Abstract
Corynebacterium glutamicum ATCC 13032 was found to be able to utilize a broad range of sulfonates and sulfonate esters as sulfur sources. The two gene clusters potentially involved in sulfonate utilization, ssuD1CBA and ssuI-seuABC-ssuD2, were identified in the genome of C. glutamicum ATCC 13032 by similarity searches. While the ssu genes encode proteins resembling Ssu proteins from Escherichia coli or Bacillus subtilis, the seu gene products exhibited similarity to the dibenzothiophene-degrading Dsz monooxygenases of Rhodococcus strain IGTS8. Growth tests with the C. glutamicum wild-type and appropriate mutant strains showed that the clustered genes ssuC, ssuB, and ssuA, putatively encoding the components of an ABC-type transporter system, are required for the utilization of aliphatic sulfonates. In C. glutamicum sulfonates are apparently degraded by sulfonatases encoded by ssuD1 and ssuD2. It was also found that the seu genes seuA, seuB, and seuC can effectively replace ssuD1 and ssuD2 for the degradation of sulfonate esters. The utilization of all sulfonates and sulfonate esters tested is dependent on a novel putative reductase encoded by ssuI. Obviously, all monooxygenases encoded by the ssu and seu genes, including SsuD1, SsuD2, SeuA, SeuB, and SeuC, which are reduced flavin mononucleotide dependent according to sequence similarity, have SsuI as an essential component. Using real-time reverse transcription-PCR, the ssu and seu gene cluster was found to be expressed considerably more strongly during growth on sulfonates and sulfonate esters than during growth on sulfate.
Corynebacterium glutamicum, a gram-positive, nonsporulating soil bacterium, was identified in 1957 as an l-glutamic acid producer (19). Some information has been obtained recently about the general sulfur metabolism of this industrially important, amino acid-producing microorganism, especially concerning the biosynthesis of the sulfur-containing amino acids l-cysteine and l-methionine (12, 29). Still, little is known about the sulfur supply of C. glutamicum. Among the possible sulfur sources used by microorganisms, sulfate esters and sulfonates play a special role since they represent the most common sulfur sources in soils (2). Of these two groups, sulfate esters can easily be cleaved through hydrolysis by sulfatases (20), enzymes which have been identified in many different species and seem to be ubiquitous (17). For the utilization of sulfonates, a specific uptake and utilization system is needed (4).
Proteins involved in the utilization of organic sulfate esters or sulfonate compounds can be found among the sulfate starvation-induced proteins, which are induced in the absence of inorganic sulfate as a sulfur source, as has been shown, for instance, in experiments with Pseudomonas aeruginosa (26). The genetic background for utilization of sulfonates has been identified in the last few years. A cluster of genes involved in sulfonate utilization, the so-called ssu genes (sulfonate-sulfur utilization), was described for Bacillus subtilis (36), Escherichia coli (35), and Pseudomonas putida (15). In all species, the ssu genes are organized as an operon which is responsible for the specific uptake and utilization of aliphatic sulfonates. The ssu operon consists of three genes encoding an ABC-type transporter, ssuA, ssuB, and ssuC, and the ssuD gene directing the synthesis of a reduced flavin mononucleotide (FMNH2)-dependent sulfonatase. In many organisms, ssu operons also contain an ssuE gene encoding an NAD(P)H-dependent reductase that contributes to the cleavage process mediated by the sulfonatase (18).
Besides the main sulfonate utilization system, some organisms are equipped with additional systems which are responsible for the degradation of a certain subgroup of sulfonates, like the E. coli Tau system encoded by the tauABCD cluster for the utilization of taurine (33) or the P. aeruginosa Msu system encoded by the msuEDC cluster directing the utilization of methanesulfonate (18).
In the actinobacteria, the only sulfur utilization system which has been studied in detail is the dsz system of Rhodococcus sp. strain IGTS8 (5, 24). The Dsz system consists of the dszA, dszB, dszC, and dszD genes. The first three of these genes encode enzymes that are responsible for the sequential cleavage of the aromatic sulfur compound dibenzothiophene, which leads to the release of sulfite. Similar to ssuD genes, the dszA and dszC genes encode FMNH2-dependent monooxygenases whose reactions are essentially dependent on a reductase, which is encoded by dszD (22).
In this study we analyzed the utilization of sulfonates and sulfonate esters as sole sulfur sources in C. glutamicum. We also characterized C. glutamicum genes involved in the utilization of these compounds. In addition, we studied the expression of the genes identified with dependence on different sulfur sources.
MATERIALS AND METHODS
Chemicals.
