Abstract
In this paper we describe the bacterial communities associated with natural hydrocarbon seeps in nonthermal soils at Rainbow Springs, Yellowstone National Park. Soil chemical analysis revealed high sulfate concentrations and low pH values (pH 2.8 to 3.8), which are characteristic of acid-sulfate geothermal activity. The hydrocarbon composition of the seep soils consisted almost entirely of saturated, acyclic alkanes (e.g., n-alkanes with chain lengths of C15 to C30, as well as branched alkanes, predominately pristane and phytane). Bacterial populations present in the seep soils were phylogenetically characterized by 16S rRNA gene clone library analysis. The majority of the sequences recovered (>75%) were related to sequences of heterotrophic acidophilic bacteria, including Acidisphaera spp. and Acidiphilium spp. of the α-Proteobacteria. Clones related to the iron- and sulfur-oxidizing chemolithotroph Acidithiobacillus spp. were also recovered from one of the seep soils. Hydrocarbon-amended soil-sand mixtures were established to examine [14C]hexadecane mineralization and corresponding changes in the bacterial populations using denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene fragments. Approximately 50% of the [14C]hexadecane added was recovered as 14CO2 during an 80-day incubation, and this was accompanied by detection of heterotrophic acidophile-related sequences as dominant DGGE bands. An alkane-degrading isolate was cultivated, whose 16S rRNA gene sequence was identical to the sequence of a dominant DGGE band in the soil-sand mixture, as well as the clone sequence recovered most frequently from the original soil. This and the presence of an alkB gene homolog in this isolate confirmed the alkane degradation capability of one population indigenous to acidic hydrocarbon seep soils.
The occurrence of natural hydrocarbon seeps in thermal areas of northwestern Wyoming, including the Yellowstone National Park (YNP) region, was reported by explorers as early as 1807 (33). Three hydrocarbon seeps located near Calcite Springs, Rainbow Springs, and Tower Bridge in the northeast corner of YNP were described by Love and Good (32). All of the seeps had abundant sulfur and were surrounded and underlain by volcanic rocks. Thermally active springs and vents were closely associated with the seeps at the time of investigations by Love and Good in 1963 and 1968 (33). The Calcite Springs oils are composed primarily of aromatic hydrocarbons, polar compounds, and asphaltenes and contain less than 1% saturated hydrocarbons, whereas the oils from Rainbow Springs consist almost entirely of n-alkanes (6). The chemical composition of Tower Bridge oils has not been reported. Of the three seeps, Rainbow Springs is the least accessible, and it is located about 32 km northeast of Lake Village, YNP.
The association of aliphatic hydrocarbons with acidic, sulfate-rich geothermal environments distinguishes the Rainbow Springs sites from other natural hydrocarbon seeps. The extreme acidity (pH <3) associated with geothermal areas of YNP is due to the oxidation of reduced S species derived from the condensation of volcanic gases, such as H2S and SO2. Many other acidic environments in nature are created as a result of human activities, such as mining and industrial wastewater treatment. Microorganisms that are metabolically active in these extremely acidic environments have attracted interest for their unique ecology and physiology (2, 3, 5, 17, 18, 28, 31), as well as for their potential applications in biotechnology (36, 39). The presence of diverse acidophilic populations in both natural and man-made acidic environments has been demonstrated by cultivation-dependent and -independent approaches (2, 3, 5, 17, 18, 28, 31). Iron- and sulfur-oxidizing chemolithotrophic acidophiles, such as Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans, often are the predominant primary producers, and heterotrophic acidophiles may coexist by metabolizing organic compounds (23, 25). Some heterotrophic acidophiles may also contribute to iron and sulfur cycling via oxidization of reduced inorganic sulfur compounds (Acidiphilium acidophilum) or ferrous iron (“Ferrimicrobium acidiphilum”) (27) and via reduction of ferric iron (Acidiphilium spp.) (24).
Contamination of acidic environments with hydrocarbons is not uncommon (e.g., contamination of acidic wastewaters from mine drainage and industries by oil spills), and as a result, there has been significant interest in the potential bioremediation of polluted extreme environments (34). However, most studies on hydrocarbon degradation have focused on neutral pH conditions, and little is known about microbial hydrocarbon metabolism in acidic environments. To date, there have been only a few studies on hydrocarbon degradation by acidophilic isolates (13, 16, 20) or in acidic soil samples (44). Twenty-three heterotrophic acidophiles isolated from acidic mine effluent have been shown to degrade a range of aliphatic hydrocarbons (16, 20); two of these strains (LGS-3 and WJB-3) were closely related to the genus Acidocella (20). A naphthalene-utilizing isolate obtained from an acidic coal storage pile was also identified as an Acidocella sp. (13). The role of these acidophilic isolates in natural environments has not been described. Stapleton et al. (44) demonstrated that aromatic hydrocarbons were degraded by indigenous microorganisms present in acid soils of a coal pile storage basin, and a consortium of bacteria and fungi may have been responsible. Since hydrocarbon-degrading acidophiles are potential candidates for bioremediation of contaminated acidic environments, a better understanding of their fitness and function in natural environments is necessary for successful application.
In this paper, we describe the diversity and potential function of bacterial populations associated with unique terrestrial hydrocarbon seeps of Rainbow Springs, YNP. Bacterial community structure was phylogenetically characterized using 16S rRNA gene clone library analysis. To link phylogenetically defined populations with potential physiological functions, we examined 16S rRNA sequence fingerprints during hydrocarbon degradation in soil-sand mixtures and also isolated a hydrocarbon-degrading organism relevant in the hydrocarbon-rich acidic soil environment.
