Abstract
Protein purification of recombinant proteins constitutes a significant cost of biomanufacturing and various efforts have been directed at developing more efficient purification methods. We describe a protein purification scheme wherein Ralstonia eutropha is used to produce its own “affinity matrix,” thereby eliminating the need for external chromatographic purification steps. This approach is based on the specific interaction of phasin proteins with granules of the intracellular polymer polyhydroxybutyrate (PHB). By creating in-frame fusions of phasins and green fluorescent protein (GFP) as a model protein, we demonstrated that GFP can be efficiently sequestered to the surface of PHB granules. In a second step, we generated a phasin-intein-GFP fusion, wherein the self-cleaving intein can be activated by the addition of thiols. This construct allowed for the controlled binding and release of essentially pure GFP in a single separation step. Finally, pure, active β-galactosidase was obtained in a single step using the above described method.
We have previously reported the development of a novel high cell density protein expression platform based on the gram-negative bacterium Ralstonia eutropha (22, 23). This system has been developed to overcome some of the shortcomings associated with recombinant protein expression in Escherichia coli (e.g., poor fermentation performance at high cell density, and inclusion body formation). Expression of organophosphohydrolase, an enzyme originally isolated from Pseudomonas diminuta (20) and prone to inclusion body formation in Escherichia coli (4, 28, 29), was demonstrated at high levels. Titers of active, soluble organophosphohydrolyase, in excess of 10 g/liter were obtained in high cell density fermentation (3), representing at least a 100-fold increase over those previously reported in E. coli.
While the successful expression of a recombinant protein is a necessary requirement, recovery and purification still remain a significant cost in recombinant protein production. We thus sought to integrate the existing R. eutropha protein expression platform with a protein purification strategy to simplify the expression and purification of recombinant proteins. This specific approach uses the natural ability of R. eutropha to produce a polymer known as polyhydroxybutyrate (PHB), which accumulates as insoluble granules within the cell. PHB is a member of the polyhydroxyalkanoate class of polymers, synthesized by many bacteria, as carbon storage compounds (2, 15, 16, 26, 30, 31, 32). Polyhydroxyalkanoates have received attention as biodegradable polymers and can be obtained by fermentation processes utilizing cheap, abundant renewable carbon sources (2, 24). Polyhydroxyalkanoates have been produced industrially by ZENECA Bioproducts (26) and Monsanto (10).
PHB synthesis in R. eutropha has been the model system for studying polyhydroxyalkanoate biosynthesis in bacteria (10, 15, 16, 18, 26). The biogenesis of polyhydroxyalkanoate granules involves two distinct proteins, the polyhydroxyalkanoate synthase (PhaC) and phasins (PhaP). Phasins are low-molecular-weight proteins whose role in polyhydroxyalkanoate formation is not well understood (10, 11, 15, 16, 18, 24, 26, 30, 31, 32). Phasins accumulate during PHB synthesis, bind to PHB granules, and promote further PHB synthesis (32). It has been shown that phaP mutants form only one large PHB granule and that up-regulating the phaP gene increases the number of PHB granules while reducing their size (15, 26). Phasins accumulate at high levels in cells that are synthesizing PHB, and as much as 5% of total cellular protein can be PhaP (16). Phasins have high affinity for PHB granules and are the predominant protein present on the granule surface (24, 26).
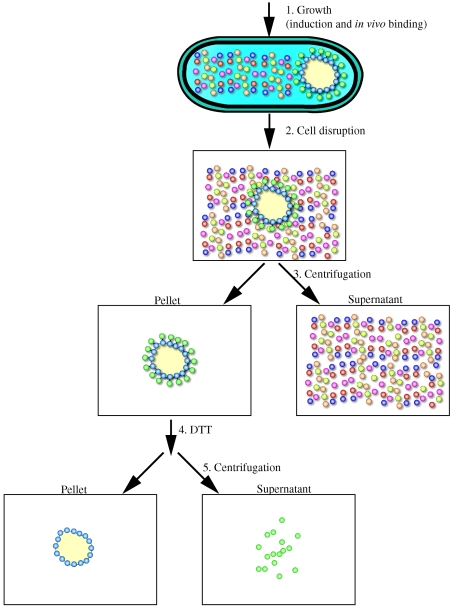
In this study, we exploit the specific affinity between PhaP and the PHB granules for the purpose of purifying a recombinant model protein (GFP). In essence, this yields an affinity-based purification scheme wherein the cell synthesizes its own chromatography matrix and PhaP is used as the affinity tag to sequester a protein of interest to the PHB granule surface. The recombinant protein can then be recovered by cell disruption followed by a centrifugation step that separates the insoluble high-density polymer from other soluble cellular components. In an improved version, the protein of interest is linked to PhaP through an intein allowing its release by thiol induced cleavage (see Fig. 3).
FIG.3.
