatments.
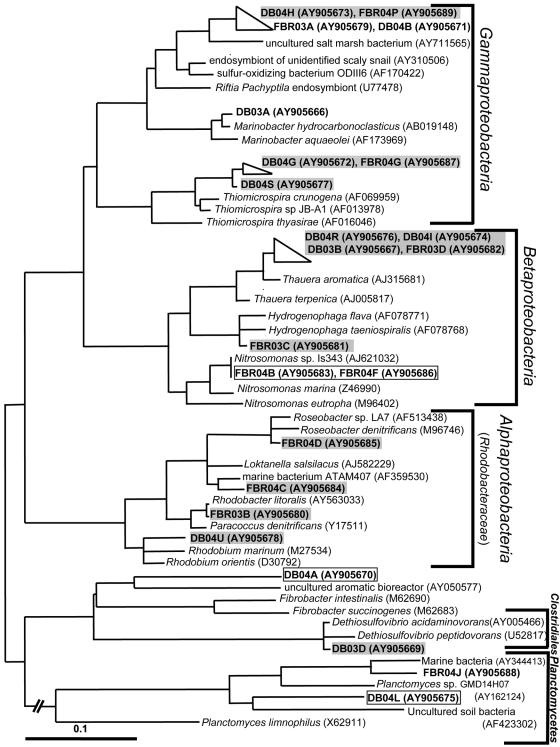
FIG. 2.
Phylogenetic tree showing gene sequences from excised DT-DGGE bands relative to closely related reference species. Neighbor-joining, maximum likelihood, and parsimony trees were constructed using only >1,000-bp sequences with the Felsenstein correction method, after which DT-DGGE band sequences were added (∼550 bp) to each tree by parsimony. The consensus tree was constructed based on the merged topologies of the three trees. Dark shading, phylotypes showing increased relative intensity in anoxic (nitrate-amended) sulfidic incubation treatments; light shading; phylotypes showing increased relative intensity in oxic thiosulfate-amended incubation treatments; no shading (boxes), phylotypes showing decreased relative intensity in sulfide-rich anoxic incubation treatments.
RT-PCR DGGE profiles were subjected to image analysis in order to compare relative band intensities of identical ribotypes in the various incubation treatments. Phylotypes that displayed substantial variation in band intensity as a result of specific incubation treatments are summarized in Table 1. Values represent the percent intensity of the excised bands, relative to selected corresponding reference bands from the same lane (DB03C, FBR03A, DB04B, and FBR04Q, which showed relatively constant intensity in both PCR and RT-PCR DGGE analyses), and the percent change in RBI units relative to the original untreated biofilter samples (in parentheses). These phylotypes were divided into three main categories based on the DT-DGGE data: (i) phylotypes whose relative band intensities increased under anoxic sulfidic conditions, including those showing increased band intensity under high sulfidic (5,000 μM H2S) anoxic conditions, (ii) phylotypes whose relative band intensities increased under oxic, sulfidic (thiosulfate-amended) conditions, and (iii) phylotypes whose relative band intensities declined following exposure to high sulfide concentrations. In anoxic, sulfide-rich (5,000 μM) incubations, the number of detected RT-PCR DGGE ribotypes decreased by approximately 25% and the relative intensity of specific bands increased. This phenomenon was more distinct in the FBR than in the DB samples.
TABLE 1.