MATERIALS AND METHODS
Sample collection.
Two hydrocarbon-contaminated soils separated by approximately 20 m (RH1 and RH2) were sampled in August 2003 in the Rainbow Springs area in Yellowstone National Park (44°46′07.2"N, 110°16′14.9"W). The RH1 and RH2 seep soils were identified by a strong petroleum odor and brown staining; however, these sites were not thermal at the time of sampling in summer 2003. The soils were collected from the surface to a depth of 5 cm with sterile spatulas, transported on ice to the laboratory, and stored at 4°C for geochemical analysis and hexadecane mineralization assays. Subsamples were stored at −80°C for molecular biological analysis.
Geochemical analysis.
Hydrocarbons were analyzed by capillary gas chromatography mass spectrometry (GC-MS) after solvent extraction. Soil samples (1 g) were spiked with a methyl stearate standard and extracted with 10 ml of dichloromethane (high-performance liquid chromatography grade; 99.9%; Acros Organics, Morris Plains, NJ) by vigorous shaking for 1 h at room temperature. The mixtures were allowed to sit overnight to settle soil particles. Aliquots (1 ml) of the extracts were spiked with an internal standard (hexachlorobenzene) at a final concentration of 20 mg/liter and analyzed by GC-MS (HP 5890 series II GC equipped with an HP 5971 series MS; Hewlett-Packard). The GC was equipped with a DB-1 column (60 m by 0.25 mm; film thickness, 0.25 μm) and a flame ionization detector (with He as the carrier gas). The temperature program was as follows: the initial oven temperature, 30°C, was held for 5 min, and then the temperature was ramped to 300°C at a rate of 3°C min−1 and finally held for 15 min at 300°C. The injector and detector temperatures were maintained at 260 and 280°C, respectively. Samples (1 μl) were injected in splitless mode. The mass range for MS was 35 to 450, and the scan rate was 0.50 s per mass decade. Various hydrocarbon components in the samples were quantified by comparing the integrated individual peak areas to the peak areas of internal standards in each sample. The total petroleum hydrocarbon content was calculated as the sum of all hydrocarbon components detected and integrated. Extractions were conducted in triplicate.
The dissolved inorganic constituents of soil-water extracts (1:3) were determined using inductively coupled plasma emission spectrometry and ion chromatography (38, 43).
DNA extraction.
Soil samples were homogenized individually. DNA was extracted from 0.5-g soil samples by the method described by Bürgmann et al. (4), with the following minor changes: for mechanical lysis, 0.75 g zirconium beads (diameter, 0.1 mm; Biospec Products Inc., Bartlesville, OK) was used, and samples were processed in a bead beater (FastPrep FP120; Bio 101/Savant, Farmingdale, NY) for 45 s at 6 m/s. Soil DNA extracts were purified further by polyvinylpolypyrrolidone (PVPP) spin column chromatography as described by Cullen and Hirsch (9). The acid-washed PVPP was prepared as described by Evans et al. (14). DNA extraction was conducted in triplicate for each soil, and the pooled extracts were used for molecular analysis. DNA was extracted from an isolate (see below) by the same protocol without the PVPP spin column purification step.
Cloning and sequencing of 16S rRNA genes.
Bacterial 16S rRNA genes were PCR amplified with primers 27F and 1392R (30). The PCR mixtures (final volume, 50 μl) contained 2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.5 μM, 5% (wt/vol) acetamide, ∼10 ng of DNA, 1.25 U of Taq polymerase, and 1× buffer (AmpliTaq Gold; Applied Biosystems, Foster City, CA). Amplifications were performed (GeneAmp 2700; Applied Biosystems) using the following program: (i) 10 min at 94°C; (ii) 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and (iii) final extension for 7 min at 72°C. PCR products were purified with a QIAquick gel extraction kit (QIAGEN, Chatsworth, CA) and cloned into the pGEM-T Easy vector (Promega, Madison, WI) by following the directions of the manufacturer. Sixty-eight and 70 clones from the RH1 and RH2 clone libraries, respectively, were screened with the enzymes HaeIII and RsaI (Promega) for restriction fragment length polymorphism (RFLP) groups. Selected clones from each RFLP group (multiple clones were selected from RFLP groups that had more than two members) were completely sequenced using an ABI Prism BigDye Terminator cycle sequencing reaction kit and an ABI 310 DNA sequencer (Applied Biosystems). The primers used for sequencing included the amplification primers mentioned above in addition to 357F, 1100R (30), and vector primers.
Phylogenetic analysis.
Sequences were assembled using Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, MI) and were compared to the GenBank database using BLAST (1). Sequences were checked for chimeras using the CHECK_CHIMERA program of the Ribosomal Database Project (7). Alignment was performed with ClustalX (version 1.81) using default values (46), and the alignments were edited manually. Distance analysis was performed using the Jukes-Cantor correction (29), followed by phylogenetic tree construction using the neighbor-joining method (41) of the PAUP*4.0 software (Sinauer Associates, Sunderland, MA).
Hexadecane mineralization in RH soil-sand mixtures.
