Abstract
Nine biological species, or mating populations (MPs), denoted by letters A to I, and at least 29 anamorphic Fusarium species have been identified within the Gibberella fujikuroi species complex. Members of this species complex are the only species of the genus Fusarium that contain the gibberellin (GA) biosynthetic gene cluster or at least parts of it. However, the ability of fusaria to produce GAs is so far restricted to Fusarium fujikuroi, although at least six other MPs contain all the genes of the GA biosynthetic gene cluster. Members of Fusarium proliferatum, the closest related species, have lost the ability to produce GAs as a result of the accumulation of several mutations in the coding and 5′ noncoding regions of genes P450-4 and P450-1, both encoding cytochrome P450 monooxygenases, resulting in metabolic blocks at the early stages of GA biosynthesis. In this study, we have determined additional enzymatic blocks at the first specific steps in the GA biosynthesis pathway of F. proliferatum: the synthesis of geranylgeranyl diphosphate and the synthesis of ent-kaurene. Complementation of these enzymatic blocks by transferring the corresponding genes from GA-producing F. fujikuroi to F. proliferatum resulted in the restoration of GA production. We discuss the reasons for Fusarium species outside the G. fujikuroi species complex having no GA biosynthetic genes, whereas species distantly related to Fusarium, e.g., Sphaceloma spp. and Phaeosphaeria spp., produce GAs.
Gibberella fujikuroi is a species complex of at least nine different biological Fusarium species, frequently named mating populations (MPs) (20, 70), and at least 29 closely related Fusarium species mostly corresponding to Fusarium section Liseola (38). These Fusarium species have been isolated from different host plants (24) and are the causative agents of numerous serious plant diseases. Furthermore, these species have been demonstrated to produce a broad variety of secondary metabolites, such as gibberellins (GAs) (58), the pigment bikaverin (21, 29), and the mycotoxins moniliformin (11, 26), fumonisin (9, 26, 42), fusaric acid (2), fusarin C (52), and beauvericin (11, 30), that in some cases may affect human and animal health. Among these mycotoxins, fumonisins are the most prominent toxins, primarily produced by members of the maize pathogens Fusarium verticillioides (MP-A) and Fusarium proliferatum (MP-D) but also by some Fusarium fujikuroi (MP-C) strains (9, 25, 42). In contrast, production of gibberellic acid (GA3) is so far restricted to F. fujikuroi (33), which is responsible for the well-known bakanae disease of rice, which causes the characteristic overgrowth symptom. F. fujikuroi is used commercially for the large-scale production of GAs, mainly GA3 and its precursors, GA4 and GA7, which have applications in agriculture and horticulture as plant growth regulators (43).
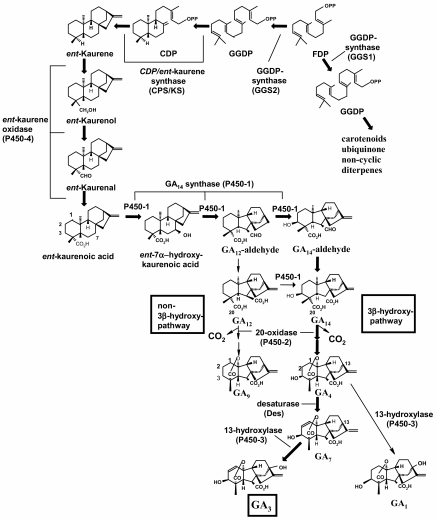
We have previously characterized the seven GA biosynthesis genes and the encoded enzymes in F. fujikuroi (46, 58, 62-64). Besides four cytochrome P450 monooxygenase genes (P450-1 to P450-4), which in most cases encode multifunctional enzymes catalyzing several biosynthetic steps, the GA gene cluster contains genes encoding a pathway-specific geranylgeranyl diphosphate (GGDP) synthase (ggs2), the ent-copalyl diphosphate/ent-kaurene synthase (cps/ks), and GA4-desaturase (des), responsible for early and late steps of the GA biosynthesis pathway (Fig. 1). GGDP is a common precursor for carotenoids, ubiquinones, and GAs (Fig. 1). In F. fujikuroi, two GGDP synthase-encoding genes, ggs1 (36) and ggs2 (58), the latter located in the GA gene cluster, have been found. Interestingly, GGDP for GA biosynthesis is specifically produced by GGS2, as disruption of the corresponding gene, ggs2, resulted in the total loss of GA production, while ggs1, which is responsible for general terpenoid biosynthesis, was unable to complement the block at GGS2 (B. Tudzynski, unpublished data).
FIG. 1.
Gibberellin biosynthesis pathway indicating the genes, enzymes, and final products. The major pathway is denoted by enlarged arrows and letters.
A single-copy cytochrome P450 reductase gene, cpr-Gf, is required as an electron donor for the activity of the GA-specific as well as all the other NADPH-dependent cytochrome P450 monooxygenases of F. fujikuroi. Deletion of the cpr-Gf gene resulted in substantial loss of GA production (32). Previous expression studies revealed that all GA cluster genes except for P450-3 and cpr-Gf are coregulated; they are repressed by high amounts of nitrogen and up-regulated under nitrogen-limiting conditions. Nitrogen regulation in Gibberella is mediated by AREA-Gf, a general transcription factor, which is involved in both derepression of several nitrogen utilization genes and GA biosynthesis genes under nitrogen starvation (37, 60, 61).
Recently, many extended phylogenetic studies of Fusarium species based on the sequences of several protein-encoding nuclear genes clearly demonstrated that the species belonging to the G. fujikuroi species complex are closely related and have a monophyletic origin (39, 40, 42). Our own recent data on the distribution of GA biosynthetic genes and GA production inside and outside the G. fujikuroi species complex clearly demonstrated that only the nine sexually fertile species and the anamorphic Fusarium species inside the species complex contain the entire GA biosynthetic gene cluster, or at least parts of it (33). Surprisingly, none of the other species of the genus Fusarium, such as F. graminearum (http://www.broad.mit.edu/annotation/fungi/fusarium/), F. oxysporum, F. sporotrichoides, F. avenaceum, or F. culmorum, contain any of these genes (33).
All phylogenetic studies to date, including our own based on the sequences of P450-4 from the GA gene cluster and the cpr gene, revealed that F. fujikuroi is most closely related to F. proliferatum (33, 39, 40, 42). In addition, some isolates of these two biological species can still interbreed and produce viable progeny, suggesting that the genetic isolation between them is not complete (27). Despite the close relatedness between these two species, strains of F. proliferatum do not produce GAs, although they contain all seven GA biosynthetic genes (33, 34). Moreover, the GA biosynthetic genes are organized in the same order and orientation in the gene cluster and have a high degree of overall sequence identity (about 90%) with the corresponding F. fujikuroi genes (34). Therefore, we started a detailed molecular analysis of F. proliferatum to identify the reasons for the loss of GA production associated with the divergent evolution of these two species. The recent functional analysis of two cytochrome P450 monooxygenases, P450-1 (D) and P450-4 (D) (where D indicates derivation from F. proliferatum strains), in F. proliferatum revealed a block in ent-kaurene oxidase activity (P450-4 [D]), whereas GA14 synthase (P450-1 [D]) was active in both strains, although the F. proliferatum enzyme is much less active than that in F. fujikuroi (34). Incubations with radiolabeled precursors, such as ent-kaurenoic acid and GA12-aldehyde, revealed genetic and functional diversity between different F. proliferatum strains in some of the GA biosynthetic enzymes, as one strain (D00502) was able to produce GA1 and GA4 as final products, whereas the other strain (D02945) failed to produce any C19 GAs. β-Glucuronidase reporter assays showed that mutations in the GATA-binding elements in the bidirectional promoter of P450-1 and P450-4, which are recognized by AREA-Gf, resulted in low expression levels of both genes in comparison with the corresponding genes in F. fujikuroi (34).