DT-DGGE image analysis of bands showing substantial shifts in relative band intensity as a result of different incubation conditions
| Band | Closest BLAST relative (% homology) | % Intensitya (% change in RBI unitsb) under the following incubation conditions:
|
||||
|---|---|---|---|---|---|---|
| 100 μM H2S + NO3− | 1,000 μM H2S + NO3− | 5,000 μM H2S + NO3− | 1,000 μM S2O3 + NO3− | 1,000 μM S2O3 + O2 | ||
| Increased relative intensity following anoxic sulfidic incubations | ||||||
| FBR04D | Roseobacter sp. strain LA7 (98) | 61 (57) | 186 (261) | NM | 90 (67) | 100 (0) |
| FBR04C | Loktanella salsilacus (94) | 164 (191) | 296 (272) | NM | 98 (80) | 27 (−15) |
| DB04U | Rhodobium orientis (93) | NM | 76 (116) | 72 (100) | 64 (105) | 0 (ND) |
| FBR03B | Rhodobacter litoralis (99) | 32 (33) | NM | 102 (244) | NM | NM |
| Paracoccus denitrificans (97) | ||||||
| DB04R | Thauera aromatica (95) | NM | 28 (19) | 72 (300) | 40 (0) | 0 (ND) |
| DB04I | Thauera aromatica (99) | NM | 63 (−60) | 125 (58) | 71 (−37) | 36 (−79) |
| DB03B | Thauera aromatica (98) | 0 (ND) | NM | 50 (1058) | NM | NM |
| FBR03D | Thauera aromatica (98) | 12 (75) | NM | 34 (1400) | NM | NM |
| FBR03C | Hydrogenophaga taeniospiralis (98) | 21 (−6) | NM | 105 (153) | NM | NM |
| DB03D | Dethiosulfovibrio acidaminovorans (98) | 5 (100) | NM | 37 (1433) | NM | NM |
| Increased relative intensity following oxic sulfidic incubations | ||||||
| DB04H | Endosymbiont of scaly snail (95) | NM | 0 (ND) | 0 (ND) | 0 (ND) | 112 (#) |
| FBR04P | Sulfur-oxidizing bacterium ODIII6 (94) | 92 (64) | 126 (111) | NM | 70 (86) | 198 (418) |
| DB04G | Thiomicrospira sp. strain JB-A1 (92) | NM | 0 (ND) | 0 (ND) | 0 (ND) | 113 (#) |
| FBR04G | Thiomicrospira sp. strain JB-A1 (92) | 0 (ND) | 0 (ND) | NM | 0 (ND) | 241 (#) |
| DB04S | Thiomicrospira crunogena (96) | NM | 0 (ND) | 0 (ND) | 2 (#) | 25 (#) |
| Decreased relative intensity following sulfidic incubations | ||||||
| FBR04B | Nitrosomonas sp. strain Is343 (99) | 220 (−46) | 0 (ND) | NM | 241 (−43) | 281 (−51) |
| FBR04F | Uncultured Nitrosomonas strain AZP2-9 (99) | 115 (130) | 0 (ND) | NM | 165 (1.5) | 211 (4.4) |
| DB04A | Deep-branching bacterium from aromatic contaminated groundwater (80) | NM | 63 (−8.9) | 0 (ND) | 54 (5.0) | 100 (13) |
| DB04L | Planctomyces sp. strain LA4-B37N (85) | NM | 84 (−4.0) | 37 (−73) | 64 (−7.9) | 78 (−34) |
Relative to reference bands. NM, not measured.
Relative to the original untreated biofilter samples. ND, not detected; #, band not detected in the original sample.
DISCUSSION
Sulfide-oxidizing bacteria play an essential role in the oxidative side of the sulfur cycle (37). Nonetheless, the highly polyphyletic nature of sulfide-oxidizing bacteria causes extreme difficulty when one is trying to identify them in natural environments. The objective of this study was to pinpoint specific bacterial phylotypes potentially responsible for oxygen- and nitrate-dependent sulfide oxidation in organic-rich mariculture biofilters under specific environmental conditions. DT-DGGE analyses were implemented to circumvent the limitations of the standard rRNA approach, which is rarely capable of correlating between phylogeny and physiology.
Evaluation of DT-DGGE.
Comparative analyses of 16S rRNA- and 16S rRNA gene-based DGGE/temperature gradient gel electrophoresis profiles have been employed previously to identify metabolically active microbial communities in environmental samples (13, 29, 31). However, these analyses displayed a snapshot of general activity of the total bacterial community and did not target specific metabolic processes, as is the case with the DT-DGGE method. DT-DGGE is an indirect approach to identify bacterial communities that may be metabolically active under specific environmental conditions. This information can assist in inferring specific physiological properties of bacterial phylotypes in complex natural environments. Despite the considerable potential of DT-DGGE, it should be understood that rRNA synthesis is not always coherent to metabolic activity (3, 14, 48) and that levels of rRNA transcription may differ between different bacterial species (35). Therefore, by using this method, assessment of the relative activity under different environmental conditions may be distorted for certain populations (e.g., ammonia-oxidizing bacteria). Moreover, it is possible that specific incubation treatments do not significantly alter preincubation conditions for selected functional groups of bacteria. In our study, for example, certain heterotrophic bacteria may continue to utilize endogenous organic carbon, even in the absence of an externally added organic carbon source. Despite these drawbacks, DT-DGGE enables targeting of specific sulfide-oxidizing bacterial phylotypes that are potentially active under different environmental conditions and are not detectable by other methods due to the highly polyphyletic nature of bacterial sulfide oxidation. In order to verify the involvement of specific bacterial phylotypes in oxic and nitrate-dependent sulfide oxidation, further DT-DGGE analyses using additional incubation conditions, as well as analysis of samples from other natural and engineered environments, are required.