The presence of indigenous alkane-degrading populations in the RH soils was examined by inoculating 1 g of an RH soil into 9 g of nonporous sand (SiO2; particle size, 75 to 600 μm; Fluka Chemica, SG, Switzerland) in 150-ml serum bottles. This approach was necessary because of the limited amounts of soil and because the soils were already contaminated with hydrocarbons. Prior to inoculation, the sand was washed with deionized distilled water three times, dried overnight, and then autoclaved three times for 1 h with 24-h intervals at room temperature. The mixtures were incubated at room temperature (25 ± 2°C) in the dark without shaking, and hexadecane mineralization was measured in triplicate. Autoclaved soils, which were prepared by autoclaving soil three times for 1 h with 24-h intervals at room temperature, were used as inocula for nonbiological controls. Nutrient solutions were added to the soil-sand mixture to achieve a water content of 20% and the following final nutrient concentrations: (NH4)2SO4, 0.7 mM; NH4NO3, 7.4 mM; KH2PO4, 0.44 mM; K2HPO4, 0.44 mM; KOH, 0.18 mM; H2SO4, 0.47 and 0.85 mM for RH1 and RH2, respectively; MgCl2, 1 mM; CaCl2, 2 mM; and FeCl2, 0.01 mM. The pH values of the nutrient solutions were adjusted to 3.8 and 2.8 for RH1 and RH2, respectively. n-Hexadecane (Sigma Chemical, St. Louis, MO) was added dropwise to each bottle using a 100-μl microsyringe to achieve a final concentration of 0.5% (wt/wt) containing 50,000 dpm [1-14C]hexadecane (purity, >98%; specific activity, 2.6 mCi mmol−1; Sigma Chemical), and each bottle was sealed with a Teflon-coated rubber septum and an aluminum crimp top. Evolved 14CO2 was measured as described previously (8). After completion of the experiment, triplicate subsamples (1 g) from each bottle were combusted in a biological oxidizer (model OX-300; R. J. Harvey Instrument, Hillsdale, NJ), and the residual radioactivity was measured using scintillation analysis. The mass balance was determined based on the sum of evolved 14CO2 plus residual 14C, and the average recovery was 92.7 ± 18.3%.
DGGE analysis.
Bacterial populations present in soil-sand mixtures were analyzed by PCR amplification of bacterial 16S rRNA gene fragments followed by denaturing gradient gel electrophoresis (DGGE) at several times during the incubation. DNA was extracted from 0.5-g subsamples removed from replicate mixtures as described above. PCR amplification was conducted with the Bacteria-specific primer 1070F and the universal primer 1392R containing a GC clamp (15), using the PCR mixtures described above. The amplification program was as follows: (i) 10 min at 94°C; (ii) initial touchdown cycles consisting of denaturation for 45 s at 94°C, annealing for 45 s at 57°C with the temperature dropping by 1°C each two cycles for eight cycles, and elongation for 90 s at 72°C; (iii) 30 cycles with annealing at 53°C; and (iv) a final extension for 7 min at 72°C. DGGE was performed with a D-Code system (Bio-Rad) as described by Muyzer et al. (35). DGGE gels containing 8% polyacrylamide with a urea-formamide gradient (40 to 70%) were run at 60 mV (constant voltage) for 16 h. The gels were stained with SYBR Green II (Molecular Probes, Eugene, OR) and photographed. The nucleotide sequences of dominant bands in DGGE gels were determined as described previously (15).
Isolation of an alkane-degrading bacterium from RH soils.
An indigenous alkane-degrading bacterium was isolated by plating serially diluted soil slurries on SXm agar plates containing (per liter of distilled water) 0.06 g of MgSO4, 0.01 g of CaCl2, 2.00 g of NH4NO3, 0.10 g of (NH4)2SO4, 0.50 g of KH2PO4, 0.10 g of KCl, 1 ml of a trace element solution (48), and 15 g of agar; the pH was adjusted to 4.5 with 1 N H2SO4 after autoclaving. This was followed by incubation at room temperature in a sealed container with n-dodecane as the sole carbon source, supplied through the vapor phase. Colonies were restreaked for isolation. PCR were performed with isolated colonies for 16S rRNA gene sequencing and for PCR-DGGE analysis as described above to confirm the purity of cultures and to determine if corresponding DGGE bands were present in the original RH soils or in the soil-sand mixture experiments. The isolate obtained and described in the current study (strain C197) was grown in liquid SXm medium containing 1% (vol/vol) n-hexadecane. Cells were collected by centrifugation at 16,000 × g for 10 min and subjected to DNA extraction as described above.
Characterization of putative alkB gene in an alkane-degrading isolate.
The putative alkane hydroxylase gene, alkB, in the alkane-degrading isolate obtained as described above (strain C197) was PCR amplified by using highly degenerate primers TS2S and deg1RE and conditions described by Smits et al. (42). A PCR product of the expected size (550 bp) was cloned into the pGEM-T Easy vector (Promega) by following the directions of the manufacturer and was sequenced using vector primers.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database under accession numbers AY678225 to AY678263 (cloned 16S rRNA genes), AY817740 to AY817745 (DGGE bands), and AY817739 (putative alkB from isolate C197).
RESULTS AND DISCUSSION
Geochemical characterization.
Hydrocarbon seeps at Rainbow Springs were associated with thermal activity in 1963 (33); however, samples collected for this study in August 2003 were not thermally active. The chemical analysis of both the RH1 and RH2 seep soils showed high concentrations of total soluble Al, Fe, Zn, and As, as well as sulfate, and low pH values (pH ∼2.8 to 3.8), which is consistent with characteristics of acid-sulfate geothermal activity (Table 1). Although both samples showed similar chemical signatures, the concentrations of most constituents were substantially higher in RH2 than in RH1. The higher concentrations of sulfate, Fe, and Al in RH2 are consistent with the lower pH of RH2 (pH 2.8) than of RH1 (pH 3.8).