Here, we describe the identification of additional enzymatic blocks in the GA biosynthesis pathway in F. proliferatum causing the loss of the initial steps that result in the formation of GGDP and ent-kaurene. Several mutations in the F. proliferatum ggs2 and cps/ks genes resulted in nonfunctional proteins, as these genes were unable to complement F. fujikuroi ggs2 and cps/ks disruption mutants. Moreover, we could show that the desaturase (DES) that catalyzes the step from GA4 to GA7 is functional in F. proliferatum strain D02945, in contrast to the homologous enzyme in strain D00502, indicating genetic and functional diversity of genes and enzymes within this species. The results indicate that the inability of strains of F. proliferatum to produce GAs results from several mutations in the coding and noncoding regions of three genes (ggs2 [D], cps/ks [D], and P450-4 [D]) that encode the enzymes that catalyze the first steps of the GA-specific pathway rather than from mutations in the regulation network. We demonstrate that complementation of F. proliferatum strains with the three homologous genes from F. fujikuroi (ggs2 [C], cps/ks [C], and P450-4 [C], where C indicates derivation from F. fujikuroi strains) restored the ability to produce GAs.
MATERIALS AND METHODS
Fungal strains and nomenclature.
In association with their classification into the different mating populations, genes and encoded proteins from different Fusarium strains are indicated with the following affixes: C if they were derived from F. fujikuroi strains and D if they were derived from F. proliferatum strains (see also reference 34). Strains IMI58289 (CABI Biosciences, Kew, United Kingdom) and m567 (Fungal Culture Collection, Weimar, Germany) are GA-producing wild-type strains of F. fujikuroi (belonging to G. fujikuroi mating population C[MP-C]). F. proliferatum strains D02945 and D00502 were kindly provided by J. F Leslie (Kansas State University). Strain Δdes (C) is a des deletion mutant of strain IMI58289, which lacks the production of GA3 and GA7 (64). Strain Δggs2 (C) (B. Tudzynski, unpublished) and strain Δcps/ks (C) T22 (59) are disruption mutants that produce no GAs or precursors.
Bacterial strains and plasmids.
Escherichia coli strain TOP10 (Invitrogen, Groningen, The Netherlands) was used for plasmid propagation. Vector pUCBM20 (Boehringer, Mannheim, Germany) was used to clone DNA fragments carrying the F. fujikuroi cluster genes and gene fragments from F. fujikuroi and F. proliferatum. Plasmid pP450-4-GK (62, 63) was used as the complementation vector. To obtain plasmid pGGS2D, carrying the entire ggs2 (D) gene, a PCR approach with primers ggs2-D-F1 and ggs2-D-R1, which were based on the ggs2 (C) sequence, was performed. The 2.3-kb PCR fragment was then cloned into vector pCR2.1-TOPO (Invitrogen). Plasmid pCKSD, containing the entire cps/ks (D) gene copy, was obtained by PCR with primers cps-D-F1 and P450-3-Fus8. The 5.2-kb fragment was cloned into vector pCR-Blunt II-TOPO (Invitrogen). For complementation of the Δdes mutant (MP-C), vector pdesGKD was used. This plasmid was produced by cloning a 3-kb SalI fragment of λ-clone DIII-1 into pUCBM20/SalI. In cotransformation experiments, either pAN7.1 (hygromycin resistance) or pNR1 (nourseothricin resistance) (34) was used for selection of transformants. The vector pNR1 was constructed by cloning the PstI/BamHI fragment of the Streptomyces noursei nat1 gene encoding the nourseothricin acetyltransferase (22) into pBluescript II KS. The gene was transcribed under the control of the Aspergillus nidulans oliC promoter (67) and terminated by the Botrytis cinerea tub1 terminator (J. A. L. van Kan, personal communication). Cosmid pGKScos1, containing both genes ggs2 and cps/ks of MP-C strain m567, was constructed by P. Linnemannstöns (unpublished data).
Media and culture conditions.
For DNA isolation, fungal strains were grown in 100 ml of liquid CM medium optimized for Fusarium spp. (41) for 3 days at 28°C on a rotary shaker at 200 rpm. The mycelium was harvested by filtration through a sterile glass filter (G2; Schott, Jena, Germany), washed with sterile distilled water, frozen in liquid nitrogen, and lyophilized for 24 h. The lyophilized mycelia were ground to a fine powder with a mortar and pestle. F. proliferatum strains were cultivated on V8 juice agar for sporulation. For RNA isolation, fungal strains were grown in 100%, 20%, or 0% ICI medium (13) containing 8% glucose, 0.5% MgSO4, 0.1% KH2PO4, and 5.0, 1.0, or 0 g/liter NH4NO3, respectively.
For analysis of gene expression and nitrogen regulation, strains were cultivated for 3 days in 10 or 20% ICI medium on a rotary shaker at 28°C. To elucidate nitrogen regulation, mycelium was washed, and 1.5 g (wet weight) each was transferred to 50 ml of 0% or 100% ICI medium. For GA production, the strains were grown for 7 to 10 days on a rotary shaker (200 rpm) at 28°C in 300-ml Erlenmeyer flasks containing 100 ml of either 20% ICI or optimized production medium containing 6% sunflower oil, 0.05% (NH4)2SO4, 1.5% corn-steep solids (Sigma-Aldrich, Taufkirchen, Germany), and 0.1% KH2PO4.
DNA and RNA isolation.
Genomic DNA was isolated from lyophilized mycelium as described previously by Doyle and Doyle (10). Lambda DNA from positive lambda clones was prepared according to a method described previously by Sambrook et al. (48). Plasmid DNA was extracted using Genomed (Bad Oeynhausen, Germany) columns according to the manufacturer's protocol. RNA was isolated using the RNAgents total RNA isolation kit (Promega, Mannheim, Germany).
PCR.
The reaction mixtures contained 25 ng DNA, 10 ng of each primer, 0.2 mM deoxynucleoside triphosphates, and 2 U Taq polymerase (Red Taq; Sigma-Aldrich, Deisenhofen, Germany) in 50 μl. PCR was carried out at 94°C for 4 min followed by 35 cycles of 94°C for 1 min, 55 to 65°C for 1 to 3 min, and 72°C for 1 min. Annealing and elongation times were applied differently, depending on the annealing temperatures of each primer and the amplified fragment. For amplification of cps/ks (D), a special high-fidelity proofreading DNA polymerase was used (Phusion; Finnzymes, Finland).
For amplification of ggs2 (D) and analysis of complementation of Δggs2 (C), primers ggs2-D-F1 (5′-CTAGAAAGCGGAGGTAATCAATAAAGT-3′) and ggs2-D-R1 (5′-GGAAACAAGACCTAGCATAAAAAGAAT-3′) were used.
For amplification of cps/ks (D) and analysis of the complementation of Δcps/ks (C), primers cps-D-F1 (5′-ATGATAATGTTGTGATGTGTTCGTT-3′) and P450-3-Fus-8 (5′-CCGAACGGACGCTGGGTAAAAA-3′) were used.
For analysis of full-length integration of vector pP450-4-GK after retransformation into strains D02945 and D00502, transformants were analyzed using primers REV (5′-GAGCGGATAACAATTTCACACAGG-3′) and P450-4-GD2 (5′-CGTGGTCTTCCTTTCCCATCTGGC-3′).
For analysis of complementation of Δdes (C), primers ORF3-1 (5′-GCCAGTGCGCAAGAGTGTCACTGC-3′) and ORF3-2 (5′-′TCTCACTTCCTCCTTGTCAGTTCC-3′) were used.
Screening the lambda DASH II library.
About 35,000 recombinant phages from a lambda DASH II library (Stratagene) prepared from genomic DNA of G. fujikuroi strain D02945 were plated and transferred to Nylon N+ membranes (Amersham Pharmacia, Freiburg, Germany). Hybridization was performed at high stringency (65°C). The blots were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C followed by a wash with 0.1× SSC-0.1% SDS at 65°C. Positive recombinant phages were used for a second round of plaque purification.
Southern and Northern blot analyses.