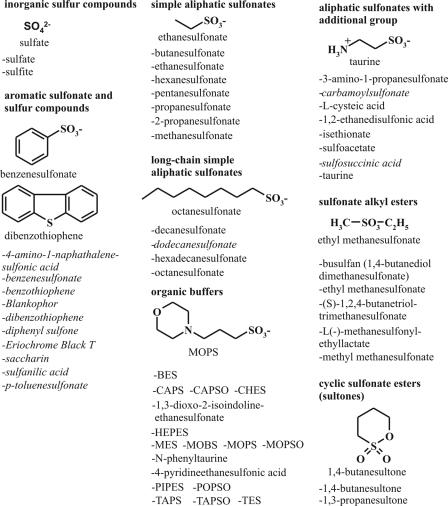
All chemicals used in this work were “for analysis” quality (purity, at least 97%). They were obtained from Sigma-Aldrich, ICN Biomedicals, Roth, Merck, or Acros. The substances tested in growth tests are listed in Fig. 1 or mentioned below. Eriochrome is a Ciba-Geigy trademark, and Blankophor is a Bayer trademark. The stability of the sulfonate esters in water was investigated by proton nuclear magnetic resonance (NMR) spectroscopy. Four-milligram portions of the sulfonate esters were dissolved in 0.6 ml of D2O at room temperature. The NMR spectra were recorded with a Bruker Advance 600 instrument at a proton resonance frequency of 600.13 MHz with reference to external trimethyl silane.
FIG. 1.
Sulfur-containing compounds tested as sulfur sources for C. glutamicum. The substances are shown in alphabetical order in groups according to their chemical structures. For each group a selected compound and its chemical structure are shown. Compounds that are not utilized as sulfur sources by the C. glutamicum wild type are indicated by italics. Abbreviations: BES, N,N-bis-(2-hydroxyethyl)taurine; CAPS, 3-cyclohexylamino-1-propanesulfonic acid; CAPSO, 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonate; CHES, 2-(cyclohexylamine)ethanesulfonic acid; MES, morpholineethanesulfonic acid; MOBS, 4-morpholino-butanesulfonic acid; MOPS, 3-morpholino-propanesulfonic acid; MOPSO, 3-morpholino-2-hydroxypropanesulfonic acid; PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid); POPSO, piperazine-N,N′-bis-(2-hydroxypropanesulfonic acid); TAPS, N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid; TAPSO, 3-[N-tris(hydroxymethyl)methylamino]-2-hydroxypropanesulfonic acid; TES, N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid.
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strains carrying plasmids were routinely grown on solid antibiotic medium no. 3 (Oxoid, Wesel, Germany) at 37°C. C. glutamicum strains were grown on solid brain heart broth (BH) (Merck, Darmstadt, Germany) at 30°C. For analysis of sulfur utilization, C. glutamicum strains were grown in sulfur-free minimal medium (MMS) (29) containing 25 g/liter glucose as a carbon source. The medium was prepared with very pure substances containing no sulfur, but no special treatment was used to remove traces of sulfur. Agarose at a concentration of 1.6% (wt/vol) was used for solid media. Sulfur sources were sterilized by filtration and added at a final concentration of 2 mM to autoclaved, cooled MMS medium. A Nephelostar Galaxy nephelometer (BMG Laboratories, Offenburg, Germany) was used to monitor growth of C. glutamicum strains in MMES medium (2.5% glucose, 17 mM NaNH4HPO4, 1 mM MgCl2, 60 mM K2HPO4, 10 mM citric acid, 37.5 μM FeCl2, 50 μM MnCl2, 67.5 μM CaCl2, 7.5 μM ZnCl2, 1 μM CuCl2, 0.1 μM NiCl2, 500 μg/liter thiamine, 50 μg/liter biotin) in order to analyze utilization of growth-limiting concentrations of sulfonate esters as sole sulfur sources. Each measurement consisted of two biological replicates, and the assay was conducted with four technical replicates per biological replicate. For clone selection, kanamycin was used at concentrations of 50 μg/ml for E. coli and 25 μg/ml for C. glutamicum strains.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant features | Source or reference |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Wild type | ATCCa |
| DK001 | Δcg1374-80 | This study |
| DK002 | Δcg1374 | This study |
| DK003 | Δcg1375 | This study |
| DK004 | ΔssuD1 | This study |
| DK005 | ΔssuC | This study |
| DK006 | ΔssuB | This study |
| DK007 | ΔssuA | This study |
| DK008 | ΔssuI | This study |
| DK009 | Δcg1148 | This study |
| DK010 | Δcg1149 | This study |
| DK011 | Δcg1150 | This study |
| DK012 | ΔseuA | This study |
| DK013 | ΔseuB | This study |
| DK014 | ΔseuC | This study |
| DK015 | ΔssuD2 | This study |
| DK016 | ΔseuABC | This study |
| DK017 | ΔssuD1 ΔseuABC ssuD2 | This study |
| DK018 | ΔssuD1 ΔseuA | This study |
| DK019 | ΔssuD1 ΔseuB | This study |
| DK020 | ΔssuD1 ΔseuC | This study |
| DK021 | ΔssuD1 ΔssuD2 | This study |
| E. coli DH5αMCR | F−endA1 supE44 mcrA thi-1 hsdR17 λ−recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 (φ80dlacZΔM15) (mrr-hsd-RMSmcrBC) | 11 |
ATCC, American Type Culture Collection, Manassas, Va.
DNA isolation, manipulation, transfer, and hybridization.
Standard procedures were employed for molecular cloning, transformation, and electrophoresis of E. coli DH5α, as well as for Southern hybridization of C. glutamicum DNA (30). Vector DNA was prepared from E. coli by the alkaline lysis technique using a QIAprep Spin miniprep kit according to the manufacturer's protocol (QIAGEN, Hilden, Germany). Transformation of C. glutamicum was performed by electroporation using the methods of Tauch et al. (32). Sequence similarity-based searches with nucleotide and amino acid sequences were performed using the basic local alignment search tool (BLAST) described by Altschul et al. (1).