Flowchart of a recombinant protein purification scheme based on PhaP mediated PHB granule sequestration. Ralstonia eutropha recombinants expressing phasin-intein-GFP (PIG) are cultivated in a shake flask or a bioreactor. Cells are harvested by centrifugation, washed and resuspended in buffer B1 prior to cell disruption. The cell lysate is centrifuged, the supernatant fraction discarded and the insoluble fraction retained. After washing the insoluble pellet, the fraction is resuspended in buffer B2. The intein is thiol activated and GFP is released from the whole cell debris into the supernatant. The pure protein is recovered by centrifugation, discarding the pellet and retaining the supernatant fraction.
Inteins are self-splicing proteins that occur as in-frame insertions in specific host proteins throughout nature, and have been adapted for use in recombinant protein expression and purification schemes (9, 27). Intein cleavage can be mediated by pH changes or the addition of thiols. The Mxe GyrA intein is a 198-amino-acid polypeptide, with N-terminal cleavage activity in the presence of thiols (25). By incorporating this intein into a PhaP-intein-GFP fusion we were able to show (i) the expression of a PhaP-intein-GFP fusion protein, (ii) its sequestration to PHB granules, and (iii) the subsequent release of GFP from the PHB granule by treating the cell debris with dithiothreitol.
MATERIALS AND METHODS
Strains, plasmids and oligonucleotides.
Strains and plasmids that were used in this study are listed in Table 1. Oligonucleotides used in this study are listed in Table 2. Standard procedures were used for the preparation and manipulation of DNA and for PCR. All PCR products were subcloned into pCR2.1-TOPO (Invitrogen), pCR4Blunt-TOPO (Invitrogen) or pCRII-Blunt-TOPO (Invitrogen) and sequence verified at the Molecular Biology Core Facility at Dartmouth College. Methods for introducing plasmids into the R. eutropha chromosome have previously been described (22, 23). In brief, all pKNOCK-Cm derived plasmids are introduced into E. coli S17 (17, 21) before being incorporated into the R. eutropha chromosome by simple biparental mating.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| R. eutropha strains | ||
| NCIMB 40124 | Wild type; gentamicin resistant | National Collections of Industrial, Food & Marine Bacteria, Aberdeen, Scotland |
| G | pG introduced into R. eutropha wt | This study |
| PG | pPG introduced into R. eutropha wt | This study |
| PIG | pPIG introduced into R. eutropha wt | This study |
| GP | pGP introduced into R. eutropha wt | This study |
| GIP | pGIP introduced into R. eutropha wt | This study |
| PIL | pPIL introduced into R. eutropha wt | This study |
| E. coli strains | ||
| TOP10 | Host strain for plasmids derived from pCR2.1-TOPO | Invitrogen |
| S-17 | Host strain for plasmids derived from pKNOCK-Cm | 8, 17, 21 |
| Plasmids | ||
| pG | phaPp::gfp transcriptional fusion introduced into pKNOCK-Cm; plasmid used to create strain R. eutropha G | This study |
| pPG | phaP ORF::gfp translational fusion introduced into pKNOCK-Cm; plasmid used to create strain R. eutropha PG | This study |
| pPIG | phaP ORF::Mxe GyrA intein::gfp translational fusion in pKNOCK-Cm; plasmid used to create strain R. eutropha PIG | This study |
| pGP | phaPp::gfp::phaP transcriptional fusion in pKNOCK-Cm; plasmid used to create strain R. eutropha GP | This study |
| pGIP | phaPp::gfp::Mxe GyrA intein::phaP transcriptional fusion in pKNOCK-Cm; plasmid used to create strain R. eutropha GIP | This study |
| pPIL | phaP ORF::Mxe GyrA intein::lacZ translational fusion in pKNOCK-Cm; plasmid used to create strain R. eutropha PIL | This study |
| pKNOCK-Cm | Suicide vector used for introducing plasmids into R. eutropha chromosome | 1 |
| pGY1a+ | Vector containing phaPp::gfpmut2 translational fusion | 30 |
| pTWIN1 | Commercially available expression vector containing Ssp DnaB intein and Mxe GyrA intein | New England Biolabs |
| pUCPPCm | Plasmid vector containing phaP promoter | 22 |
| pGB27 | pKNOCK containing the phaP promoter | This study |
| pGB73 | Mxe GyrA intein introduced into pCR2.1-TOPO | This study |
| pGB76 | phaP ORF introduced into pCR2.1-TOPO | This study |