After digestion with restriction endonucleases and electrophoresis, genomic or lambda DNA was transferred onto Hybond N+ filters (Amersham Pharmacia, Freiburg, Germany). 32P-labeled probes were prepared using the random oligomer-primer method (48). Filters were hybridized at 65°C or 56°C in 5× Denhardt's solution containing 5% dextran sulfate (48). Filters were washed at the same temperature used for hybridization in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% SDS and 1× SSPE-0.1% SDS.
Northern blot hybridizations were accomplished according to the method described previously by Church and Gilbert (8). The G. fujikuroi rRNA gene was used as a control hybridization probe to confirm RNA transfer. For quantification of expression levels, the method of densitometry was performed by using TotalLAB 1 D software, version 1.00 (Amersham, Pharmacia). Hybridizing bands were each normalized against the corresponding band of the wild-type (MP-C) gene copy.
Sequencing.
DNA sequencing of recombinant plasmid clones was accomplished with an automatic sequencer (LI-COR 4000; MWG, München, Germany). The two strands of overlapping subclones obtained from the genomic DNA clones were sequenced using the universal and the reverse primers or specific oligonucleotides obtained from MWG Biotech (München, Germany). DNA and protein sequence alignments were made with DNA Star (Madison, WI).
Transformation of Fusarium strains.
The preparation of protoplasts and the transformation procedure were performed as previously described (32, 34). For complementation experiments, 107 protoplasts (50 μl) of strain D00502, D02945, Δdes-MP-C, Δggs2 (C), or Δcps/ks (C) were transformed with up to 10 μg of the circular complementation vectors pP450-4-GK and pGKScos1. Plasmids pdes-GKD, pP450-4GK, pGGS2D, and pCKSD were cotransformed with pNR1 (32) carrying the nourseothricin resistance marker.
Transformed protoplasts were regenerated at 28°C on complete regeneration agar [0.7 M sucrose, 0.05% yeast extract, 0.1% (NH4)2SO4] containing 120 μg/ml hygromycin B (Calbiochem, Bad Soden, Germany) or 80 μg/ml nourseothricin (Werner BioAgents, Jena, Germany) for 6 to 7 days. For purification, single conidial cultures were obtained from hygromycin B- or nourseothricin-resistant transformants and used for DNA isolation and Southern blot analysis.
Gibberellin analysis.
GA3, GA4, and GA7 in the culture fluids of wild-type strain IMI58289, MD-D02945, and different transformants were analyzed by high-performance liquid chromatography (HPLC) according to a method described previously by Barendse et al. (3) using a Merck HPLC system with a UV detector and a Lichrospher 100 RP18 column (5 μm, 250 by 4 mm). These GAs were also analyzed by thin-layer chromatography (TLC) on silica gel eluted with ethyl acetate/chloroform/acetic acid (60:40:5). The complete GA complement produced by the different strains was determined by gas chromatography-mass spectrometry (GC-MS) analysis after extraction from the culture fluid as previously described (64).
Nucleotide sequence accession numbers.
The gene sequences for des (D), ggs2 (D), and cps/ks (D) of F. proliferatum have been deposited in the GenBank database under accession numbers AJ628021, AJ 810803, and AJ810802, respectively.
RESULTS
Biochemical analysis of the initial steps of GA biosynthesis in strains of F. proliferatum.
Previously, we reported that F. proliferatum strains D02945 and D00502 do not produce any biologically active C19 GAs, including GA3, GA4, GA7, and GA1 (33, 34). Moreover, we could demonstrate an enzymatic block at F. proliferatum ent-kaurene oxidase (P450-4 [D]), whereas F. proliferatum GA14 synthase (P450-1 [D]) shows low but significant enzymatic activity when the corresponding precursors were incubated with these strains (34).
In order to investigate whether there are also enzymatic blocks at the earliest steps of the GA biosynthesis pathway, the synthesis of GGDP by GGS2 and the two-step cyclization of GGDP to ent-kaurene via ent-copalyl diphosphate (Fig. 1), GC-MS analyses of culture filtrates and mycelial extracts were carried out. No GAs or the kaurenoid precursors ent-kaurene and ent-kaurenoic acid were detected in the culture fluids or the respective mycelial extracts of strains D00502 and D02945 (data not shown). These results indicate a putative block in the GA biosynthetic pathway in F. proliferatum at the stage of geranylgeranyl diphosphate synthase (GGS2 [D]) and/or ent-copalyl diphosphate/ent-kaurene synthase (CPS/KS [D]).
GA biosynthesis in F. proliferatum is blocked at the initial steps of the pathway.
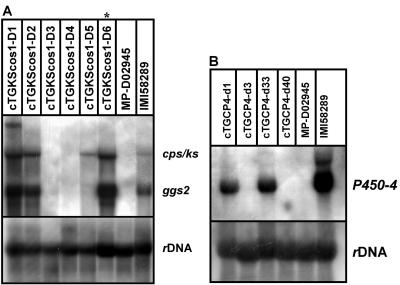
To confirm the proposed block in GGS2 and/or CPS/KS activities in F. proliferatum, we transformed the cosmid pGKScos1, carrying ggs2 (C) and cps/ks (C) genes of F. fujikuroi (strain m567), into F. proliferatum (D02945). Four positive transformants were confirmed by Southern blot analysis and shown to contain one to three copies of the entire genomic fragment (data not shown). Northern blot analysis clearly demonstrated high transcript levels for both genes, comparable to those in F. fujikuroi wild-type strain IMI58289 (Fig. 2A). Analysis of transformants cTGKScos1-D2 and -D6 revealed high levels of ent-kaurene in the mycelia, whereas the recipient strain, D02945, as well as strain D00502 did not accumulate ent-kaurene, as expected (data not shown). Therefore, we were able to restore the initial enzymatic steps of GA biosynthesis in F. proliferatum.
FIG. 2.
Restoration of GA production in F. proliferatum strain D02945. (A) Northern blot analysis after transformation of strain D02945 with pGKScos1. (B) Northern blot analysis after transformation of strain cTGKScos-D6 with pP450-4-GK. Strains were grown for 3 days in 20% ICI medium. *, used for further transformation experiments. rRNA (rDNA) was used as a loading control.
In order to identify the mutations in ggs2 (D) and/or cps/ks (D) that cause the enzymatic block, we cloned the ggs2 gene of strain D02945 (GenBank accession number AJ810803) and analyzed its expression and functionality in the F. fujikuroi background. Sequence alignment of the promoter regions of ggs2 from F. fujikuroi and F. proliferatum indicated 91% sequence identity. The ggs2 promoters from both species contained 10 GATA sequence elements, which are putative binding sites for the nitrogen regulator AreA-Gf (37). Sequence analysis of the coding regions revealed 94% and 92% identity at the nucleotide and amino acid levels, respectively. The positions of the introns and of five characteristic domains, representing the catalytic center (5), were found to be highly conserved in both species (Fig. 3A).
FIG. 3.
Comparison of GGS2 and CPS/KS from F. fujikuroi and F. proliferatum (D02945). (A) Alignment of GGS2 (C) and GGS2 (D). Conserved amino acid residues of the five catalytic domains (5) are marked in boldface type and are overlined. Amino acid substitutions are indicated by asterisks. (B) Alignment of CPS/KS (C) and CPS/KS (D). Conserved amino acid residues of the aspartate-rich domains (54, 68, 69) are indicated in boldface type and are overlined. Amino acid substitutions are indicated by asterisks.
Mutations resulting in amino acid substitutions were identified particularly in the N-terminal region of the protein in front of domain I and between the catalytic domains (Fig. 3A). Interestingly, insertion of an additional cytosine at the 3′ coding region of the F. proliferatum ggs2 (D) led to a translational termination different from that in the F. fujikuroi ggs2 gene (Fig. 3A). In addition, only a faint band was detected for F. proliferatum ggs2 in Northern blot analysis, indicating low levels of expression (Fig. 2A).