Construction of plasmids.
Plasmids pDK001 to pDK017 were constructed using the gene-SOEing method described by Horton et al. (13) with the primers listed in Table S1 in the supplemental material. The appropriate primers were designed using the Sci Ed Central program hub with the Primer Designer 4.2 software (Sci Ed Software). Primers were purchased from SIGMA-ARK (Darmstadt, Germany). The primary products were amplified using Pwo DNA polymerase (Roche, Mannheim, Germany). The resulting products were purified using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and then used as templates for the second round of PCR. The final products were digested with restriction enzymes corresponding to cleavage sites which were introduced via the primers and ligated into appropriately digested pK18mobsacB. The ligation mixture was used to transform E. coli DH5αMCR, and the transformants were selected on antibiotic medium no. 3 plates containing 50 μg/ml kanamycin and 40 mg/liter X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Restriction endonucleases and T4 DNA ligase were obtained from Amersham-Pharmacia (Freiburg, Germany) and Roche-Diagnostics (Mannheim, Germany).
Site-specific gene disruption and gene replacement.
Site-specific gene disruption was performed using the nonreplicable integration vector pK18mobsacB, which can be used for marker-free deletion of a target gene (31). The resulting plasmids, pDK001 to pDK017, were transformed into C. glutamicum ATCC 13032 by electroporation (32). Integration of the introduced plasmids into the chromosome by single crossover was tested by selection on BH plates containing 25 μg/ml kanamycin. For deletion of the target gene, kanamycin-resistant (Kmr) cells were grown overnight in liquid BH and spread on BH plates containing 10% sucrose. Cells growing on these plates were tested for kanamycin sensitivity (Kms) by parallel picking on BH plates containing either kanamycin or sucrose. Sucrose-resistant and kanamycin-sensitive cells were then tested for the deletion by PCR, and, if necessary, the results were validated by Southern hybridization. Therefore, isolated chromosomal DNA from a putative mutant and the wild type were digested using an appropriate restriction enzyme. An agarose gel containing the digested mutant DNA and similarly digested wild-type DNA was hybridized with a digoxigenin-labeled probe complementary to the genes analyzed (binding outside the deletion region). The probe was generated with a digoxigenin DNA labeling and detection kit (nonradioactive) obtained from Roche (Mannheim, Germany).
RNA preparation and real-time RT-PCR.
For real-time reverse transcription (RT)-PCR measurements, cultures were grown in MMS with a sulfur source at a concentration of 2 mM and, in parallel, with 2 mM sulfate as a reference. A total of 1 × 109 C. glutamicum cells from a culture in the early logarithmic phase were mixed with killing buffer (10 mM sodium azide, 10 mM Tris, 5 mM MgCl2) at 4°C at a 1:1 ratio and put on ice for 10 min before centrifugation. The cell pellets were dissolved in 700 μl RLT buffer, and total RNA was isolated as described by Hüser et al. (14). The real-time RT-PCR was performed with a LightCycler machine (Roche, Mannheim, Germany) with a QuantiTect SYBR Green RT-PCR kit (QIAGEN). Oligonucleotides used for real-time RT-PCR were constructed to amplify intragenic regions (length, about 150 bp) of the genes analyzed. The primers (Table S2 in the supplemental material) were designed using the Primer Designer 4.2 software (Sci Ed Software) and were purchased from SIGMA-ARK (Darmstadt, Germany). The reverse transcriptase reaction was carried out at 50°C for 20 min, and this was followed by denaturation at 95°C for 15 min, which was used to activate the HotStarTaq DNA polymerase and to inactivate the reverse transcriptase. This was followed by 55 PCR cycles of 10 s at 95°C, 20 s at 55°C, and 12 s at 72°C. The melting curve was recorded over a range from 65 to 95°C with a heating rate of 0.1°C per s by continuous fluorescence measurement, and the reaction mixtures were finally cooled to 40°C. The crossing point (CP) for each gene and condition was determined using the second-derivative maximum data analysis method (LightCycler software, version 3.5). This algorithm measures the CP at the maximum increase or acceleration of fluorescence (27). The CPs obtained with RNA of cultures grown on MMS with sulfate were used as a reference. The nonnormalized relative expression ratios were calculated using the following equation: ratio = Etarget(CP of control − CP of sample) (25), where E is the PCR efficiency (at 100% efficiency, E is 2). Experiments with differentially diluted RNA indicated that the PCR efficiency was 100% (data not shown). Thus, the expression ratios obtained in the experiments can be considered the actual ratios of the mRNAs of the genes analyzed.
Nucleotide and amino acid sequence accession numbers.
The nucleotide sequences of ssuI, seuA, seuB, seuC, ssuD2, ssuD1, ssuC, ssuB, and ssuA are available in the GenBank database; the accession number for all these nucleotide sequences is BX927151. The amino acid sequences of the corresponding proteins can be retrieved from the TrEMBL database; the accession numbers are CAF19713 (SsuI), CAF19717 (SeuA), CAF19718 (SeuB), CAF19719 (SeuC), CAF19720 (SsuD2), CAF19924 (SsuD1), CAF19925 (SsuC), CAF19926 (SsuB), and CAF19927 (SsuA).