Bacterial phylotypes showing substantial differences in band intensities as a result of the different treatments were divided into three major groups (Table 1). The first contains phylotypes from both the DB and the FBR that displayed enhanced relative band intensities following anoxic sulfidic incubations. Four of these were affiliated with the family Rhodobacteraceae (Fig. 2). Several phylotpes, Rhodobacteraceae, including some highly similar (>98%) to those detected in this study, have been detected previously in the DB and FBR, where they were found to constitute a substantial fraction of the bacterial consortium (12). The Rhodobacteraceae are characterized by a highly diverse metabolism, and several species are capable of oxidizing reduced sulfur species under both oxic and anoxic conditions (15, 23, 37, 38, 40, 45, 46). All of the Rhodobacteraceae-like phylotypes in the system showed enhanced band intensity in all anoxic nitrate- and sulfide (or thiosulfate)-amended incubations. This suggests that the phylotypes of Rhodobacteraceae are involved in nitrate-mediated sulfide oxidation in the DB and FBR.
Phylotypes highly similar to Thauera (FBR03D, DB03B, DB04I, and DB04R), Hydrogenophaga (DB03C), and Dethiosulfovibrio (DB03D) species showed enhanced band intensity in the sulfide-rich (5,000 μM H2S) anoxic incubations. However, in these cases the correlation to nitrate-dependent sulfide oxidation cannot be unambiguously established. The enhanced relative band intensities may indicate greater tolerance to high sulfide concentrations than other phylotypes. Thauera aromatica-related phylotypes (>96% sequence similarity) were ubiquitous in all of the 5,000 μM H2S incubations from both the DB and the FBR, and highly similar strains (>98%) have been identified previously in both of these biofilters (12; E. Cytryn, unpublished data). Thauera spp. are generally denitrifying bacteria specifically recognized for their capability for anaerobic degradation of aromatic organic compounds (2, 36). The ubiquitous enhancement of Thauera-related phylotypes in the anoxic sulfide-rich incubations may suggest that they are also capable of anoxic sulfide oxidation. However, sulfide oxidation has not been associated with Thauera species before, and additional studies are required to confirm these findings. Current research is focusing on isolation of these Thauera strains in order to assess their capacity for anoxic sulfide oxidation.
Hydrogenophaga spp. are facultative heterotrophs frequently associated with wastewater treatment (5, 20, 22). Certain Hydrogenophaga species are capable of thiosulfate oxidation (18); thus, the Hydrogenophaga-related phylotype detected in this study and a previous study (12) may participate in sulfide oxidation in the FBR.
Dethiosulfovibrio spp. are obligately anaerobic fermentative bacteria known to utilize proteins, peptides, amino acids, and some organic acids. Concomitantly with organic matter degradation, Dethiosulfovibrio species are known to be capable of reducing sulfur and thiosulfate to hydrogen sulfide, which enables them to increase cell yields and growth rates (42, 43). Dethiosulfovibrio species have been detected previously in sulfide-rich, low-redox zones of the DB, where they may degrade protein-rich particulate organic matter such as uneaten food and feces (11). Based on previous studies, these organisms do not appear to be capable of chemolithotrophic sulfide oxidation (42, 43).
Incubation in aerated, thiosulfate-amended medium resulted in substantial enhancement of two prominent DT-DGGE phylotype groups in both the DB and the FBR samples. The first (phylotypes DB04G, FBR04G, and DB04S) clustered within the Thiomicrospira genus of the Gammaproteobacteria. This group of Thiomicrospira spp. consists primarily of aerobic chemolithotrophic sulfur oxidizers (7, 8, 39), supporting our findings that they participate in oxic sulfide oxidation in the FBR and DB. The second phylotype group (DB04H and FBR04P) was affiliated with a cluster of Gammaproteobacteria containing various symbiotic and free-living oxic, sulfide-oxidizing bacteria (17, 39). As in the case of the Thiomicrospira-like species, this phylogenetic information supports the notion that these phylotypes are involved in oxygen-dependent sulfide oxidation in our system.