In order to investigate the functionality of ggs2 (D), we transformed vector pGGS2D into the F. fujikuroi disruption mutant Δggs2 (C), which fails to produce GAs. Transformants were screened for correct integration of the entire ggs2 (D) gene by PCR with primers ggs2-D-F1 and ggs2-D-R1 and by Southern blot analysis (data not shown). Ten transformants, all carrying two or three entire gene copies, were chosen for Northern blot analysis. Interestingly, only a faint, hardly detectable transcript was found for ggs2 (D) in the transformants, and a smear of hybridizing RNA fragments appeared, starting from the postulated size of ggs2 (data not shown). Analysis of transformants by TLC and HPLC revealed no GA production.
Similarly, F. proliferatum cps/ks (D) was cloned by PCR. The nucleotide sequence of the cps/ks (D) gene can be accessed as GenBank accession number AJ810802. Sequencing of three independent clones indicated 89% identity with the F. fujikuroi cps/ks (C), at both the nucleotide and the amino acid levels. Typical aspartate-rich domains (see references 1, 68, and 69) are conserved between CPS/KS from F. fujikuroi and that from F. proliferatum (Fig. 3B). Amino acid substitutions were distributed over the entire protein. As cps/ks (D) expression was not detectable in Northern blot analyses of strain D02945 (Fig. 2A), functionality of the corresponding CPS/KS (D) enzyme was investigated by transformation of the corresponding gene into F. fujikuroi Δcps/ks deletion strain T22 (58). Putative transformants were checked for full-length integration by PCR and Southern blot analysis. cps/ks (D) expression levels in 11 transformants were determined by Northern blot analysis. In contrast to the F. proliferatum ggs2 gene, significant expression was detected for some high-copy transformants, but in each case, the level was only about 50% of that of cps/ks in F. fujikuroi. Analysis of extracts of culture filtrates by TLC and HPLC revealed that there was no GA production in these transformants.
In summary, neither ggs2 (D) nor cps/ks (D) was able to restore GA production in the corresponding F. fujikuroi Δggs2 and Δcps/ks deletion strains. Therefore, GGS2 (D) and CPS/KS (D) were not functional. Additionally, RNA decay in the case of ggs2 (D) and the generally low expression levels of both genes in the F. proliferatum background could also contribute to the failure to produce GAs.
Restoration of GA production in F. proliferatum by complementation of two strains with ggs2 (C), cps/ks (C), and P450-4 (C).
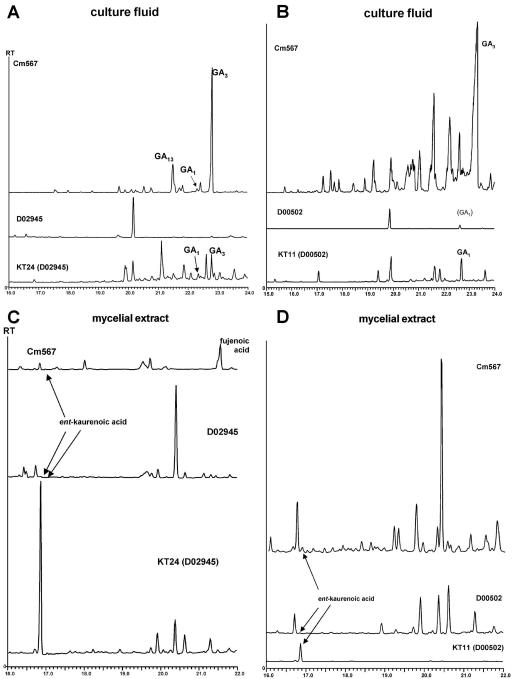
In order to restore GA production in both F. proliferatum strains, we first transformed plasmid pP450-4-GK, carrying the entire F. fujikuroi P450-4 gene copy, into D02945 transformant cTGKScos-D6, which was shown to accumulate ent-kaurene after complementation with the F. fujikuroi ggs2 (C) and cps/ks (C) genes (see above and Fig. 2A). Twenty positive transformants were confirmed by PCR and Southern blot analysis and shown to carry one to three entire P450-4 (C) gene copies. However, Northern blot analysis revealed a significant expression level of P450-4 (C) in only 10 transformants (shown for cTGCP4-d1 and -33 in Fig. 2B), whereas most transformants did not show any expression by Northern analysis, despite carrying multiple gene copies. Analysis of several independent transformants by TLC, HPLC, and GC-MS showed that GA production capacity was restored in these lines. Interestingly, GA amounts corresponded closely with the gene expression levels, although total GA amounts in the transformants were significantly less than those in F. fujikuroi wild-type strain m567 (Table 1 and Fig. 4A and C). For example, as shown for transformant KT24 (D02945), significant amounts of GA1 and GA3 and several other unidentified compounds were detected in the culture filtrate. In addition, high levels of the intermediate ent-kaurenoic acid were found in the mycelial extract from KT24, whereas the wild-type strains of F. fujikuroi and F. proliferatum contain only traces of this compound (Fig. 4C).
TABLE 1.
Qualitative analysis of GAs and related metabolites in F. fujikuroi wild-type strain m567, F. proliferatum strains D02945 and D00502, and transformants of both F. proliferatum strains after complementation of the blocks in the first three enzymes of the pathwaya
| Strain | Products
|
|
|---|---|---|
| Culture filtrate | Mycelial extract | |
| m567 (MP-C) | GA3; GA13; fujenoic acid; GA3 isolactone; 7β,18-dihydroxykaurenolide; GA16; gibberellenic acid; GA7; GA25; GA1; GA9; GA4 | ent-kaurenoic acid; fujenoic acid; GA25; GA9; ent-kaurene; ent-kaurenal; ent-kaurenol |
| D02945 (MP-D) | ent-kaurenoic acid (trace) | |
| KT24 (D02945) | GA3; GA1; GA4; GA3 isolactone; fujenoic acid; ent-kaurenoic acid; GA13 | ent-kaurenoic acid; ent-kaurenal; ent-kaurenol; ent-kaurene |
| D00502 (MP-D) | GA1 (trace) | ent-kaurenoic acid (trace) |
| KT11 (D00502) | GA1; GA13; fujenoic acid | ent-kaurenoic acid; ent-kaurene; ent-kaurenal (trace) |
See the text for details. Strains were cultivated for 10 days in 20% ICI. Substances are listed in decreasing order of amounts. Experiments were repeated with similar results.
FIG. 4.
Qualitative GC-MS analysis of F. proliferatum strains D02945 and D00502 before and after complementation with ggs2 (C) and cps/ks (C) in comparison with MP-C wild-type strain m567. Strains were incubated for 10 days in 20% ICI. For full details, see Materials and Methods. (A) Culture filtrate of m567, D02945, and transformant KT24; (B) culture filtrate of m567, D00502, and transformant KT11; (C) mycelial extract of m567, D02945, and transformant KT24; (D) mycelial extract of m567, D00502, and transformant KT11.
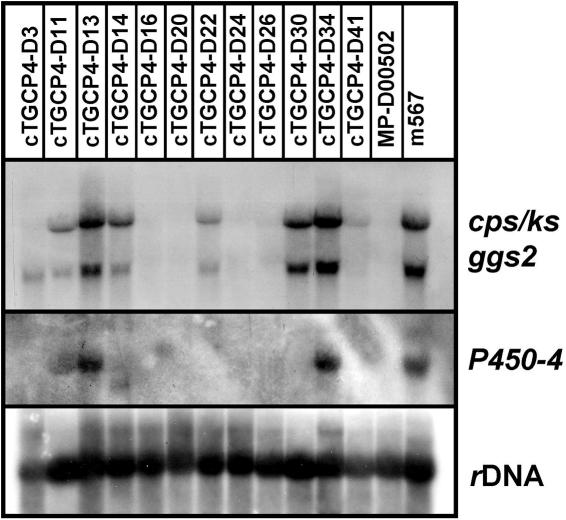
Strain D00502 was shown to metabolize ent-[14C]kaurenoic acid to [14C]GA1, demonstrating that enzymes P450-1, P450-2, and P450-3 are active in this strain but indicating a block at DES (see reference 34 and Fig. 1). Therefore, in a second experiment, we transformed pGKScos1 and pP450-4-GK into this strain in one step, producing 12 transformants, all carrying one to three entire copies of ggs2 (C), cps/ks (C), and P450-4 (C) (data not shown). Northern blot analysis revealed that ggs2 (C) and cps/ks (C) were expressed in all the transformants but at different levels. Three transformants, D11, D13, and D34, also showed a transcript for P450-4 (C) by Northern blot analysis (Fig. 5).