RESULTS
Sulfur source utilization by C. glutamicum ATCC 13032.
To analyze the spectrum of sulfonates that could be utilized as sulfur sources by C. glutamicum, wild-type strain ATCC 13032 was grown on minimal agar plates containing one of the compounds shown in Fig. 1 as the sole sulfur source at a concentration of 2 mM. For comparison, the wild-type strain was also grown on minimal agar plates containing 2 mM inorganic sulfate, which is a preferred sulfur source for most bacteria (17), as a positive control, or without any added sulfur source as a negative control. This assay yielded information about whether a deleted gene could be considered essential for utilization of the compound tested as a sulfur source and resulted in growth of an appropriate mutant comparable to the growth of the negative control.
In total, more than 50 different sulfur sources were tested (Fig. 1), including one amino acid (l-cysteic acid), simple and complex aliphatic sulfonates (like ethanesulfonate and taurine), organic buffers (like morpholinepropanesulfonic acid [MOPS]), sulfonates with one or more esterifications, cyclic sultones, aromatic sulfonates (benzenesulfonate), aromatic sulfides (e.g., benzothiophene), and, in addition, the inorganic sulfur sources sulfate and sulfite. Of the 36 aliphatic sulfonates tested, 33 could be utilized by the C. glutamicum wild type (Fig. 1). Carbamoylsulfonate, sulfosuccinate, and dodecanesulfonate could not be utilized. All sulfonate alkyl esters tested could be utilized, but none of the aromatic sulfonates tested (with a sulfonate group linked directly to an aromatic ring), none of the aromatic sulfonate derivatives (saccharin), and none of the aromatic sulfides (benzo- and dibenzothiophene) could be utilized (Fig. 1).
Identification of genes potentially involved in sulfonate utilization.
The recent availability of the complete C. glutamicum ATCC 13032 genome sequence (16) allowed identification of possible candidate genes for sulfonate utilization by similarity searches. To identify possible homologues of known ssu genes, the sequences of the encoded proteins of B. subtilis, E. coli, and P. putida were retrieved from the Swiss-Prot protein database (3). With these sequences, similarity-based searches were performed with the program BLASTP (1) and the amino acid sequences predicted from the C. glutamicum genome sequence. This approach resulted in a cluster of coding sequences (CDS) whose gene products showed highly significant hits to SsuA, SsuB, SsuC, or SsuD (Table 2). Additionally, a second CDS whose gene product exhibited a high level of similarity to SsuD was found at a different genomic locus. A candidate gene encoding SsuE was not detected in the C. glutamicum genome by similarity searches. Proteins similar to a transport system for aromatic sulfonates, the AsfC/AtsR-AtsB-AtsC system of P. putida (37), were also not detected in the amino acid sequences deduced from the C. glutamicum genome.
TABLE 2.
C. glutamicum candidate coding sequences possibly involved in the utilization of sulfonates and their esters, based on similarity searches
| C. glutamicum coding sequence | Similar gene and/or functiona | Organism | E value | No. of identical amino acids/total no. (%) |
|---|---|---|---|---|
| cg1147 (ssuI) | actI, actinorhodin polyketide dimeraseb | Streptomyces coelicolor | 8e-11 | 39/132 (29) |
| cg1148 | Hypothetical protein | No significant hit | ||
| cg1149 | Hypothetical protein | No significant hit | ||
| cg1150 | Putative reductasec | S. coelicolor | 1e-52 | 102/184 (55) |
| cg1151 (seuA) | dszA, dibenzothiophene desulfurization enzyme Ab | Rhodococcus sp. strain IGTS8 | 3e-77 | 173/443 (39) |
| cg1152 (seuB) | dszC, dibenzothiophene desulfurization enzyme Cb | Rhodococcus sp. strain IGTS8 | 3e-13 | 94/365 (25) |
| cg1153 (seuC) | dszC, dibenzothiophene desulfurization enzyme Cb | Rhodococcus sp. strain IGTS8 | 5e-30 | 105/384 (27) |
| cg1156 (ssuD2) | ssuD, sulfonataseb | B. subtilis | 2e-92 | 179/359 (49) |
| cg1376 (ssuD1) | ssuD, sulfonataseb | B. subtilis | 6e-94 | 181/358 (50) |
| cg1377 (ssuC) | ssuC, transporter (transmembrane)b | E. coli | 8e-53 | 98/240 (40) |
| cg1379 (ssuB) | ssuB, transporter (ATP binding)b | E. coli | 1e-49 | 115/228 (50) |
| cg1380 (ssuA) | ssuA, transporter (periplasmatic)b | B. subtilis | 1e-30 | 104/328 (31) |
Only the genes that represent the best hits for the C. glutamicum coding sequences analyzed in similarity searches on the protein level and the corresponding organisms are shown.
Proteins deduced from the coding sequences analyzed were compared with the Swiss-Prot database.
Proteins deduced from the coding sequences analyzed were compared with the NCBI nonredundant database.