The intensities of several phylotypes decreased substantially in response to specific incubation conditions (Table 1). Phylotypes DB04A and FBR04F completely disappeared from both PCR and RT-PCR analyses following incubation in media containing elevated sulfide concentrations (5,000 and 1,000 μM for the DB and FBR samples, respectively). The disappearance of these strains in the PCR-amplified DNA samples suggests that the high sulfide concentrations affected the vitality of these phylotypes.
Conclusion.
Bacterial communities responsible for sulfide oxidation are important in natural sulfide-rich marine environments as well as in recirculating mariculture systems, because they safeguard marine organisms from sulfide toxicity. Consequently, greater phylogenetic and physiological understanding of these communities is imperative. Based on the DT-DGGE analyses presented here, we propose that specific members of the family Rhodobacteraceae participate in nitrate-dependent sulfide oxidation and that the gammaproteobacterium Thiomicrospira and phylotypes related to sulfide-oxidizing symbiotic bacteria participate in oxygen-dependent sulfide oxidation in the biofilters of our mariculture system. The physiology of their closest relatives makes the DT-DGGE-inferred activity of these detected phylotypes highly probable. In other cases, when characterized relatives are too distantly related for conclusions to be drawn (e.g., for phylotypes FBR04C and DB04A), or when the phylotypes detected are not compatible with the established physiology (e.g., for the Thauera-, Hydrogenophaga-, and Dethiosulfovibrio-like phylotypes), DT-DGGE can at least provide a good starting point for further experiments to confirm or reject the proposed metabolic functions of the bacterial populations identified. We anticipate that DT-DGGE will also be useful in identifying metabolically active sulfide-oxidizing bacteria in other natural environments, such as marine sediments (39, 45), sulfidic oil wells (47), soils (18), and wastewater treatment facilities (21). Furthermore, DT-DGGE can be employed to trace other functionally distinct yet phylogenetically diverse microbial populations in nature.
Acknowledgments
This research was supported by a grant (research project 1-723-165.8/01) from the German Israeli Foundation for Scientific Research (GIF).
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, Y. J., Y. H. Joo, I. Y. Hong, H. W. Ryu, and K. S. Cho. 2004. Microbial characterization of toluene-degrading denitrifying consortia obtained from terrestrial and marine ecosystems. Appl. Microbiol. Biotechnol. 65:611-619. [DOI] [PubMed] [Google Scholar]
- 3.Araki, N., T. Yamaguchi, S. Yamazaki, and H. Harada. 2004. Quantification of amoA gene abundance and their amoA mRNA levels in activated sludge by real-time PCR. Water Sci. Technol. 50:1-8. [PubMed] [Google Scholar]
- 4.Binder, B. J., and Y. C. Liu. 1998. Growth rate regulation of rRNA content of a marine synechococcus (Cyanobacterium) strain. Appl. Environ. Microbiol. 64:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl. Environ. Microbiol. 66:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bower, C. E., and T. Holm-Hansen. 1980. A salicylate-hypochlorate method for determining ammonia in seawater. Can. J. Fish Aquat. Sci. 37:794-798. [Google Scholar]
- 7.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkhoff, T., G. Muyzer, C. O. Wirsen, and J. Kuever. 1999. Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol. 49:385-392. [DOI] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 10.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cytryn, E., I. Gelfand, Y. Barak, J. van Rijn, and D. Minz. 2003. Diversity of microbial communities correlated to physiochemical parameters in a digestion basin of a zero-discharge mariculture system. Environ. Microbiol. 5:55-63. [DOI] [PubMed] [Google Scholar]
- 12.Cytryn, E., D. Minz, I. Gelfand, A. Neori, A. Gieseke, D. de Beer, and J. van Rijn. 2005. Sulfide-oxidizing activity and bacterial community structure in a fluidized bed reactor from a zero-discharge mariculture system. Environ. Sci. Technol. 39:1802-1810. [DOI] [PubMed] [Google Scholar]
- 13.Felske, A., A. D. Akkermans, and W. M. de Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flardh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 16.Gelfand, I., Y. Barak, Z. Even-Chen, E. Cytryn, J. van Rijn, M. D. Krom, and A. Neori. 2003. A novel zero discharge intensive seawater recirculating system for the culture of marine fish. J. World Aquacult. Soc. 34:344-358. [Google Scholar]
- 17.Goffredi, S. K., A. Waren, V. J. Orphan, C. L. Van Dover, and R. C. Vrijenhoek. 2004. Novel forms of structural integration between microbes and a hydrothermal vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 70:3082-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff, A., and S. Stubner. 2003. Isolation and molecular characterization of thiosulfate-oxidizing bacteria from an Italian rice field soil. Syst. Appl. Microbiol. 26:445-452. [DOI] [PubMed] [Google Scholar]