| pGB80 | phaPp::gfp::phaP transcriptional fusion in pKNOCK-Cm | This study |
| pGB82 | phaPp::gfp::NEB Mxe GyrA intein::phaP transcriptional fusion in pKNOCK-Cm | This study |
| pGB85 | phaP::gfp transcriptional fusion in pCR4-TOPO | This study |
| pGB91 | phaP::gfp transcriptional fusion in pKNOCK-Cm | This study |
| pGB93 | phaP::NEB Mxe GyrA intein::gfp transcriptional fusion in pKNOCK-Cm | This study |
| pGB96 | Peptide linker::phaP transcriptional fusion in pCR2.1-TOPO | This study |
| pGB97 | phaP::peptide linker transcriptional fusion in pCR2.1-TOPO | This study |
| pGB470 | lacZ ORF fragment cloned introduced into pCR4Blunt-TOPO | This study |
wt, wild type.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Location and orientationb |
|---|---|---|
| oGB41 | GAAGAAGAGCTCATGAGTAAAGGAGAAGAACTTTTC | 5′ end of gfpmut2 ORF (+) |
| oGB42 | AAGAACTCGAGGAGTCGATTATTTGTATAGTTCATCCATGCCATG | 3′ end of gfpmut2 ORF (−) |
| oGB176 | GGCGCGCCTATGTGCATCACGGGAGATGCACTAGTT | 5′ end of Mxe GyrA ORF (+) |
| oGB177 | TCTAGAAGCGTGGCTGACGAACCCGTTCGT | 3′ end of Mxe GyrA ORF (−) |
| oGB183 | GATATCGGCAGCATCGGCGCGCCTGCTTCTAGAGGCAGCATCATGATCCTCACCCCGGAACAA | 5′ end of phaP ORF (+) |
| oGB44 | GAAGAAGGTACCTCAGGCAGCCGTCGTCTTCTTTGCCGT | 3′ end of phaP ORF (−) |
| oGB185 | GCGGCCGCGAGCTCATGATCCTCACCCCGGAACAAGTT | 5′ end of gfp ORF (+) |
| oGB186 | GGCAGCCGTCGTCTTCTTTGCCGT | 3′ end of gfp ORF (−) |
| oGB187 | GCAAAGAAGACGACGGCTGCCGGCAGCATCGGCGCGCCTGCTTCTAGAGGCAGCATCATGAGTAAAGGAGAAGAACTT | 5′ end of phaP ORF (+) including overlap region of gfp ORF |
| oGB40 | GAAGAACTCGAGTTATTTGTATAGTTCATCCATGCCATG | 3′ end of phaP ORF (−) |
| oGB208 | GGCGCGCCGGATCCGCTGCCGCCCCCGCCGCTGCCGCCCCCGCCGCTGCCGCCCCCGCCGGCAGCCGTCGTCTTCTTTGC | 3′ end of phaP ORF (−) containing peptide linker |
| oGB209 | TCTAGAGGATCCGGCGGGGGCGGCAGCGGCGGGGGCGGCAGCGGCGGGGGCGGCAGCATGATCCTCACCCCGGAACAAGT | 5′ end of phaP ORF (+) containing peptide linker |
| oGB216 | GAAGAATCTAGAGGCAGCATCATGACCATGATTACGGATTCACTG | 5′ end of lacZ ORF (+) containing peptide linker |
| oGB217 | GAAGAACTCGAGTTATTTTTGACACCAGACCAACTGGTA | 3′ end of lacZ ORF (−) |
Restriction sites engineered into the sequences are underlined.
Forward (+) or reverse (−) orientation relative to ORF is indicated.
Growth media, antibiotics and cultivation conditions.
E. coli strains were grown in Luria-Bertani (LB) medium (13). R. eutropha strains were grown in one of the following media depending on the application: LB medium or Lee medium (20 g/liter glucose, 3 g/liter Na2HPO4 · 7H2O, 1 g/liter KH2PO4, 2 g/liter NH4Cl, 0.2 g/liter MgSO4 · 7H2O, 1 ml/liter Corn Steep Liquor [Sigma], 2.4 ml/liter trace element solution [22]). Antibiotics were added to the growth medium to the following concentrations depending on the application: chloramphenicol (50 μg/ml), kanamycin (50 μg/ml), and gentamicin (10 μg/ml). R.eutropha and E. coli strains were cultivated at 30°C and 37°C, respectively.
Fluorescence microscopy.
To prepare cells for fluorescence microscopy, cells were transferred from LB agar plates into 200 μl of buffer (phosphate-buffered saline) and resuspended thoroughly; 10 μl of this cell suspension were transferred to a single well in a 15-well slide pretreated with 1% poly-l-lysine. Microscopy was carried out using a Leica epifluorescence light microscope. An ORCA-ER charge-coupled device camera (Hamamatsu) and OPENLAB software (Improvision) were used for all image acquisition and processing.
Sucrose gradient fractionation.
Strains were cultivated in 50 ml of Lee medium to an approximate optical density at 600 nm of 10. The cultures were centrifuged and the cells resuspended in 2 ml of buffer B1 (20 mM Tris, 500 mM NaCl, 1 mM EDTA, pH 8.5). Cells were sonicated in a Fisher Scientific Sonic Dismembrator 550 in ten pulsed cycles (2 seconds ON, 0.5 second off, 30 second duration, 5 min cooling on ice between cycles); 1 ml of the lysate was loaded onto a sucrose density gradient. The sucrose density gradient consists of nine layered 1 ml fractions of buffer B1 containing 0 to 2 M sucrose (0.25 M increments). The 10-ml solutions were spun at 1,500 × g for 3 h. Ten 1 ml fractions were collected with a syringe and needle.