To verify the proposed functions, reverse searches with the Ssu proteins predicted from the C. glutamicum genome were carried out with the Swiss-Prot protein sequence database. Additionally, proteins predicted from the neighboring CDS were analyzed (Table 2). No further CDS with potential importance for sulfur metabolism were found near the ssuD1CBA gene cluster. In contrast, upstream of the CDS designated ssuD2, a number of CDS were found to encode proteins possibly involved in sulfur utilization. Thus, the similarity searches revealed two clusters of interest consisting of the CDS ssuD1CBA (cg1376 to cg1380) and ssuI seuABC ssuD2 (cg1147 and cg1151 to cg1156) (Fig. 2). For these CDS, the following functions in utilization of sulfur sources could be proposed. The ssuC, ssuB, and ssuA genes encode proteins with similarity to the ABC-type sulfonate transporter SsuABC from E. coli or B. subtilis. The ssuD1 and ssuD2 genes both encode proteins exhibiting high levels of similarity to known SsuD sulfonatases, which are FMNH2-dependent monooxygenases. The CDS designated seuA, seuB, and seuC encode proteins similar to the Dsz monooxygenases from Rhodococcus sp. strain IGTS8, where “seu” indicates sulfonate ester utilization. The involvement of the seu genes in sulfonate ester utilization is described below. While SeuA resembles DszA, the proteins encoded by seuB and seuC exhibit levels of similarity of 25% and 27%, respectively, to the DszC protein. Searches using the NCBI nonredundant protein database also revealed similarities for both deduced proteins to a putative dehydrogenase from Pseudomonas syringae. The protein encoded by ssuI exhibits similarity to a putative oxidoreductase from Mycobacterium tuberculosis and thus is a possible functional replacement for SsuE.
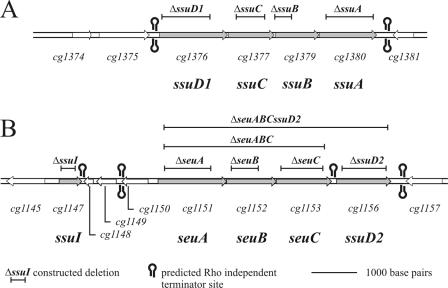
FIG. 2.
Physical map of the DNA regions of the C. glutamicum ATCC 13032 genome carrying genes possibly involved in utilization of sulfonates and their esters. (A) Chromosomal region containing the genes cg1374 to cg1381; (B) region containing the genes cg1145 to cg1157. The extent and position of an introduced deletion for a given gene are indicated, as are sites for predicted Rho-independent terminators.
The deduced protein sequences of the genes identified were also used in protein domain similarity searches of the NCBI conserved domain database (21). SsuD1, SsuD2, and SeuA exhibited significant hits to the strictly FMNH2-dependent alkanesulfonate monooxygenase protein family (6). Furthermore, the residues known for FMNH2 binding and catalysis in this family were found to be conserved in the deduced SsuD1, SsuD2, and SeuA amino acid sequences (data not shown). A similar situation was found for SeuB and SeuC, which exhibited significant hits to the strictly FMNH2-dependent dibenzothiophene desulfurization enzyme DszC, and the amino acid residues described as essential for FMNH2 binding of DszC were identified as conserved residues in SeuB and SeuC (data not shown). These findings provided strong bioinformatic evidence that the enzymes SsuD1, SsuD2, SeuA, SeuB, and SeuC are FMNH2-dependent monooxygenases. In addition, the predicted protein domain structure of the putative flavin mononucleotide (FMN) reductase SsuI exhibited significant similarity to the structure of FMN-binding proteins, with high levels of conservation in the residues important for FMN binding (data not shown).
The genomic locations of the genes described here and also of neighboring CDS are shown in Fig. 2. This figure also shows rho-independent transcription terminators found by analysis with the TransTerm software (9). The ssuD1, ssuC, ssuB, and ssuA genes form a dense cluster with proposed terminators up- and downstream, which supports the hypothesis that there is an operon structure. The ssuI, seuA, seuB, seuC, and ssuD2 genes are interrupted by the CDS cg1148, cg1149, and cg1150, which are located downstream of ssuI and are oriented in the direction opposite the orientation of the ssu and seu genes. For these CDS, only the protein encoded by cg1150 exhibited a significant hit in similarity searches, and it resembled a putative reductase. The genes in the cluster seuABC also seem to form an operon due to the lack of intergenic space between them and the presence of a predicted terminator downstream of the cluster. There are also terminators with the appropriate orientation located downstream of ssuI, ssuD2, and cg1150, indicating that these CDS are transcribed monocistronically, while no predicted terminator was found between cg1148 and cg1149, indicating that there is polycystronic transcription.
Analysis of ssuC, ssuB, and ssuA deletion mutants.
To analyze the importance of the CDS identified for sulfonate utilization, defined deletion mutants were constructed for each CDS (Fig. 2). To determine the relevance of the CDS in the ssu and seu gene clusters for sulfonate utilization, the growth of the C. glutamicum wild-type strain was compared with the growth of each constructed deletion mutant on various sulfur sources. The wild type and the mutants were grown on minimal agar plates containing one of the compounds shown in Fig. 1 as the sole sulfur source at a concentration of 2 mM. The results of the growth tests for all strains tested are summarized in Table 3 and given in detail in Table S3 in the supplemental material.