TABLE 1.
Concentrations of total soluble constituents, pH values, and total petroleum hydrocarbons of Rainbow Springs soilsa
| Soil | pH | TPH (%)b | Anion and weak acid concn (μM)
|
Cation concn (μM)
|
Total charge (mM)
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SO4-S | F | Cl | B | As | PO4-P | NO3-N | NH4-N | Ca | Fe | Mg | K | Na | Al | Mn | Zn | Cu | Cd | Pb | Anions | Cations | |||
| RH1 | 3.8 | 5.5 | 1,219 | 90 | 26 | 48 | 1.5 | 28.1 | <2.0 | <3.5 | 454 | 144 | 263 | 258 | 544 | 71 | 4.2 | 5.5 | <0.2 | <0.2 | <0.5 | 2.6 | 2.9 |
| RH2 | 2.8 | 8.3 | 7,938 | 151 | 143 | 143 | 26.7 | 5.5 | 5.5 | 70.0 | 2,098 | 1,023 | 704 | 491 | 38 | 937 | 37.7 | 9.5 | 0.3 | 0.2 | <0.5 | 16.2 | 15.4 |
Concentrations and pH values were measured in 1:3 soil-water extracts.
TPH, total petroleum hydrocarbon content, calculated as the sum of all hydrocarbon components detected and integrated by GC-MS analysis.
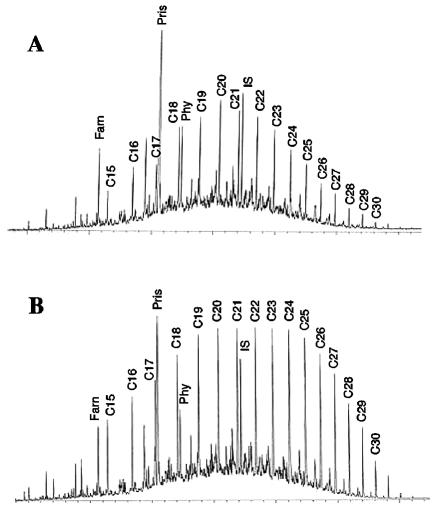
The hydrocarbons of RH1 and RH2 consisted almost entirely of saturated, acyclic alkanes with chain lengths of C15 to C30, as well as branched alkanes, predominately pristane and phytane (Fig. 1). The gas chromatography profiles of the n-alkanes in RH1 and RH2 were centered at C20 to C22, and 80% of the total n-alkanes ranged from C16 to C24 and from C18 to C26, respectively. Pristane and phytane comprised 35% of the total hydrocarbons in RH1, in contrast to 11% in RH2. The n-C17/pristane and n-C18/phytane ratios were 0.11 and 0.61, respectively, for RH1, and 0.52 and 2.35, respectively, for RH2. Assuming that there were similar hydrocarbon sources in the two soils, the hydrocarbon signatures showing lower n-C17/pristane and n-C18/phytane ratios in RH1 than in RH2 may indicate that there was greater n-alkane decomposition via microbial activity, since, in general, hydrocarbon-degrading bacterial populations preferentially utilize n-alkanes relative to branched alkanes (22, 47). The hydrocarbon compositions of RH1 and RH2 are consistent with the results of previous analyses conducted with oil collected from Rainbow Springs in 1963 (6), which showed that these sites contain a highly paraffinic low-S crude oil in which lower-molecular-weight hydrocarbons are depleted.
FIG. 1.
Gas chromatograms of hydrocarbon components in Rainbow Springs soils RH1 (A) and RH2 (B). Hydrocarbon components: C15 to C30, n-alkanes (the numbers indicate chain lengths); Farn, farnesane; Pris, pristane; Phy, phytane; IS, added internal standard.
Bacterial community characterization.
Bacterial populations present in acidic hydrocarbon-contaminated soils were characterized by 16S rRNA gene clone library analysis. Of 68 and 70 clones analyzed from the RH1 and RH2 libraries, 15 and 24 different clone groups were identified by RFLP patterns, respectively. One chimeric sequence was identified and excluded from further analysis. The majority of the clones recovered from both sites (65 of 67 RH1 clones and 56 of 70 RH2 clones) were affiliated with the Acetobacteraceae group, including Acidisphaera spp. and Acidiphilium spp., of the α-Proteobacteria (Fig. 2). With only one exception, the RH1 and RH2 clones in this Acetobacteraceae group each formed distinct clusters. The largest group of 43 clones from RH1 (represented by RH1-A) formed a cluster with three other clone groups from RH1 (RH1-i3, RH1-E2, and RH1-iG2, total of seven clones) that was clearly distinct from sequences available in the GenBank database. The closely related cultured relatives to this clone group include moderately acidophilic heterotrophs, such as Acidomonas methanolica (94.4% similarity) (50) and Acidisphaera rubrifaciens (92.9% similarity) (21), which is consistent with the moderately acidic nature of the RH1 soil (pH 3.8). The largest clone group from RH2 (30 of 70 RH2 clones, represented by RH2-A) and two other related groups from RH2 (RH2-R and RH2-C2, total of six clones) formed a cluster closely related to Acidiphilium acidophilum (99% similarity). Relatively little is known about the diversity and distribution of heterotrophic acidophiles. Recently, the presence of diverse heterotrophic acidophilic populations in acid mine drainage (28) and in geothermal acidic sites in YNP (26) was established using cultivation approaches. This, along with the current results, suggests that the significant microbial diversity present in acidic environments is far from thoroughly characterized. Only two clones from the RH1 soil (RH1-E and RH1-C2) were not affiliated with the acidophilic bacterial group, and these clones were distantly related to a facultative photoheterotroph, Paracraurococcus ruber (92% similarity), which was reported to show optimum growth at neutral pH (pH 6.6 to 6.8) (40).