FIG. 5.
Northern blot analysis of ggs2, cps/ks, and P450-4 gene expression in F. fujikuroi (m567) and F. proliferatum (D00502) strains and in cTGKScos-D2 after transformation with pP450-4-GK. Strains were grown for 3 days in 20% ICI medium. rRNA (rDNA) was used as a loading control.
Analysis of these transformants by GC-MS revealed significant levels of GA1 in the culture fluids, whereas recipient strain D00502 accumulated only traces of this compound (Table 1 and Fig. 4B). Moreover, no GA3 was found in any of the D00502 transformants (shown for transformant KT11 [D00502] in Fig. 4B and D), in contrast to the D02945 transformants discussed above.
Thus, complementation of blocks in the first three enzymes of the pathway restored GA production to both F. proliferatum strains, although strain D00502 appears to lack DES activity since no GA3 was found in any transformant from this strain.
Functional analysis of des (D), encoding the GA4 desaturase.
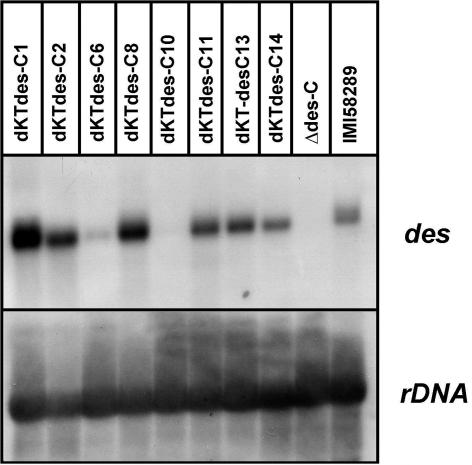
The des gene encodes the GA4 desaturase, which catalyzes the conversion of GA4 to GA7. As mentioned above, strain D00502 accumulates [14C]GA1 and [14C]GA4 rather than [14C]GA3 when incubated with precursors such as [14C]GA12-aldehyde, suggesting a defect in the GA4 DES (34). For strain D02945, incubations with ent-[14C]kaurenoic acid or [14C]GA12-aldehyde did not result in the production of any C19 GAs. We obtained the corresponding des (D) gene from F. proliferatum after screening the lambda DASH II genomic library of strain D02945 with a cDNA clone of F. fujikuroi des (C) as a probe. Sequence analysis of des from D02945 (GenBank accession number AJ628021) shows 92% and 96% identity at the nucleotide and amino acid levels, respectively, with des of F. fujikuroi. As for P450-1 (D) and P450-4 (D) (34), des expression levels were very low in F. proliferatum. In order to analyze the functionality of the encoded GA4 desaturase in detail, we transformed the entire des (D) gene into a F. fujikuroi Δdes deletion strain of IMI58289, which is unable to produce GA7 and GA3 and, like strain D00502, accumulates GA1 and GA4 instead (64; Fig. 1). The gene was integrated with different copy numbers into the genome of strain Δdes (C) and was highly expressed in some of the transformants (Fig. 6). To investigate the functionality of DES (D), the concentrations of C19 GAs in culture filtrates, determined by HPLC, were compared in strains Δdes (C) and three of the transformants (dKTΔdes-C1, -C2, and -C10) (Table 2). As expected, F. fujikuroi mutant strain Δdes (C) accumulated GA4, which is partially metabolized to GA1 by the 13-hydroxylase P450-3 (64) (data not shown; Table 2). In contrast, transformants dKTΔdes-C1 and -C2 showed the wild-type GA pattern: the main component was GA3 rather than GA1 and GA4. Thus, des (D) fully restored the ability to produce GA3 via GA7, overcoming the block between GA4 and GA7. Therefore, des (D) of strain D02945 encodes a functional protein, in contrast to the corresponding gene of D00502 (see above).
FIG. 6.
Northern blot analysis of des gene expression in F. fujikuroi IMI58289 (wild type), the deletion mutant Δdes, and transformants of strain Δdes complemented with the F. proliferatum des gene. Strains were grown for 3 days in 20% ICI medium. rRNA (rDNA) was used as a loading control.
TABLE 2.
Production of GAs in F. fujikuroi strains IMI58289, Δdes, and different transformants of Δdes complemented with the F. proliferatum des geneb
| Strain | Concn (mg/liter)c
|
||
|---|---|---|---|
| GA3 | GA4 | GA7 | |
| IMI58289 | 100 | 45 | 78 |
| Δdes (C) | −a | 340 | — |
| dKTΔdes-C1 | 175 | 7 | 35 |
| dKTΔdes-C2 | 85 | 9 | 130 |
| dKTΔdes-C10 | −a | 365 | — |
GA1, rather than GA3, is the final product.
Strains were harvested after cultivation for 6 days in optimized production medium.
Given values are the averages of two independent measurements. Experiments were repeated three times with similar results. —, less than 1 mg/liter.
DISCUSSION
Recent results demonstrated the highest phylogenetic relatedness between F. proliferatum and F. fujikuroi within the G. fujikuroi species complex (27, 33, 40, 42). Moreover, F. proliferatum contains all of the GA biosynthesis genes, arranged in the same pattern as found in F. fujikuroi, but does not produce any GAs (33, 34). Functional analysis of two of the cytochrome P450 monooxygenase genes, P450-4 (D) and P450-1 (D), and the encoded enzymes in two F. proliferatum strains revealed a block at ent-kaurene oxidase, P450-4 (D), which catalyzes the conversion of ent-kaurene to ent-kaurenoic acid (34) (Fig. 1). Further biochemical analysis of the initial steps of GA biosynthesis in both strains D00502 and D02945 clearly indicated additional blocks at ggs2 (D) and cps/ks (D), encoding GGS2 and CPS/KS, respectively, as no ent-kaurene accumulated in these strains. Complementation of strain D02945 with a cosmid carrying both ggs2 (C) and cps/ks (C) from F. fujikuroi resulted in ent-kaurene accumulation, confirming the conclusion that there are additional blocks in one or both of these genes.
Prenyltransferases such as GGS2 are key enzymes in many essential biosynthetic pathways in which they are necessary for the formation of chlorophyll (49) and ubiquinones (55) in plants as well as carotenoids in plants (16) and fungi (28). In F. fujikuroi, two GGDP synthase-encoding genes, ggs1 (36) and ggs2 (58), have been characterized. One of them, ggs2, is located in the GA biosynthetic gene cluster and is specifically involved in GA production, whereas no pathway-specific ggs gene has been found so far in the F. fujikuroi carotenoid gene cluster (J. Avalos, personal communication).
In the present study, we demonstrated that the GA-specific enzyme GGS2 is not functional in F. proliferatum since complementation of an F. fujikuroi ggs2 (C) mutant with the F. proliferatum ggs2 (D) gene did not restore GA production. There are several reasons for this. Despite the fact that the amino acid sequence of GGS2 (D) is unchanged in the five highly conserved prenyltransferase domains that are important for catalytic activity (1, 5, 6, 35, 51), several mutations between the catalytic domains were found. Furthermore, a substitution of nine consecutive amino acid residues (Fig. 3) was found in the N terminus, which may affect protein folding and, consequently, enzyme activity. In addition to these mutations within the coding region, the low expression level of the ggs2 (D) gene can also contribute to the loss of GGS2 activity. Sequence analysis of the promoter region revealed a 7-nucleotide deletion in front of a GATA element, the binding site of the general transcription factor AREA-Gf (37). Mutations in the region of GATA-binding elements may affect the expression level of AreA-regulated genes, as was shown previously for the bidirectional promoter of P450-1/P450-4 genes in F. fujikuroi (37) and F. proliferatum (34) and for the avirulence gene Avr9 of Cladosporium fulvum (50).