TABLE 3.
Growth of C. glutamicum strains with deletions in ssu and seu genes on different classes of sulfonates and sulfonate estersa
| Compound | Growth of strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | ΔssuA or ΔssuB or ΔssuC (transporter)b,c | ΔssuD1 (monooxy- genase)c | ΔssuD2 (monooxy- genase)c | ΔssuD1 ΔssuD2 (monooxy- genases)c,d | ΔseuA or ΔseuB (monooxy- genase)b,c | ΔseuC (monooxy- genase)c | ΔssuD1 ΔssuD2 ΔseuABC (monooxy- genases)c,d | ΔssuI (reductase)c | |
| Inorganic sulfur compounds | |||||||||
| Sulfate, sulfite | + | + | + | + | + | + | + | + | + |
| Sulfonates | |||||||||
| Simple aliphatic sulfonates (e.g., ethanesulfonate) | + | − | + | + | − | + | + | − | − |
| Long-chain simple aliphatic sulfonates (e.g., octanesulfonate) | + | + | + | + | − | + | + | − | − |
| Organic buffers (e.g., MOPS) | + | − | + | + | − | + | + | − | − |
| Aliphatic sulfonates with additional group (e.g., taurine) | + | − | —e | + | − | + | + | − | − |
| Sulfonate esters | |||||||||
| Busulfan | + | + | + | + | + | + | + | − | − |
| Butanesultone | + | + | + | + | + | − | + | − | − |
| (S)-1,2,4-Butanetriol-trimethane-sulfonate | + | − | + | + | − | + | + | − | − |
| Ethyl methanesulfonate | + | + | + | + | + | + | + | − | − |
| l-(−)-Methanesulfonylethyllactate | + | + | + | + | + | + | + | − | − |
| Methyl methanesulfonate | + | + | + | + | + | + | + | − | − |
| Propanesultone | + | − | + | + | − | + | + | − | − |
+, compound can be used as a sole sulfur source; −, growth was the same as the growth with no added sulfur after 72 h of incubation at 30°C. The test substances are shown in Fig. 1. Only compounds that could be utilized as sulfur sources by the wild type were used for the growth tests.
Strains having a deletion in any of the genes had the same phenotype.
The data in parentheses are the proposed functions of the deleted genes.
A strain having multiple deletions was constructed and tested.
Deletion of ssuD1 resulted in a mutant able to utilize all sulfonates except l-cysteic acid, 1,2-ethanedisulfonate, and sulfoacetate.
The growth tests with the deletion mutants showed that deletion of either ssuC, ssuB, or ssuA always resulted in the same phenotype: no growth on most aliphatic sulfonates. Thus, these genes are essential for utilization of nearly all aliphatic sulfonates except sulfonates with a chain length of eight carbon atoms or more. Based on the results of the growth tests and the similarity searches, these CDS were considered genes that encode an ABC-type sulfonate transporter and were designated ssuC, ssuB, and ssuA. As far as sulfonate esters are concerned, the phenotypes of the mutants were less homogeneous. The ssuC, ssuB, and ssuA deletion mutants could utilize all sulfonate esters tested with exception of (S)-1,2,4-butanetriol trimethanesulfonate and propanesultone, for which the SsuABC transporter was required (Table 3). The phenotypes obtained can be explained by the presence of an additional, as-yet-unidentified transporter for sulfonate esters which is not encoded in the ssu or seu gene cluster.
Analysis of ssuD1 and ssuD2 deletion mutants.
The similarity searches revealed two candidate genes, ssuD1 and ssuD2, that may encode sulfonatases. Deletion of ssuD1 resulted in a mutant which was unable to utilize l-cysteic acid, 1,2-ethanedisulfonate, and sulfoacetate (Table 3). The results of the growth tests and similarity searches led to the conclusion that the ssuD1 gene encodes a sulfonatase, but SsuD1 is not the only broad-range sulfonatase in C. glutamicum.
A mutant with a deletion only in the second possible sulfonatase gene, ssuD2, exhibited no growth defect on any of the substances tested. In contrast, deletion of both ssuD1 and ssuD2 resulted in a mutant that was not able to grow on any aliphatic sulfonate tested, including long-chain aliphatic sulfonates (Table 3). Thus, both of these genes can be considered genes that encode broad-range sulfonatases that together cover the complete spectrum of sulfonates with their enzymatic activities. However, a mutant having the ssuD1 ssuD2 double deletion was still able to grow on most sulfonate esters, as were mutants having a deletion in either ssuA, ssuB, or ssuC. These results clearly indicate that there is a separate pathway for the degradation of sulfonate esters in C. glutamicum.
Analysis of seuA, seuB, and seuC deletion mutants.
An ssuD1 ssuD2 double-deletion mutant is still able to grow on most sulfonate esters, suggesting that other genes must be involved in the degradation of this class of compounds. Interestingly, mutants having a single deletion of either seuA or seuB (sulfonate ester utilization) were unable to grow on minimal agar plates containing butanesultone as the sole sulfur source (Table 3). In contrast, a mutant with a deletion in the seuC gene did not show this effect. However, the phenotypes observed occurred only if the medium containing butanesultone was fresh.