FIG. 2.
Neighbor-joining tree showing phylogenetic positions of 16S rRNA gene sequences cloned from Rainbow Springs soils. Sequences corresponding to nucleotide positions 112 to 1337 of the Escherichia coli 16S rRNA gene were used for the analysis. Sequences from the RH1 soil are indicated by boldface type, sequences from the RH2 soil are indicated by boldface type followed by an asterisk, and the boldface numbers in parentheses indicate the number of clones obtained for each sequence. The arrows indicate the largest clone groups recovered from the RH1 and RH2 libraries. Bootstrap values (per 100 trials) for major branch points are indicated. Bar = 0.01 substitution per sequence position.
While all sequences recovered from the RH1 soil were affiliated with the α-Proteobacteria, sequences recovered from the RH2 soil were affiliated with diverse phylogenetically distinct groups; of 70 clones, 56 α-proteobacterial clones, 10 γ-proteobacterial clones, 2 Acidobacteria-related clones, and 2 Firmicutes-related clones were recovered. Within the α-Proteobacteria, there were two clone groups, RH2-P3 and RH2-T3 (total of three clones), that clustered with methanotrophic bacteria, including Methylocella tundrae (10) and Methylocapsa acidiphila (11), which are recently cultivated moderately acidophilic methanotrophs. This result suggests that methane may be present in this seep site, which is rather likely since low-molecular-weight hydrocarbon gases have been reported for numerous locations in YNP (12, 19). Three clone types, represented by RH2-B, RH2-O2, and RH2-U3 (total of eight clones), were clustered with the γ-proteobacterial acidophilic chemolithotroph Acidithiobacillus spp. Two clones (RH2-D and RH2-J) related to Legionella spp., two clones (RH2-F2 and RH2-L3) related to the Acidobacteria group, and two clones (RH2-T2 and RH2-M) related to the Firmicutes group were also recovered.
The distinct microbial community compositions of RH1 and RH2 soils may reflect differences in geochemical properties of the two hydrocarbon-contaminated soils (Table 1). Conversely, activities of different microbial populations may also contribute to differences in the chemical properties of the two soils. For instance, higher concentrations (approximately sevenfold higher) of soluble Fe and SO4-S in RH2 than in RH1 (Table 1) may explain the recovery of 16S rRNA gene sequences affiliated with the Fe- and S-oxidizing chemolithotroph Acidithiobacillus ferrooxidans (23) and the S-oxidizing mixotroph Acidiphilium acidophilum (27) from the RH2 soil, and these populations may have contributed to the greater acidity in the RH2 soil than in the RH1 soil. Various Acidiphilium spp. are also capable of reducing Fe(III) in the absence or even in the presence of 20 to 40% oxygen (24) and may establish mutualistic relationships with Fe(II)-oxidizing chemolithotrophs via Fe cycling (23, 37). Considering the Fe- and hydrocarbon-rich nature of the RH2 soil, we speculate that Fe cycling in this environment may be linked with hydrocarbon metabolism. The chemolithotrophic organism Acidithiobacillus ferrooxidans could supply Fe(III) to the Acidiphilium spp., which may utilize hydrocarbons as a carbon and energy source and Fe(III) as an electron acceptor. However, more comprehensive physiological and geochemical characterization is required to elucidate the ecological processes mediated by these organisms in the RH soils.
In summary, phylogenetic analysis showed the presence of diverse acidophile-related bacterial populations in these hydrocarbon-contaminated acid-sulfate soils. However, a physiological association of these populations with hydrocarbon metabolism cannot be inferred from phylogenetic analysis alone.
Hexadecane mineralization by RH soil-sand mixtures.
To examine the presence of indigenous hydrocarbon-degrading bacteria in the RH soils, soil-sand mixtures were amended with hexadecane to monitor the emergence of specific populations concomitant with hydrocarbon degradation. After a lag period of 5 to 10 days, both RH1 and RH2 mixtures readily mineralized hexadecane, although the RH1 mixtures showed a higher rate and greater extent of hexadecane mineralization than the RH2 mixtures (Fig. 3). Approximately 56 and 39% of the added [14C]hexadecane were recovered as 14CO2 during an 80-day incubation in RH1 and RH2 mixtures, respectively. Control treatments inoculated with autoclaved RH soils showed no production of 14CO2, confirming the biological conversion of hexadecane in these experiments.
FIG. 3.

Mineralization of [1-14C]hexadecane in RH1 (circles) and RH2 (triangles) soil-sand mixtures. Solid symbols indicate soil-inoculated samples, and open symbols indicate autoclaved control soil-inoculated samples. Each point represents the average of triplicate bottles; the error bars indicate standard deviations.