A third reason for loss of enzyme activity could be the nonsense mutation we observed in the F. proliferatum ggs2 (D) gene (Fig. 3A). This mutation is probably the reason for the decay of the corresponding mRNA demonstrated in Northern blot analysis. In filamentous fungi, not much is known about mechanisms and reasons for targeted RNA degradation. Nonsense-mediated RNA decay, the loss of mRNA carrying premature stop codons, is a process by which cells recognize and degrade nonsense mRNAs to prevent possible toxic effects of truncated peptides (44). This process was also found, e.g., for the plant flavonoid 3′ hydroxylase gene in Ipomoea (17) and human cancer cells (18).
Besides P450-4 (D) and GGS2 (D), CPS/KS (D) was also found to be blocked in F. proliferatum. This last enzyme is responsible for ent-kaurene synthesis in the GA biosynthesis pathway (Fig. 1). Domain analyses of CPS/KS (D) clearly showed the existence of several aspartate-rich regions, which were also found to be typical for plant terpene cyclases, including the plant ent-kaurene synthases (54, 58, 68, 69). Thus, CPS/KS (D), like CPS/KS (C) from F. fujikuroi, has the characteristics of a bifunctional enzyme. Besides these two CPS/KS enzymes in different fusaria and FCPS/KS in Phaeosphaeria spp. (19, 56), only the aphidicolin-16β-ol synthase in Phoma betae (57) and abietadiene synthase from Abies grandis (66) have been shown to be bifunctional diterpene cyclases. The terpene cyclase motif D(I/V)DDTA, which is missing in most plant ent-kaurene synthases, was shown to be particularly important for ring closure reactions of terpenoid compounds (53). On the other hand, the DDXXD motif, which is not found in plant CPS enzymes, mediates the binding of the divalent metal ion phosphate complex of the prenyl substrate (1). Attempts to complement the cps/ks (C) deletion mutant of F. fujikuroi with cps/ks (D) failed to restore GA production in the mutant, indicating that cps/ks (D) does not encode a functional enzyme. As shown in Fig. 3B, there are several amino acid substitutions relative to CPS/KS (C) throughout the CPS/KS (D) protein sequence, which may produce incorrect folding or affect substrate binding to the enzyme active site.
One of the aims of this work was to determine if it is possible to overcome all enzymatic blocks and thus restore GA biosynthesis in both F. proliferatum strains. Indeed, complementation of all three enzymatic blocks (at GGS2, CPS/KS, and P450-4) found in these strains resulted in the restoration of GA production, although the nature of the end products differed in the two complemented F. proliferatum strains (Fig. 4). Recently, it was shown for strain D00502 that incubations with radiolabeled substrates for the P450-1 monooxygenase (GA14 synthase) led to the formation of significant but low amounts of the C19 GAs GA1 and GA4, whereas incubation of the same precursors with cultures of strain D02945 resulted in the formation of unidentified oxidation products but no C19 GAs (34). However, significant P450-1 (D) activity was observed in microsomal fractions from strain D02945, which partially converted the precursor [14C]GA12-aldehyde to [14C]GA14 (34). These results indicated that the strains, particularly D02945, contained only low P450-1 activities. The lower P450-1 activity in D02945 compared with D00502 would also explain the failure of strain D02945 to produce GAs after 3 days of incubation with [14C]GA12-aldehyde, although significant amounts of GAs were detected in the strain after genetic complementation of the enzymatic blocks and a longer incubation period of 10 days. The much higher amounts of ent-kaurenoic acid that accumulate in the mycelia of the complemented D02945 strain compared with complemented D00502 (Fig. 4C and D) also suggest very low P450-1 activity in D02945. The activity of P450-1 (D) seems to be rate limiting for the formation of C19 GAs, such as GA1, GA3, or GA4, in the complemented F. proliferatum mutants. It is noteworthy that after 10 days, in contrast to shorter cultivation periods, traces of GA1 were detected in strain D00502, indicating a residual activity for all the GA biosynthetic enzymes in this strain. However, a cross-contamination of corresponding D00502 culture filtrates cannot be excluded, as it seems unlikely that the F. proliferatum P450-4 or CPS/KS has any activity. In complementation transformants of both F. proliferatum strains, high amounts of unidentified ent-kaurenoid-like compounds were detected in addition to the GAs and ent-kaurenoids that are present in the F. fujikuroi strains (Fig. 4). These unidentified compounds, which are not present in the untransformed strains, may be formed by the unspecific action of endogenous enzymes on ent-kaurenoic acid, for which they compete with the relatively low-abundance GA biosynthetic enzymes. The low activity of the GA enzymes in F. proliferatum is evident from the much lower amounts of GAs found in the complemented strains compared with those in F. fujikuroi wild-type strain m567.
In the complementation experiments, the site of integration of the introduced gene seems to be important not only for its expression but also for functionality of the encoded enzyme. Integration of the entire P450-4 gene of either F. fujikuroi or F. proliferatum into the genome of strain D02945 did not always result in detectable expression of the transgene (34). Similar results were obtained in experiments with Aspergillus parasiticus, in which β-glucuronidase reporter gene assays with the aflatoxin biosynthetic genes ver1 and nor1 clearly demonstrated that only integration into the homologous site resulted in expression of the fused uidA gene (7). Studies with Aspergillus spp. showed the existence of a global transcriptional regulator, LaeA, for secondary metabolism that is specific for a gene cluster, as transcription of genes upstream and downstream of secondary metabolism gene clusters was unaffected (4). In addition, some transformants of strain D02945 with high expression levels of P450-4 (C) exhibited low levels of activity of the encoded ent-kaurene oxidase, resulting in the formation of low amounts of ent-kaurenoic acid. Therefore, the genetic environment seems to play a fundamental role not only for gene expression but also for activity and functionality of the encoded protein, although the exact reasons for this are still not identified.
Complementation of the F. fujikuroi desaturase deletion strain Δdes (C) with the corresponding desaturase gene from F. proliferatum strain D02945 resulted in the restoration of GA3 and GA7 production (Table 2), demonstrating the functionality of des in strain D02945. In contrast, strain D00502 produces only [14C]GA1 and [14C]GA4 (as observed for the des [C] deletion strain) when incubated with precursors such as ent-[14C]kaurenoic acid or [14C]GA12-aldehyde, indicating that the des gene in this strain does not encode a functional enzyme and/or is poorly expressed (34). Thus, genetic diversity between GA biosynthesis genes occurs not only between isolates of different species but even between sexually compatible strains of one species.
In summary, it is noteworthy that several mutations in coding and 5′ noncoding regions of different genes of the GA cluster led to the total loss of GA production in F. proliferatum. Major blocks were determined at the first three enzymes of the GA biosynthesis pathway: GGS2, CPS/KS, and ent-kaurene oxidase (P450-4). It was clearly shown by expression studies that the regulatory mechanisms are present in F. proliferatum and were not affected during evolution. Thus, the F. fujikuroi ggs2, cps/ks, and P450-4 genes were highly expressed in the F. proliferatum background, allowing the restoration of GA production.
The results presented here raise questions about the origin and evolution of fungal GA gene clusters. After completion of the cloning and characterization of GA biosynthetic genes in F. fujikuroi (reviewed in reference 65), and to a large extent in Arabidopsis thaliana and other plants (reviewed in reference 14), we can exclude the possibility of horizontal gene transfer from higher plants to the fungus for several reasons (reviewed in reference 15). Thus, several oxidation steps are catalyzed by dioxygenases in plants but by P450 monooxygenases in the fungus. Furthermore, the fungal GA biosynthetic genes are organized in a gene cluster but are dispersed in the genome of higher plants.