Sulfonate anions are weak bases and therefore groups that can be easily replaced (23). Therefore, sulfonate esters can generally be regarded as chemically unstable in the medium used, and the ester bond might be cleaved by spontaneous hydrolysis, leading to the release of a sulfonate and an alcohol. For example, it can be assumed that propanesultone is converted mainly to the appropriate 3-hydroxy-1-propanesulfonate after it is added to the medium, since this compound is known to be unstable in aqueous solution (10). Also intra- or extracellular C. glutamicum esterases might cleave sulfonate esters, leading to the same situation. To obtain information about the chemical stability of the sulfonate esters tested in aqueous solution, H1-NMR studies were performed. The samples were analyzed after 24 h of incubation in D2O, and the NMR data were compared to data for freshly dissolved samples (Table 4). Butanesultone and l-(−)-methanesulfonyl-ethyllactate were found to be completely stable for 24 h under these conditions, while for methyl methanesulfonate, propanesultone, and ethyl methanesulfonate, the remaining amounts of the original substances were 72%, 22%, and 90%, respectively. (S)-1,2,4-Butanetriol-trimethanesulfonate was completely converted to 1,2-butanediol-dimethanesulfonate, releasing an equimolar amount of methanesulfonic acid, while busulfan was completely converted to an unidentified substance. Although these experiments were conducted in cell-free aqueous solutions and not under exactly the same conditions as the biological growth tests, they showed that for most sulfonate esters tested, a significant amount of methanesulfonate was released by spontaneous hydrolysis, which eventually led to misinterpretation of growth test results.
TABLE 4.
Stability of sulfonate esters measured by H1-NMR spectroscopy
| Substance | Stability of compound (%)a | Released sulfonic acid |
|---|---|---|
| Busulfan | 0 | Unknown |
| Butanesultone | 100 | |
| (S)-1,2,4-Butanetriol-trimethane-sulfonate | 0 | Methane sulfonic acid |
| Ethyl methanesulfonate | 90 | Methane sulfonic acid |
| l-(−)-Methanesulfonyl-ethyllactate | 100 | |
| Methyl methanesulfonate | 72 | Methane sulfonic acid |
| Propanesultone | 22 | 3-Hydroxypropane sulfonic acid |
The values indicate the amount of original substance remaining after 24 h of incubation in D2O.
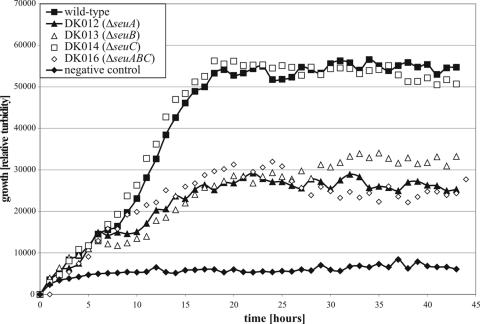
To avoid such a misinterpretation, all growth tests with sulfonate esters were repeated for the mutants with deletions in the seu genes. The growth tests were conducted in liquid media with lower, growth-limiting concentrations so that possible sulfonate contamination alone could not result in complete growth of the seu deletion mutants. This led to significant reductions in the growth rate and final cell density for strains DK012 (ΔseuA), DK013 (ΔseuB), and DK016 (ΔseuABC) during growth with 25 μM ethyl methanesulfonate as the sulfur source, in contrast to the results obtained for DK014 (ΔseuC), which showed growth comparable to that of the wild type (Fig. 3). When growth-limiting concentrations were used, all sulfonate esters except propanesultone and butanetriol-trimethanesulfonate resulted in growth phenotypes for mutants with deletions in seuA or seuB that differed from the growth phenotype of the wild type (data not shown). However, the strains tested did not have different phenotypes when they were grown on limiting concentrations of sulfate or methanesulfonate, demonstrating that the mutants did not have a general growth deficiency in the minimal media used and that involvement of the seuA and seuB genes in the utilization of methanesulfonate is unlikely (data not shown).
FIG. 3.
Growth of the C. glutamicum wild-type and mutant strains. Growth in liquid minimal medium containing ethyl methanesulfonate at a concentration of 25 μM as the sole source of added sulfur was monitored with a nephelometer and is expressed as relative turbidity. Growth of the wild type without added sulfur was used as a negative control.
The results of the growth assays can be simply explained by the stability of the sulfonate esters tested in aqueous environments. The growth phenotypes of the seuA and seuB mutant strains were negative in plate tests when the stable sulfonate ester butanesultone was used. In addition, growth-limiting concentrations of the partially stable sulfonate esters resulted in significantly reduced growth of these mutants, which was only due to sulfonates resulting from partial hydrolysis.
Mutants with the complete seuABC gene cluster deleted exhibited the same phenotype as mutants with a deletion in seuA or seuB in all growth tests. These findings can be explained by the assumption that SeuA and SeuB form an enzyme pair, which might work in an enzyme complex or degrade sulfonate esters sequentially as single enzymes. A specific role for SeuC in this degradation process could not be deduced.