Bacterial populations in the mixtures were examined by DGGE analysis of PCR-amplified 16S rRNA gene fragments. DGGE profiles were obtained using replicate samples to demonstrate the reproducibility of the techniques (Fig. 4). In both RH1 and RH2 mixtures, distinct bacterial populations were established during the degradation of hexadecane. In RH1 mixtures, approximately 20% of the added hexadecane was mineralized to CO2 by day 24, which was accompanied by detection of three dominant DGGE bands (bands 1, 2, and 3 in Fig. 4). These three bands remained relatively dominant throughout the incubation period, while multiple minor bands emerged later (days 56 and 88). The DNA sequence of DGGE band 1 was identical to the sequence of the largest clone group from RH1, represented by RH1-A, and RH1-i3 (Table 2). The other two DGGE bands in the RH1 mixtures, bands 2 and 3, were closely related to each other (96.3% similarity) and clustered with their close relatives Acidocella sp. strain LGS-3 and Acidocella facilis (data not shown). Identical clone sequences corresponding to bands 2 and 3 were not detected in clones retrieved from the original RH1 soil (Table 2). The absence of corresponding clone sequences identical to these DGGE bands may have been due to the low levels of these populations in the original sample, to methodological biases during DNA extraction (49) and PCR amplification (45, 49), and/or to the relatively small size of the clone libraries evaluated. It is also possible that the microenvironmental conditions in soil-sand mixtures, which may not exactly mimic the original soil environments, selected for different microbial populations than present in the original soils.
FIG. 4.
DGGE profiles of 16S rRNA gene fragments during hexadecane mineralization by RH1 and RH2 soil-sand mixtures compared with profiles obtained for the original RH soils and isolate C197. The nucleotide sequences of the labeled bands (bands 1 to 6) were determined and are described in Table 2.
TABLE 2.
Sequence analysis of 16S rRNA gene DGGE bands obtained from RH1 and RH2 soil-sand mixtures
| Soil | DGGE banda | Length (bp) | Most closely related organism (accession no.) | % Similarity | Identical clone group(s) |
|---|---|---|---|---|---|
| RH1 | 1 | 323 | Acidisphaera sp. strain NO-15 (AF376024) | 96.6 | RH1-A, RH1-i3 |
| 2 | 321 | Acidocella sp. strain LGS-3 (AF253413) | 97.5 | ||
| 3 | 320 | Acidocella facilis (D30774) | 99.7 | ||
| RH2 | 4 | 321 | Acidiphilium acidophilum (D86511) | 100.0 | RH2-A, RH2-R, RH2-C2, RH2-P2, RH2-N2 |
| 5 | 321 | Acidiphilium sp. strain NO-13 (AF376022) | 100.0 | RH2-J2 | |
| 6 | 261 | Acidobacteriaceae isolate Ellin5241 (AY234592) | 97.6 | RH2-L3 |
The DGGE band numbers correspond to the numbers in Fig. 4.
In RH2 mixtures, three major DGGE bands (bands 4, 5, and 6 in Fig. 4) were detected throughout the incubation period. The DNA sequences of these three dominant DGGE bands in RH2 mixtures were identical to clone sequences from two distinct phylogenetic groups (Table 2). The DGGE band 4 sequence was identical to the sequence of the largest clone group, represented by RH2-A, and the band 5 sequence was identical to the clone representative RH2-J2. Bands 4 and 5 clustered with major clone groups (representing 63% of the clones recovered from RH2) and their close α-proteobacterial relatives Acidiphilium acidophilum and Acidiphilium sp. strain NO-13. The sequence of another dominant DGGE band in RH2 mixtures, band 6, was identical to the clone RH2-L3 sequence, which was associated with the heterotrophic acidophile Acidobacterium sp.
In both RH1 and RH2 mixtures, sequences of the predominant DGGE bands matched sequences of the largest clone groups recovered from 16S rRNA gene clone libraries (RH1-A and RH2-A, respectively), suggesting that these heterotrophic acidophile-related populations are relevant in these soils and may degrade hydrocarbons in the original soil environments.
Isolation and characterization of an alkane-degrading bacterium relevant in RH soil.
Definitive evidence for the central role of heterotrophic acidophiles in hydrocarbon metabolism, at least in RH1 soil, was obtained by successful isolation of a hexadecane-degrading isolate relevant in the original soil. One isolate, strain C197, produced a DGGE band that comigrated with band 1, a dominant DGGE band in the original RH1 soil, as well as in the soil-sand mixtures (Fig. 4); the sequence of band 1 was 100% identical to the short-fragment 16S rRNA gene sequence of isolate C197. Furthermore, the nearly full-length 16S rRNA gene sequence of isolate C197 was identical to the sequence of the largest clone group from the RH1 library, represented by RH1-A. Isolate C197 showed relatively low sequence similarities to previously cultured close relatives, including the heterotrophic acidophile Acidisphaera sp. strain NO-15 (94.1%), the methanol-utilizing organism Acidomonas methanolica (94.4%), the acetic acid bacterium Gluconacetobacter sacchari (94.0%), and the bacteriochlorophyll-containing acidophile Acidisphaera rubrifaciens (92.9%). In addition, isolate C197 was rather distantly related to the thermophilic acidophilic Acidisphaera-like isolate Y008 (91.7%), which was recently cultivated from Norris Geyser Basin, YNP (26), as well as the hydrocarbon-utilizing acidophilic isolate Acidocella sp. strain LGS-3 (92.6%) (16, 20). Although the cultivation strategy employed in the current study was successful in retrieving one organism corresponding to a predominant clone group (RH1-A), it is clear that additional strategies are necessary to isolate other relevant organisms from this environment.