- 19.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 2000. Use of combined microautoradiography and fluorescence in situ hybridization to determine carbon metabolism in mixed natural communities of uncultured bacteria from the genus Achromatium. Appl. Environ. Microbiol. 66:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gumaelius, L., G. Magnusson, B. Pettersson, and G. Dalhammar. 2001. Comamonas denitrificans sp. nov., an efficient denitrifying bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 51:999-1006. [DOI] [PubMed] [Google Scholar]
- 21.Ito, T., K. Sugita, and S. Okabe. 2004. Isolation, characterization, and in situ detection of a novel chemolithoautotrophic sulfur-oxidizing bacterium in wastewater biofilms growing under microaerophilic conditions. Appl. Environ. Microbiol. 70:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampfer, P., R. Schulze, U. Jackel, K. A. Malik, R. Amann, and S. Spring. 2005. Hydrogenophaga defluvii sp. nov. and Hydrogenophaga atypica sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 55:341-344. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, D. P., J. K. Shergill, W. P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 24.Kjeldgaard, N. O., O. Maaløe, and M. Schaechter. 1958. The transition between different physiological states during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:607-616. [DOI] [PubMed] [Google Scholar]
- 25.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matin, A. 1978. Organic nutrition of chemolithotrophic bacteria. Annu. Rev. Microbiol. 32:433-468. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, C. A., A. Hudson, A. Konopka, and C. H. Nakatsu. 2002. Analyses of microbial activity in biomass-recycle reactors using denaturing gradient gel electrophoresis of 16S rDNA and 16S rRNA PCR products. Can. J. Microbiol. 48:333-341. [DOI] [PubMed] [Google Scholar]
- 30.Muyzer, G., T. Brinkhoff, U. Nubel, C. M. Santegoeds, H. Schafer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oved, T., A. Shaviv, T. Goldrath, R. T. Mandelbaum, and D. Minz. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38:749-770. [DOI] [PubMed] [Google Scholar]
- 35.Pernthaler, A., J. Pernthaler, H. Eilers, and R. Amann. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness? Appl. Environ. Microbiol. 67:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, L. A., and J. G. Kuenen. 1992. The colorless sulfur bacteria, p. 385-411. In A. Balows, H. G. Truper, M. H. W. Dworkin, and K. H. Schleifer (ed.), The Prokaryotes. Springer Press, New York, N.Y.
- 38.Rother, D., H. J. Henrich, A. Quentmeier, F. Bardischewsky, and C. G. Friedrich. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 183:4499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievert, S. M., T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorokin, D. Y., T. P. Tourova, A. N. Antipov, G. Muyzer, and J. G. Kuenen. 2004. Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thioalkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology 150:2435-2442. [DOI] [PubMed] [Google Scholar]
- 41.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 42.Surkov, A. V., M. E. Bottcher, and J. Kuever. 2000. Stable sulfur isotope fractionation during the reduction of thiosulfate by Dethiosulfovibrio russensis. Arch. Microbiol. 174:448-451. [DOI] [PubMed] [Google Scholar]
- 43.Surkov, A. V., G. A. Dubinina, A. M. Lysenko, F. O. Glockner, and J. Kuever. 2001. Dethiosulfovibrio russensis sp. nov., Dethiosulfovibrio marinus sp. nov. and Dethiosulfovibrio acidaminovorans sp. nov., novel anaerobic, thiosulfate- and sulfur-reducing bacteria isolated from ‘Thiodendron’ sulfur mats in different saline environments. Int. J. Syst. Evol. Microbiol. 51:327-337. [DOI] [PubMed] [Google Scholar]
- 44.Takai, K., H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Sulfurihydrogenibium subterraneum gen. nov., sp. nov., from a subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 53:823-827. [DOI] [PubMed] [Google Scholar]
- 45.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timer-ten Hoor, A. 1981. Cell yield and bioenergetics of Thiomicrospira denitrificans compared with Thiobacillus denitrificans. Antonie Leeuwenhoek 47:231-243. [DOI] [PubMed] [Google Scholar]
- 47.Voordouw, G., S. M. Armstrong, M. F. Reimer, B. Fouts, A. J. Telang, Y. Shen, and D. Gevertz. 1996. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl. Environ. Microbiol. 62:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K. H. Schleifer. 1995. In-situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 49.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]