Fluorometry.
Fluorescence was measured using the SpectraMax Gemini spectrophotometer (Molecular Devices). Excitation and emission wavelengths of 360 nm and 509 nm, respectively, were used.
PHB analysis.
The concentration of PHB was quantified by the sulfuric acid-HPLC method of Karr et al. (12) with modifications (30).
Intein mediated cleavage.
A published intein mediated cleavage protocol (14) has been adapted for this study; 300 μl of the lysate generated from the sonication was centrifuged, the supernatant discarded and the insoluble pellet retained. The pellet was washed three times by resuspension in 1 ml of buffer B1 followed by centrifugation. The pellet was then resuspended in 500 μl of buffer B2 (buffer B1 containing 40 mM dithiothreitol). The pellet was incubated overnight at 37°C. After incubation, the solution was centrifuged and the supernatant and pellet retained. The pellet was again washed as described above and resuspended in the original volume (500 μl). Samples were subjected to fluorometry and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a 12% Tris-HCl polyacrylamide gel (Bio-Rad), stained with SimplyBlue SafeStain (Invitrogen).
β-Galactosidase assay.
We added 70 μl of an aqueous 4 mg/ml ortho-nitrophenyl-β-d-galactopyranoside (ONPG) solution to 200 μl of 1X cleavage buffer (0.6 M Na2HPO4·7H2O, 0.4 M NaH2PO4·H2O, 0.1 M KCl, 0.01 M MgSO4·7H2O, 2.7 ml/liter β-mercaptoethanol) and incubated in a 30°C water bath for 10 min; 5 or 10 μl of sample (diluted to an appropriate concentration in Buffer B1) was added to the cleavage buffer containing ONPG and incubated at 30°C for 30 min. The reaction was quenched by adding 500 μl Stop Buffer (1 M Na2CO3). The amount of ONPG hydrolyzed was determined by measuring optical density at 420 nm and using the molar extinction coefficient of ONPG (4,500 M−1 cm−1). Total protein concentrations were measured using a Bradford protein assay kit (Bio-Rad, Hercules, CA). 1 unit of galactosidase activity was defined as the cleavage of 1 nmol of ONPG to o-nitrophenol and galactose in 1 min at 30°C.
Construction of plasmid pG (phaPp::gfp transcriptional fusion in pKNOCK-Cm).
The phaP promoter from pUCPPCm was inserted into pKNOCK-Cm as a EcoRI/EcoRV fragment yielding pGB27. The gfpmut2 open reading frame (ORF) was PCR amplified from plasmid pGY1a+ using oligonucleotides oGB41 and oGB42 and cloned into pCR2.1-TOPO (Invitrogen). The gfp ORF was cloned from the resultant TOPO derivative into pGB27 as a SacI/XhoI fragment yielding pG.
Subcloning an intein (pGB73).
The Mxe GyrA intein was PCR amplified from plasmid pTWIN1 (New England Biolabs) with oGB176 and oGB177 and cloned into pCR2.1-TOPO to yield pGB73.
Construction of plasmids pGB80 and pGB82 (phaPp::gfp::phaP and phaPp::gfp::intein::phaP transcriptional fusions in pKNOCK-Cm).
The phaP ORF was PCR amplified from R. eutropha genomic DNA with oligonucleotides oGB183 and oGB44 and subcloned into pCR2.1-TOPO yielding pGB76. Plasmid pGB76 was digested with EcoRV and KpnI and cloned into pG digested with MlyI and KpnI to generate plasmid pGB80. The intein from plasmid pGB73 was cloned into plasmid pGB80 as an AscI/XbaI fragment, yielding vector pGB82.
Construction of plasmids pGB91 and pGB93 (phaP::gfp and phaP::intein gfp translational fusions in pKNOCK-Cm).
A translational fusion of the phaP ORF and the gfp ORF was constructed by overlap PCR. The phaP ORF was PCR amplified from R. eutropha genomic DNA using oligonucleotides oGB185 and oGB186. The gfp ORF was PCR amplified from pGY1a+ using oligonucleotides oGB187 and oGB40. The resultant PCR products were purified and used as templates in a subsequent PCR. The resultant 1.2-kb PCR fragment was cloned into pCR4Blunt-TOPO yielding pGB85. The 1.2-kb phaP::gfp translational fusion was cloned into pKNOCK-Cm as a NotI/XhoI fragment yielding pGB91. Plasmid pGB93 was created by introducing the intein from pGB73 into pGB91 as an AscI/XbaI fragment.