Isolate C197 was able to grow in minimal medium (pH 4.5) with 1% (vol/vol) n-dodecane or n-hexadecane as the sole carbon source. When grown on solid medium having the same composition, C197 formed salmon-pink colonies, which has also been observed with Acidisphaera rubrifaciens as a result of carotenoid production (21). Isolate C197 was capable of mineralizing [14C]hexadecane, and 10.0 ± 0.2% and 6.7 ± 1.8% of the added radiolabel was recovered as 14CO2 under illuminated and nonilluminated conditions, respectively, after a 30-day incubation (data not shown).
The presence of an alkB gene coding for alkane hydroxylase (a key enzyme in alkane oxidation) in isolate C197 was examined by the PCR method developed by Smits et al. (42). A PCR product of the expected size was obtained from isolate C197, and the sequence of the fragment was determined. The deduced peptide sequence encoded by the PCR product (185 amino acids) from isolate C197 showed high levels of sequence similarity to a putative AlkB fragment from Xanthobacter flavus (92.5%) and AlkB from Alcanivorax borkumensis AP1 (89.7%) (Fig. 5).
FIG. 5.
Phylogenetic position of the deduced amino acid sequence encoded by the putative alkane hydroxylase gene (alkB) cloned from isolate C197. The neighbor-joining tree was generated based on partial gene sequences (550 bp) of previously described and putative alkane hydroxylases; the xylM sequence was used as an outgroup. Bootstrap values (per 100 trials) are indicated at the nodes. Bar = 0.05 substitution per sequence position.
The physiological and molecular mechanisms of hydrocarbon degradation by acidophilic bacteria have not been elucidated. Two previous studies using DNA from an acidic environment (44) or naphthalene-utilizing acidophilic isolates (13) were unable to detect genes commonly associated with aromatic hydrocarbon degradation using specific PCR primers or hybridization probes. To our knowledge, there has been no description of genes coding for alkane degradation in acidic environments or in acidophilic bacteria. The detection of a putative alkB gene in isolate C197 extends the collection of alkB homologues from neutrophilic bacteria to acidophilic or acidotolerant bacteria. Further molecular and physiological characterization is required to confirm the function of the putative alkane hydroxylase in isolate C197. The evidence that isolate C197 was a relevant bacterial population in the alkane-contaminated seep soil strongly suggests its importance in hydrocarbon metabolism in this acid-sulfate soil.
This study of natural hydrocarbon seeps in YNP provided insight into the biodegradation potential of indigenous bacterial populations in acidic environments. The presence of diverse acidophile-related populations and their potential function in situ were shown by both cultivation-independent and cultivation-dependent approaches. The isolation of an acidophile-related alkane-degrading organism, which was also shown to be a relevant environmental population using molecular methods, provides strong support for the hypothesis that this organism is involved in alkane degradation in situ. More detailed community characterization and hydrocarbon degradation assays, as well as additional isolation strategies, are necessary to obtain a complete understanding of the microbial ecology of hydrocarbon seeps in acid-sulfate soils.
Acknowledgments
We greatly thank Mary Bateson and Katharine Schultz for technical assistance and Joe Sears for the GC-MS analysis. We also appreciate the support of the Yellowstone Park Division of Resources (J. Varley and C. Hendrix), which permitted this work in Yellowstone National Park.
This work was supported by the USEPA (project 829357-01-0), by the Thermal Biology Institute via a grant from the National Aeronautic and Space Administration (NASA project NAG5-8807), and by the Montana Agricultural Experiment Station (projects 911398 and 911352).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 4.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 5.Burton, N. P., and P. R. Norris. 2000. Microbiology of acidic, geothermal springs of Montserrat: environmental rDNA analysis. Extremophiles 4:315-320. [DOI] [PubMed] [Google Scholar]
- 6.Clifton, C. G., C. C. Walters, and B. R. T. Simoneit. 1990. Hydrothermal petroleums from Yellowstone National Park, Wyoming, USA. Appl. Geochem. 5:169-191. [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colores, G. M., R. E. Macur, D. M. Ward, and W. P. Inskeep. 2000. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 66:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993. [Google Scholar]
- 10.Dedysh, S. N., Y. Y. Berestovskaya, L. V. Vasylieva, S. E. Belova, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, W. Liesack, and G. A. Zavarzin. 2004. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54:151-156. [DOI] [PubMed] [Google Scholar]
- 11.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 12.Des Marais, D. J., J. H. Donchin, N. L. Nehring, and A. H. Truesdell. 1981. Molecular carbon isotopic evidence for the origin of geothermal hydrocarbons. Nature 292:826-828. [Google Scholar]
- 13.Dore, S. Y., Q. E. Clancy, S. M. Rylee, and C. F. Kulpa. 2003. Naphthalene-utilizing and mercury-resistant bacteria isolated from an acidic environment. Appl. Microbiol. Biotechnol. 63:194-199. [DOI] [PubMed] [Google Scholar]
- 14.Evans, H. J., B. Koch, and R. Klucas. 1972. Preparation of nitrogenase from nodules and separation into components. Methods Enzymol. 24:470-476. [DOI] [PubMed] [Google Scholar]
- 15.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gemmell, R. T., and C. J. Knowles. 2000. Utilisation of aliphatic compounds by acidophilic heterotrophic bacteria. The potential for bioremediation of acidic wastewaters contaminated with toxic organic compounds and heavy metals. FEMS Microbiol. Lett. 192:185-190. [DOI] [PubMed] [Google Scholar]
- 17.Goebel, B. M., and E. Stackebrandt. 1994. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl. Environ. Microbiol. 60:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunter, B. D., and B. C. Musgrave. 1971. New evidence on the origin of methane in hydrothermal gases. Geochim. Cosmochim. Acta 35:113-118. [Google Scholar]
- 20.Hallberg, K. B., Å. K. Kolmert, D. B. Johnson, and P. A. Williams. 1999. A novel metabolic phenotype among acidophilic bacteria: aromatic degradation and the potential use of these organisms for the treatment of wastewater containing organic and inorganic pollutants, p. 719-728. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century. Elsevier, Amsterdam, The Netherlands.