Surprisingly, only the species in the G. fujikuroi species complex contain the entire gene cluster or parts of it, whereas other species, such as F. graminearum, F. oxysporum, F. sporotrichoides, and F. culmorum, do not contain any of the GA biosynthetic genes in their genomes (33). On the other hand, members of at least two other fungal genera, Sphaceloma spp. and Phaeosphaeria spp., neither of which are closely related to Fusarium, and also bacteria such as Rhizobium phaseoli, Azospirillum lipoferum, and Azospirillum brasilense were also found to produce GAs (reviewed in reference 31). So far, only one gene, cps/ks, has been characterized from Phaeosphaeria species (56). As in F. fujikuroi, the encoded enzyme CPS/KS is bifunctional and catalyzes the first two cyclization steps to produce ent-kaurene via ent-copalyl diphosphate.
Recently, a new diterpene gene cluster has been identified in the fungus Phoma betae which is responsible for the production of aphidicolin, a compound with a GA-like structure (57). This gene cluster contains two P450 monooxygenase genes and a cps/ks-like bifunctional diterpene cyclase gene encoding aphidicolan-16β-ol synthase. Again, the diterpene cyclase gene acts as a functional unit together with a pathway-specific GGDP synthase gene. All these data indicate that different fungal genomes contain basic units of a diterpene biosynthetic gene cluster with a diterpene cyclase-encoding gene linked to a pathway-specific ggs gene by a bidirectional promoter region.
The availability of an increasing amount of fungal sequence data makes it possible to study the distribution and evolution of secondary metabolite gene clusters in general. Thus, a sirodesmin gene cluster was identified in Leptosphaeria maculans (12), and very similar gene clusters have been found in at least four additional distantly related ascomycetes, such as Aspergillus fumigatus, Chaetomium globosum, Magnaporthe grisea, and F. graminearum.
The reasons for the organization of fungal genes involved in secondary metabolism in clusters and the continued retention of such large gene units even if there is no obvious or essential function in the corresponding organism are interesting questions. If such cooperating genes are conditionally dispensable but have adaptive value for colonizing certain ecological niches, the incipient cluster would be maintained by positive selection in those environments (23, 45, 47). The increasing number of sequenced fungal genomes will give us the opportunity to study the evolutionary origin and advantages of secondary metabolite gene clusters in the near future.
Acknowledgments
We thank Sabine Richter for excellent technical assistance.
The project was funded by the DFG (Tu101/9-1) and Fondo Nacional de Desarrollo Cientifico y Tecnologico (grants 1020140 and 7040117). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
REFERENCES
- 1.Ashby, M. N., and P. A. Edwards. 1990. Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 265:13157-13164. [PubMed] [Google Scholar]
- 2.Bacon, C. W., J. K. Porter, W. P. Norred, and J. F. Leslie. 1996. Production of fusaric acid by Fusarium species. Appl. Environ. Microbiol. 62:4039-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barendse, G. W. M., P. H. van de Werken, and N. Takahashi. 1980. High-performance liquid chromatography of gibberellins. J. Chromatogr. 198:449-455. [Google Scholar]
- 4.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli, A., N. Romano, P. Ballario, G. Morelli, and G. Macino. 1991. The Neurospora crassa carotenoid biosynthetic gene (Albino-3) reveals highly conserved regions among prenyltransferases. J. Biol. Chem. 266:5854-5859. [PubMed] [Google Scholar]
- 6.Chen, A., P. A. Kroon, and C. D. Poulter. 1994. Isoprenyl diphosphate synthases: protein sequence comparison, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 3:600-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiou, C. H., M. Miller, D. L. Wilson, F. Trail, and J. E. Linz. 2002. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl. Environ. Microbiol. 68:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desjardins, A. E., R. D. Plattner, T. C. Nelsen, and J. F. Leslie. 1995. Genetic analysis of fumonisin production and virulence of Gibberella fujikuroi mating population A (Fusarium moniliforme) on maize (Zea mays) seedlings. Appl. Environ. Microbiol. 61:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12:13-15. [Google Scholar]
- 11.Fotso, J., J. F Leslie, and J. S. Smith. 2002. Production of beauvericin, moniliformin, fusaproliferin, and fumonisins B1, B2, and B3 by fifteen ex-type strains of Fusarium species. Appl. Environ. Microbiol. 68:5195-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner, D. M., A. J. Cozijnsen, L. M. Wilson, M. S. Pedras, and B. J. Howlett. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53:1307-1318. [DOI] [PubMed] [Google Scholar]
- 13.Geissman, T. A., A. J. Verbiscar, B. O. Phinney, and B. Cragg. 1966. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry 5:933-947. [Google Scholar]
- 14.Hedden, P., and A. L. Phillips. 2000. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5:523-530. [DOI] [PubMed] [Google Scholar]
- 15.Hedden, P., A. L. Phillips, M. C. Rojas, E. Carrera, and B. Tudzynski. 2001. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution. J. Plant Growth Regul. 20:319-331. [DOI] [PubMed] [Google Scholar]
- 16.Hirschberg, J. 2001. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4:210-218. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino, A., Y. Morita, J. D. Choi, N. Saito, K. Toki, Y. Tanaka, and S. Iida. 2003. Spontaneous mutations of the flavonoid 3′-hydroxylase gene conferring reddish flowers in the morning glory species. Plant Cell Physiol. 44:990-1001. [DOI] [PubMed] [Google Scholar]
- 18.Ionov, Y., N. Nowak, M. Perucho, S. Markowitz, and J. K. Cowell. 2004. Manipulation of nonsense mediated decay gene mutations in colon cancer cells with microsatellite instability. Oncogene 23:639-645. [DOI] [PubMed] [Google Scholar]
- 19.Kawaide, H., R. Imai, T. Sassa, and Y. Kamiya. 1997. ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J. Biol. Chem. 272:21706-21712. [DOI] [PubMed] [Google Scholar]
- 20.Kerényi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:4071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjaer, D., A. Kjaer, C. Pedersen, J. D. Bulock, and J. R. Smith. 1971. Bikaverin and norbikaverin, benzoxanthentrione pigments of Gibberella fujikuroi. J. Chem. Soc. C 16:2792-2794. [DOI] [PubMed] [Google Scholar]
- 22.Krügel, H., G. Fiedler, C. Smith, and S. Baumberg. 1993. Sequence and transcriptional analysis of nourseothricin acetyltransferase-encoding gene nat-1 from Streptomyces noursei. Gene 127:127-131. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5:355-359. [DOI] [PubMed] [Google Scholar]
- 24.Leslie, J. F. 1995. Gibberella fujikuroi: available populations and variable traits. Can. J. Bot. 73:S282-S291. [Google Scholar]
- 25.Leslie, J. F., F. J. Doe, R. D. Plattner, D. D. Shackelford, and J. Jonz. 1992. Fumonisin B1 production and vegetative compatibility of strains from Gibberella fujikuroi mating population “A” (Fusarium moniliforme). Mycopathologia 117:37-46. [DOI] [PubMed] [Google Scholar]
- 26.Leslie, J. F., W. F. O. Marasas, G. S. Shephard, E. W. Sydenham, S. Stockenström, and P. G. Thiel. 1996. Duckling toxicity and the production of fumonisin and moniliformin by isolates in the A and F mating populations of Gibberella fujikuroi (Fusarium moniliforme). Appl. Environ. Microbiol. 62:1182-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie, J. F., K. A. Zeller, M. Wohler, and B. A. Summerell. 2004. Interfertility of two mating populations in the Gibberella fujikuroi species complex. Eur. J. Plant Pathol. 110:611-618. [Google Scholar]
- 28.Linnemannstöns, P., M. M. Prado, R. Fernandez-Martin, B. Tudzynski, and J. Avalos. 2002. A carotenoid biosynthesis gene cluster in Fusarium fujikuroi: the genes carB and carRA. Mol. Genet. Genom. 267:593-602. [DOI] [PubMed] [Google Scholar]