Construction of plasmids pGP and pGIP (phaPp::gfp::peptide linker::phaP and phaPp::gfp::intein::peptide linker::phaP transcriptional fusions in pKNOCK-Cm).
The phaP ORF was PCR amplified from genomic DNA using oligonucleotides oGB209 and oGB44. The resultant 0.6 kb fragment was cloned into pCR2.1-TOPO to yield pGB96. The phaP ORF in plasmids pGB80 and pGB82 was excised as an XbaI/KpnI fragment and replaced with the 0.6-kb XbaI/KpnI fragment of pGB96 to yield plasmids pGP and pGIP, respectively.
Construction of plasmids pPG and pPIG (phaP::peptide linker::gfp and phaP::peptide linker::intein::gfp translational fusions in pKNOCK-Cm).
The phaP ORF was PCR amplified from genomic DNA using oligonucleotides oGB185 and oGB208. The resultant 0.6 kb fragment was cloned into pCR2.1-TOPO to yield pGB97. The phaP ORF in plasmids pGB91 and pGB93 was excised as a SacI/AscI fragment and replaced with the 0.6-kb SacI/AscI fragment of pGB97 to yield plasmids pPG and pPIG, respectively.
Construction of plasmid pPIL (phaP::peptide linker::intein::lacZ translational fusion in pKNOCK-Cm).
The lacZ ORF was PCR amplified from E. coli S17 genomic DNA using oligonucleotides oGB216 and oGB217. The 3.0-kb fragment was subcloned into pCRII-Blunt-TOPO to generate pGB470. The lacZ ORF from pGB470 was cloned into pPIG as an XbaI/XhoI fragment, yielding plasmid pPIL.
R. eutropha strain generation.
Ralstonia eutropha recombinant strains were generated according to methods previously described (3, 22, 23). R. eutropha G, was generated using plasmid pG, which carries a transcriptional fusion between the phaP promoter and the gfpmut2 ORF (6) (phaPp::gfp). Plasmid pG is a suicide plasmid and is integrated at the phaP promoter locus of the R. eutropha chromosome. Since integration occurs within the promoter region, the wild-type phaP gene remains intact. R. eutropha PG and R. eutropha PIG were generated using plasmids pPG and pPIG, respectively. Plasmid pPG contains an in-frame translational fusion between the phaP ORF and gfpmut2 ORF (phaP::gfp). Plasmid pPIG is isogenic to pPG, with the exception of the in-frame insertion of the Mxe GyrA intein between the two open reading frames (phaP::Mxe GyrA intein::gfp). Plasmids pPG and pPIG do not contain the phaP promoter and the phaP ORF serves as the homologous recombination locus. Therefore in R.eutropha PG and R. eutropha PIG, the wild-type phaP gene has been replaced by a translational fusion encoding phaP::gfp and phaP::intein::gfp, respectively.
RESULTS
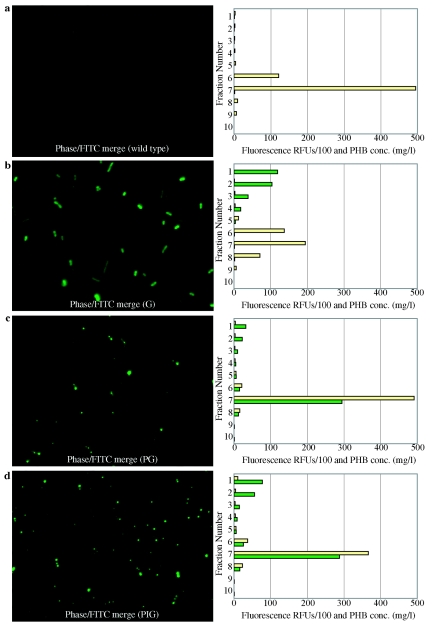
Both fluorescence microscopy and sucrose density gradient fractionation of cell lysates were used to examine localization of GFP in R. eutropha strains. The fluorescence microscopy images in Fig. 1 show that the wild type exhibited no autofluorescence and that GFP is evenly distributed throughout the cell in R.eutropha G. However, Fig. 1 shows fluorescent foci throughout the cells in R. eutropha PG and R. eutropha PIG, presumably where GFP is localized on the surface of PHB granules.
FIG. 1.
Representative GFP localization. Fluorescence microscopy and sucrose density gradient fractionation of cell lysates. a) Wild-type R.eutropha. b) R. eutropha G (expressing GFP). c) R. eutropha PG (expressing PhaP-GFP). d) R. eutropha PIG (expressing PhaP-intein-GFP). Cell lysates were generated by sonication and added to the top of a sucrose density gradient. Fluorescence of individual sucrose density gradient fractions is expressed in relative fluorescence units divided by 100 and shown in green. Corresponding PHB concentrations are expressed in mg/liter and shown in tan.