- 21.Hiraishi, A., Y. Matsuzawa, T. Kanbe, and N. Wakao. 2000. Acidisphaera rubrifaciens gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium isolated from acidic environments. Int. J. Syst. Evol. Microbiol. 50:1539-1546. [DOI] [PubMed] [Google Scholar]
- 22.Hostettler, F. D., and K. A. Kvenvolden. 1994. Geochemical changes in crude oil spilled from the Exxon Valdez supertanker into Prince William Sound, Alaska. Org. Geochem. 21:927-936. [Google Scholar]
- 23.Johnson, D. B. 1998. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 27:307-317. [Google Scholar]
- 24.Johnson, D. B., and T. A. M. Bridge. 2002. Reduction of ferric iron by acidophilic heterotrophic bacteria: evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J. Appl. Microbiol. 92:315-321. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, D. B., and K. B. Hallberg. 2003. The microbiology of acidic mine waters. Res. Microbiol. 154:466-473. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. B., N. Okibe, and F. F. Roberto. 2003. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch. Microbiol. 180:60-68. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, D. B., and F. F. Roberto. 1997. Heterotrophic acidophiles and their roles in the bioleaching of sulfide minerals, p. 259-280. In D. E. Rawlings (ed.), Biomining: theory, microbes and industrial processes. Springer-Verlag/Landes Bioscience, Georgetown, Tex.
- 28.Johnson, D. B., S. Rolfe, K. B. Hallberg, and E. Iversen. 2001. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3:630-637. [DOI] [PubMed] [Google Scholar]
- 29.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 31.López-Archilla, A. I., I. Marín, and R. Amils. 2001. Microbial community composition and ecology of an acidic aquatic environment: the Tinto River, Spain. Microb. Ecol. 41:20-35. [DOI] [PubMed] [Google Scholar]
- 32.Love, J. D., and J. M. Good. 1970. Hydrocarbons in thermal areas, northwestern Yellowstone National Park, Wyoming. U.S. Geological Survey Professional Paper 644-B. U.S. Geological Survey, Washington, D.C.
- 33.Love, J. D., and J. M. Good. 1982. Hydrocarbons in thermal areas, northwestern Wyoming, p. 265-288. In Wyoming Geological Association guidebook. Thirty-Third Annual Field Conference 1982. Wyoming Geological Association Publications, Casper, Wyo.
- 34.Margesin, R., and F. Schinner. 2001. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 56:650-663. [DOI] [PubMed] [Google Scholar]
- 35.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehaus, F., C. Bertoldo, M. Kähler, and G. Antranikian. 1999. Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 51:711-729. [DOI] [PubMed] [Google Scholar]
- 37.Norris, P. R., and D. B. Johnson. 1998. Acidophilic microorganisms, p. 133-154. In K. Horikoshi and W. D. Grant (ed.), Extremophiles: microbial life in extreme environments. Wiley-Liss, Inc., New York, N.Y.
- 38.Page, A. L., R. H. Miller, and D. R. Keeney. 1982. Methods of soil analysis, part 2. Soil Science Society of America, Madison, Wis.
- 39.Rawlings, D. E. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65-91. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh, S., T. Suzuki, and Y. Nishimura. 1998. Proposal of Craurococcus roseus gen. nov., sp. nov. and Paracraurococcus ruber gen. nov., sp. nov., novel aerobic bacteriochlorophyll a-containing bacteria from soil. Int. J. Syst. Bacteriol. 48:1043-1047. [DOI] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 42.Smits, T. H. M., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ. Microbiol. 1:307-317. [DOI] [PubMed] [Google Scholar]
- 43.Sparks, D. L., A. L. Page, P. A. Helmke, and R. H. Loeppert. 1996. Methods of soil analysis, part 3. Soil Science Society of America, Madison, Wis.
- 44.Stapleton, R. D., D. C. Savage, G. S. Sayler, and G. Stacey. 1998. Biodegradation of aromatic hydrocarbons in an extremely acidic environment. Appl. Environ. Microbiol. 64:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, D. M., R. M. Atlas, P. D. Boehm, and J. A. Calder. 1980. Microbial biodegradation and chemical evolution of oil from the Amoco spill. AMBIO 9:277-283. [Google Scholar]
- 48.Wiegant, W. W., and J. A. M. de Bont. 1980. A new route for ethylene glycol metabolism in Mycobacterium E44. J. Gen. Microbiol. 120:325-331. [Google Scholar]
- 49.Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita, S., T. Uchimura, and K. Komagata. 2004. Emendation of the genus Acidomonas Urakami, Tamaoka, Suzuki and Komagata 1989. Int. J. Syst. Evol. Microbiol. 54:865-870. [DOI] [PubMed] [Google Scholar]