- 29.Linnemannstöns, P., J. Schulte, M. M. Prado, R. H. Proctor, J. Avalos, and B. Tudzynski. 2002. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet. Biol. 37:143-148. [DOI] [PubMed] [Google Scholar]
- 30.Logrieco, A., A. Moretti, G. Castella, M. Kostecki, P. Golinski, A. Ritieni, and J. Chelkowski. 1998. Beauvericin production by Fusarium species. Appl. Environ. Microbiol. 64:3084-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacMillan, J. 2001. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20:387-442. [DOI] [PubMed] [Google Scholar]
- 32.Malonek, S., M. C. Rojas, P. Hedden, P. Gaskin, and B. Tudzynski. 2004. The NADPH: cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem. 279:25075-25084. [DOI] [PubMed] [Google Scholar]
- 33.Malonek, S., C. Bömke, M. C. Rojas, P. Hedden, P. Gaskin, E. Bornberg-Bauer, and B. Tudzynski. 2005. Distribution of gibberellin biosynthetic genes and gibberellin production in the Gibberella fujikuroi species complex. Phytochemistry 66:1296-1311. [DOI] [PubMed] [Google Scholar]
- 34.Malonek, S., M. C. Rojas, P. Hedden, P. Gaskin, P. Hopkins, and B. Tudzynski. 2005. Functional characterization of two cytochrome P450 monooxygenase genes, P450-1 and P450-4, of the gibberellic acid gene cluster in Fusarium proliferatum (Gibberella fujikuroi MP-D). Appl. Environ. Microbiol. 71:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marrero, P. F., C. D. Poulter, and P. A. Edwards. 1992. Effects of site-directed mutagenesis of the highly conserved aspartate residues in domain II of farnesyl diphosphate synthase activity. J. Biol. Chem. 267:21873-21878. [PubMed] [Google Scholar]
- 36.Mende, K., V. Homann, and B. Tudzynski. 1997. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol. Gen. Genet. 255:96-105. [DOI] [PubMed] [Google Scholar]
- 37.Mihlan, M., V. Homann, T. W. D. Liu, and B. Tudzynski. 2003. AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol. Microbiol. 47:975-991. [DOI] [PubMed] [Google Scholar]
- 38.Nirenberg, H. I., and K. O'Donnell. 1998. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90:434-458. [Google Scholar]
- 39.O'Donnell, K., and E. Cigelnik. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7:103-116. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell, K., H. Nirenberg, T. Aoki, and E. Cigelnik. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41:61-78. [Google Scholar]
- 41.Pontecorvo, G. V., J. A. Poper, L. M. Hemmonns, K. D. Mac Donald, and A. W. J. Buften. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 141:141-238. [DOI] [PubMed] [Google Scholar]
- 42.Proctor, R. H., R. D. Plattner, D. W. Brown, J. A. Seo, and Y. W. Lee. 2004. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycol. Res. 108:815-822. [DOI] [PubMed] [Google Scholar]
- 43.Rademacher, W. 1997. Gibberellins, p. 193-205. In T. Anke (ed.), Fungal biotechnology. Chapman & Hall, London, United Kingdom.
- 44.Rajavel, K. S., and E. F. Neufeld. 2001. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 21:5512-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reams, A. B., and E. L. Neidle. 2004. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 58:119-142. [DOI] [PubMed] [Google Scholar]
- 46.Rojas, M. C., P. Hedden, P. Gaskin, and B. Tudzynski. 2001. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA 98:5828-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosewich, U. L., and H. C. Kistler. 2000. Role of horizontal gene transfer in the evolution of fungi. Annu. Rev. Phytopathol. 38:325-363. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schledz, M., A. Seidler, P. Beyer, and G. Neuhaus. 2001. A novel prenyltransferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett. 499:15-20. [DOI] [PubMed] [Google Scholar]
- 50.Snoeijers, S. S., A. Perez-Garcia, T. Goosen, and P. J. De Wit. 2003. Promoter analysis of the avirulence gene Avr9 of the fungal tomato pathogen Cladosporium fulvum in the model filamentous fungus Aspergillus nidulans. Curr. Genet. 43:96-102. [DOI] [PubMed] [Google Scholar]
- 51.Song, L. S., and C. D. Poulter. 1994. Yeast farnesyl diphosphate synthase. Site-directed mutagenesis of residues in highly conserved prenyltransferase domain I and domain II. Proc. Natl. Acad. Sci. USA 91:3044-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song, Z., R. Cox, C. M. Lazarus, and T. J. Simpson. 2004. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. ChemBioChem 5:1196-1203. [DOI] [PubMed] [Google Scholar]
- 53.Sun, T. P., and Y. Kamiya. 1994. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6:1509-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, T. P., and Y. Kamiya. 1997. Regulation and cellular localization of ent-kaurene synthesis. Physiol. Plant 101:701-708. [Google Scholar]
- 55.Swiezewska, E., G. Dallner, B. Andersson, and L. Ernster. 1993. Biosynthesis of ubiquinones and plastoquinone in the endoplasmatic reticulum-Golgi membranes of spinach leaves. J. Biol. Chem. 268:1494-1499. [PubMed] [Google Scholar]
- 56.Toyomasu, T., H. Kawaide, A. Ishizaki, S. Shinoda, M. Otsuka, W. Mitsuhashi, and T. Sassa. 2000. Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional diterpene cyclase. Biosci. Biotechnol. Biochem. 64:660-664. [DOI] [PubMed] [Google Scholar]
- 57.Toyomasu, T., K. Nakaminami, H. Toshima, T. Mie, K. Watanabe, H. Ito, H. Matsui, W. Mitsuhashi, T. Sassa, and H. Oikawa. 2004. Cloning of a gene cluster responsible for the biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha. Biosci. Biotechnol. Biochem. 68:146-152. [DOI] [PubMed] [Google Scholar]
- 58.Tudzynski, B., and K. Hölter. 1998. The gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 59.Tudzynski, B., H. Kawaide, and Y. Kamiya. 1998. Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr. Genet. 34:234-240. [DOI] [PubMed] [Google Scholar]
- 60.Tudzynski, B. 1999. Biosynthesis of gibberellins in Gibberella fujikuroi: biomolecular aspects. Appl. Microbiol. Biotechnol. 52:298-310. [DOI] [PubMed] [Google Scholar]
- 61.Tudzynski, B., V. Homann, B. Feng, and G. A. Marzluf. 1999. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol. Gen. Genet. 261:106-114. [DOI] [PubMed] [Google Scholar]
- 62.Tudzynski, B., P. Hedden, E. Carrera, and P. Gaskin. 2001. The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthetic pathway. Appl. Environ. Microbiol. 67:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tudzynski, B., M. C. Rojas, P. Gaskin, and P. Hedden. 2002. The Gibberella fujikuroi gibberellin 20-oxidase is a multifunctional monooxygenase. J. Biol. Chem. 277:21246-21253. [DOI] [PubMed] [Google Scholar]
- 64.Tudzynski, B., M. Mihlan, M. C. Rojas, P. Linnemannstöns, P. Gaskin, and P. Hedden. 2003. Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J. Biol. Chem. 278:28635-28643. [DOI] [PubMed] [Google Scholar]
- 65.Tudzynski, B. 2005. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 66:597-611. [DOI] [PubMed] [Google Scholar]
- 66.Vogel, B. S., M. R. Wildung, G. Vogel, and R. Croteau. 1996. Abietadiene synthase from grand fir (Abies grandis). cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J. Biol. Chem. 271:23262-23268. [DOI] [PubMed] [Google Scholar]
- 67.Ward, M., and G. Turner. 1986. The ATP synthase subunit-9 of Aspergillus nidulans—sequence and transcription. Mol. Gen. Genet. 205:331-338. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi, S., T. Saito, H. Abe, H. Yamane, N. Murofushi, and Y. Kamiya. 1996. Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme. ent-kaurene synthase B from pumpkin (Cucurbita maxima L.). Plant J. 10:203-213. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi, S., T. P. Sun, H. Kawaide, and Y. Kamiya. 1998. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 116:1271-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeller, K. A., B. A. Summerell, and J. F. Leslie. 2003. Gibberella konza (Fusarium konzum) sp. nov. from prairie grasses, a new species in the Gibberella fujikuroi species complex. Mycologia 95:943-954. [PubMed] [Google Scholar]