Sucrose density gradient fractionation of cell lysates was performed to further examine GFP localization (see Materials and Methods). R. eutropha strains were cultivated in Lee medium, a phosphate limited growth medium that induces both PHB formation and transcription of genes under the control of the phaP promoter. Cells were recovered, washed, resuspended in buffer B1, and sonicated. Cell lysates were loaded onto a sucrose gradient (density from 1.02 g/ml to 1.29 g/ml) and equilibrated by centrifugation. PHB granules have a density of approximately 1.20 g/ml (19) and accumulate near the bottom of the sucrose density gradient. In contrast, soluble proteins accumulate in the low density fractions at the top of the sucrose density gradient. A fluorescence spectrophotometer was used to measure the fluorescence of each individual fraction of the sucrose gradient. R. eutropha G showed fluorescence predominantly in the top fractions, consistent with fluorescence micrographs that suggest that GFP is present as a soluble protein in the cytoplasm and not localized to PHB granules.
R. eutropha PG and R. eutropha PIG showed a strong fluorescent signal in fraction 7, which coincides with the fraction containing PHB. These results strongly suggest that in R. eutropha PG and R. eutropha PIG, the GFP is localized to the PHB granules.
A fluorescent signal also appeared in the upper fractions of the R. eutropha PG and R. eutropha PIG density gradients. We propose several possible explanations for the presence of soluble GFP in these strains: (i) excess PhaP-GFP and PhaP-intein-GFP cannot bind because the PHB granule binding capacity has been exhausted, (ii) affinity is reduced due to the C-terminal fusion of GFP to the PhaP protein, or (iii) the minor fluorescence could represent a small amount of desorption that occurs during sonication and sucrose density gradient fractionation.
From the fluorescence microscopy and sucrose density fractionation, we concluded that PhaP-GFP and PhaP-intein-GFP fusions are localized in vivo to PHB granules. A cleavage experiment was designed to demonstrate the release of pure GFP from whole cell debris (see Materials and Methods). Briefly, R. eutropha strains were cultivated in Lee medium, harvested, resuspended in buffer B1 and sonicated. The lysate was centrifuged and the supernatant fraction, containing the soluble protein fraction, was discarded. The pellet was washed in buffer B1. To induce intein cleavage, the pellet was resuspended in buffer B2, and incubated overnight at 37°C. The mixture was then centrifuged and the pellet and supernatant fractions both retained. The pellet was again washed with buffer B1.
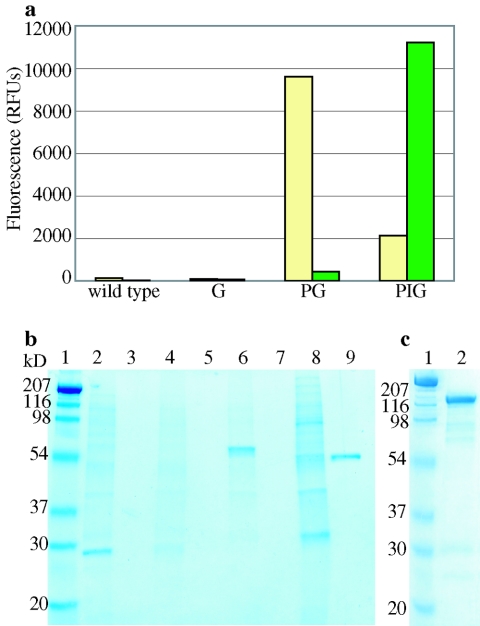
Figure 2A shows the fluorescence of the pellet and supernatant fractions. Neither the R. eutropha wild-type pellet nor the corresponding supernatant showed appreciable fluorescence. Similarly, the pellet and supernatant fractions of R.eutropha G showed no appreciable fluorescence as expected. As expected, R. eutropha PG showed strong fluorescence on the pellet with no appreciable fluorescence present in the supernatant. In contrast, R. eutropha PIG showed very strong fluorescence in the supernatant fraction, indicating that GFP had been released from the pellet into the supernatant fraction. Although the bulk of the total fluorescence was present in the supernatant, a minor amount of fluorescence remained on the PHB granule. To confirm the dithiothreitol-mediated release of GFP and the absence of pH effects, an identical experiment was performed in which the cell debris fraction generated from R. eutropha PIG was resuspended in buffer B1 (i.e., buffer B2 lacking dithiothreitol). No appreciable fluorescence was found in the supernatant fraction and the fluorescence of the cell debris remained unchanged (data not shown).
FIG. 2.
Intein mediated cleavage of GFP and beta-galactosidase from whole cell debris. R. eutropha strains were lysed by sonication, the supernatant discarded and the insoluble pellet containing PHB granules retained. Intein mediated cleavage was activated by incubating the washed pellet overnight in buffer B2 at 37°C. After incubation, the pellet and supernatant fractions were isolated. (a) Fluorometry. Green bars show the fluorescence of the supernatant fractions. Tan colored bars denote the fluorescence of the resulting pellet fraction. (b) SDS-PAGE of fractions. Lanes 2 and 3: R. eutropha wild-type pellet and supernatant following dithiothreitol treatment, respectively. Lanes 4 and 5: R. eutropha G pellet and supernatant. Lanes 6 and 7: R. eutropha PG pellet and supernatant. Lanes 8 and 9: R. eutropha PIG pellet and supernatant. (c) SDS-PAGE. Lane 2: R. eutropha PIL supernatant.
Figure 2B shows an SDS-PAGE gel for the pellet and supernatant fractions with the corresponding fluorescence data depicted in Fig. 2A. As expected, the whole cell debris for each strain contains numerous proteins. No protein is visible on the gel for the supernatant fractions of R. eutropha wild-type, R.eutropha G, and R. eutropha PG. The PhaP-intein-GFP fusion protein is expected to be 70 kDa in size. If intein-mediated cleavage occurs, a protein of 49 kDa, corresponding to an intein-GFP (IG) fusion, should be released. Figure 2B, lane 9, shows that IG was the only protein present in the supernatant fraction. This observation confirms that intein mediated cleavage, activated by thiol addition, released GFP from the granule in the cell debris of R. eutropha PIG.
To further investigate the robustness of this method we attempted to purify a relatively large, multimeric, catalytically active protein (β-galactosidase, 4 × 116 kDa). R. eutropha PIL was generated using plasmid pPIL (see Table 1). Plasmid pPIL is isogenic to plasmid pPIG with the exception that the gfpmut2 ORF in pPIG has been replaced with the lacZ ORF in pPIL. Therefore, in R. eutropha PIL, the wild-type phaP gene has been replaced by a phaP::Mxe GyrA intein::lacZ translational fusion.
Intein mediated release of pure β-galactosidase, using cell extracts of R. eutropha PIL was demonstrated. Craven et al. (7) reported a specific activity of 40,000 U/nmol (340,000 U/mg, 28°C, pH 7.0) for purified β-galactosidase obtained by three purification steps (ammonium sulfate precipitation, size exclusion chromatography and DEAE chromatography). Colby and Hu (5) reported 19,000 U/nmol (160,000 U/mg, 30°C, pH 7.0) for purified β-galactosidase obtained by five purification steps (ammonium sulfate precipitation, electrophoresis, DEAE chromatography, size exclusion chromatography and crystallization). Following the above described method, a specific activity of 53,000 U/nmol was measured in the supernatant following dithiothreitol treatment of whole cell debris. Thus, a single purification step, without external chromatography; results in a protein fraction of high purity consistent with previously reported data for pure β-galactosidase and SDS-PAGE (see Fig. 2C).
We have also explored the effect of orientation by using PhaP as the C-terminal fusion partner (i.e., creating plasmids pGIP and pLIP). Similar to results generated using PhaP as the N-terminal fusion partner, we have shown that soluble, active GFP and β-galactosidase can be recovered in a single purification step when PhaP was used as the C-terminal fusion partner (data not shown).
DISCUSSION
In this study, we report the development of an integrated protein expression and purification approach that obviates the need for external chromatography. By replacing the wild-type phaP gene with a triple translational fusion (phaP ORF, Mxe GyrA intein and gfpmut2), we were able to show that the fusion protein can be localized to the PHB granule and separated from the remaining cytosolic protein fraction by centrifugation. In a subsequent step, we were able to release pure GFP by resuspending whole cell debris (insoluble fraction of cell lysate, containing PHB granules) in a buffer containing dithiothreitol (see Fig. 3). We also expressed, recovered and purified β-galactosidase to demonstrate the applicability of this method to purify catalytically active proteins. We showed that the enzyme is essentially pure and that the specific activity is comparable to previously reported literature values for pure β-galactosidase.
The single step purification eliminates the need for elaborate and costly protein purification schemes. Moreover, adapting the use of inteins eliminates the need for specific endopeptidases, which are routinely used to release recombinant protein from affinity matrixes. It is contemplated that this system could be adapted for use in other hosts that either produce PHB granules naturally or hosts that have been engineered to produce PHB.
By integrating high-level recombinant protein expression with a simple, protein purification step, this system has the potential to improve upon current technologies for the large-scale production of commodity enzymes, therapeutic proteins, vaccines, and peptides.
Acknowledgments
This work was supported by ARO grant DAAD19-00, DOJ grant 2000-DT-CX-K001(S-1), and NIST grant 60NANB1D0064.
We thank George O' Toole (Dartmouth Medical School) for the use of his fluorescence microscopy equipment.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. BioTechniques 26:824-826. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, G. C., G. H. Henderson, S. Srinivasan, and T. U. Gerngross. 2004. High level recombinant protein expression in Ralstonia eutropha using T7 RNA polymerase based amplification, Protein Express. Purif. 38:264-271. [DOI] [PubMed] [Google Scholar]
- 4.Cha, H. J., C. F. Wu, J. J. Valdes, G. Rao, and W. E. Bentley. 2000. Observations of green fluorescent protein as a fusion partner in genetically engineered Escherichia coli: monitoring protein expression and solubility. Biotechnol. Bioeng. 67:565-574. [PubMed] [Google Scholar]
- 5.Colby, C., Jr., and A. S. Hu. 1968. Purification and comparison of the β-galactosidase synthesized by Escherichia coli F-lac+ and Proteus mirabilis F-lac+. Biochim. Biophys. Acta 157:167-177. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Craven, G. R., E. Steers, Jr., and C. B. Anfinsen. 1965. Purification, composition, and molecular weight of the β-galactosidase of Escherichia coli K12. J. Biol. Chem. 240:2468-2477. [PubMed] [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derbyshire, V., D. W. Wood, W. Wu, J. T. Dansereau, J. Z. Dalgaard, and M. Belfort. 1997. Genetic definition of a protein-splicing domain: functional min-inteins support structure predictions and a model for intein evolution. Proc. Natl. Acad. Sci. USA 94:11466-11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanley, S. Z., D. J. Pappin, D. Rahman, A. J. White, K. M. Elborough, and A. R. Slabas. 1999. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 447:99-105. [DOI] [PubMed] [Google Scholar]
- 11.Jurasek, L., and R. H. Marchessault. 2002. The role of phasins in the morphogenesis of poly(3-hydroxybutyrate) granules. Biomacromolecules 3:256-261. [DOI] [PubMed] [Google Scholar]
- 12.Karr, D. B., J. K. Waters, and D. W. Emerich. 1983. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detections. Appl. Environ. Microbiol. 60:3952-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luria, S. E., J. N. Adams, and R. C. Ting. 1960. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology 12:348-390. [DOI] [PubMed] [Google Scholar]
- 14.New England Biolabs. 2003. IMPACT-TWIN instruction manual, version 1.2, 12/03. New England Biolabs, Beverly, Mass.
- 15.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1995. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasing to polyhydroxyalkanoic acid granules. J. Bacteriol. 177:2513-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pötter, M., M. H. Madkour, F. Mayer, and A. Steinbüchel. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413-2426. [DOI] [PubMed] [Google Scholar]
- 17.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 18.Rehm, B. H., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3-19. [DOI] [PubMed] [Google Scholar]
- 19.Resch, S., K. Gruber, G. Wanner, S. Slater, D. Dennis, and W. Lubitz. 1998. Aqueous release and purification of poly(beta-hydroxybutyrate) from Escherichia coli. J. Biotechnol. 65:173-182. [DOI] [PubMed] [Google Scholar]
- 20.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic-hydrolysis of organophosphates-cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta. Biotechnology 3:567-571. [Google Scholar]
- 21.Simon, R., U. Priefer, and A. Pfihler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 22.Srinivasan, S., G. C. Barnard, and T. U. Gerngross. 2002. A novel high cell density protein expression system based on Ralstonia eutropha. Appl. Environ. Microbiol. 68:5925-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan, S., G. C. Barnard, and T. U. Gerngross. 2003. Production of recombinant proteins using multiple copy gene integration in high cell density fermentations of Ralstonia eutropha. Biotechnol. Bioeng. 84:114-120. [DOI] [PubMed] [Google Scholar]
- 24.Steinbüchel, A., R. Wieczorek, and N. Krüger. 1996. PHA biosynthesis, its regulation and application of C1-utilizing microorganisms for polyester production, p. 237-244. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds: proceedings of the 7th International Symposium on Microbial Growth on C1 Compounds, San Diego, Calif., 27 August-1 September 1995.
- 25.Telenti, A., M. Southworth, F. Alcaide, S. Daugelat, W. R. Jacobs, Jr., and F. B. Perler. 1997. The Mycobacterium xenopi GyrA protein splicing element: characterization of a minimal intein. J. Bacteriol. 179:6378-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood, D. W., W. Wu, G. Belfort, V. Derbyshire, and M. Belfort. 1999. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17:889-892. [DOI] [PubMed] [Google Scholar]
- 28.Wu, C. F., H. J. Cha, G. Rao, J. J. Valdes, and W. E. Bentley. 2000. A green fluorescent protein fusion strategy for monitoring the expression, cellular location, and separation of biologically active organophosphorus hydrolase. Appl. Microbiol. Biotechnol. 54:78-83. [DOI] [PubMed] [Google Scholar]
- 29.Wu, C. F., H. J. Cha, G. Rao, J. J. Valdes, and W. E. Bentley. 2001. Enhancement of organophosphorus hydrolase yield in Escherichia coli using multiple gene fusions. Biotechnol. Bioeng. 75:100-103. [DOI] [PubMed] [Google Scholar]
- 30.York, G. M., B. H. Junker, J. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.York, G. M., J. Stubbe, and A. J. Sinskey. 2001. New insight into the role of the PhaP phasing of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasing to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]