Destruction of all degradation pathways for sulfonates and sulfonate esters through deletion of ssuD1, ssuD2, and seuABC resulted in a mutant that was not able to grow on any sulfonate or sulfonate ester in any liquid or solid medium growth test (Table 3), further strengthening the model for the action of the seu gene products in the degradation of sulfonate esters.
ssuI gene is essential for sulfonate and sulfonate ester utilization.
Additional deletion mutants were constructed and tested to analyze the importance of the CDS upstream of seuABC ssuD2. The growth of mutants with deletions in cg1148, cg1149, or cg1150 did not differ from the growth of the wild type (data not shown). In contrast, deletion of ssuI (cg1147), which according to the similarity searches is the second CDS besides cg1150 that potentially encodes a reductase, resulted in a mutant which exhibited growth on all sulfonates and sulfonate esters tested that was comparable to the growth of the negative control (Table 3). Thus, ssuI can be considered a gene that plays an essential role in the utilization of both classes of compounds. However, when the growth tests for the ssuI deletion mutant were prolonged beyond the normal observation time (72 h), cultures grew slowly to a final density comparable to that of the sulfate-grown positive control (data not shown). This phenotype could be confirmed when the ssuI deletion mutant was grown in liquid media. Evidently, the function of SsuI can be bypassed by prolonged cultivation.
Transcriptional studies of the ssu and seu genes.
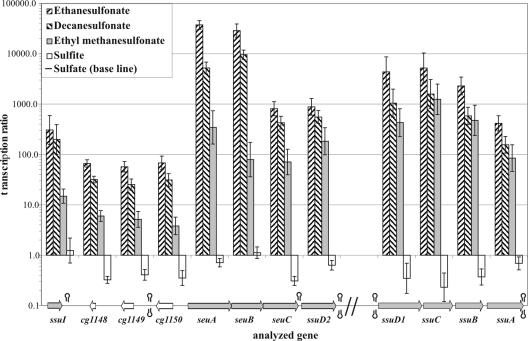
The growth tests with the deletion mutants showed that ssuD1, ssuD2, and ssuI are essential for the utilization of all aliphatic sulfonates. The ssuCBA genes can be considered genes that encode the transporter for nearly all aliphatic sulfonates, and the seu genes are involved in sulfonate ester utilization. It was of interest to analyze whether these genes are transcriptionally regulated for dependence on the sulfur source used. To answer this question, the mRNA levels of the genes studied were measured using real-time RT-PCR. Thus, C. glutamicum cultures were grown in liquid minimal medium containing different sulfur sources, including a short-chain aliphatic sulfonate (ethanesulfonate), a long-chain aliphatic sulfonate (decanesulfonate), a sulfonate ester (ethyl methanesulfonate), sulfate, or sulfite, at a concentration of 2 mM and were harvested at the logarithmic growth phase. Two independently grown cultures were used for each growth condition, and RNA was isolated, purified, and used in an automated real-time RT-PCR experiment with a LightCycler. All genes in the two gene clusters were analyzed, and the mRNA abundance ratios were determined by comparison to the values obtained from a culture grown on sulfate (reference culture).
The transcription ratios are shown in Fig. 4. Very high ratios (up to 10,000) for all genes in the ssu and seu clusters were observed for growth on a sulfonate or a sulfonate ester compared to growth on sulfate or sulfite. The CDS cg1374 and cg1375 were also tested, but they showed constant expression for all test conditions, confirming that there is no observable relevance of these genes for sulfur compound utilization (data not shown). An interesting result is the strong induction of the seu genes during growth on sulfonates, although these genes are considered genes that are not important for sulfonate utilization. This observation indicates that there is no differentiation in terms of regulation between sulfonates and sulfonate esters.
FIG. 4.
Expression of the C. glutamicum ssu and seu genes depending on the available sulfur source. The transcription rates of the genes examined for growth of C. glutamicum ATCC 13032 on sulfonates, a sulfonate ester, or sulfite as the sole sulfur source were correlated with the transcription rates for growth on sulfate (baseline). The values are the means of at least two independent experiments with two technical replicates each.
The CDS cg1148 to cg1150 were also expressed more highly during growth on a sulfonate or sulfonate ester than during growth on sulfate, albeit to a significantly lesser extent than the neighboring ssu and seu genes. Since mutants with a deletion in one of these CDS showed no growth phenotype, it can be speculated that these genes might have nonessential functions in the utilization of sulfonates and sulfonate esters.
To analyze the transcriptional organization of the ssuD1CBA and seuABC ssuD2 gene regions, primer pairs amplifying sequences of two adjacent genes, including the complete intergenic regions, were tested for both clusters by real-time RT-PCR analysis (data not shown). By analyzing total RNA from cultures grown in the presence of ethanesulfonate, the transcription of the intergenic regions of ssuD1CBA and the seuABC gene cluster was found to be indistinguishable from that of the intragenic regions. Based on the presence of putative transcriptional terminators in front of as well as downstream of the gene clusters analyzed, operon structures for ssuD1CBA and seuABC seem likely. Interestingly, the intergenic region between seuC and ssuD2 also showed the same expression pattern, indicating that there is a polycystronic seuABC ssuD2 mRNA, despite the predicted rho-independent transcriptional terminator in front of ssuD2.
DISCUSSION
The purposes of this study were elucidation and characterization of the genes involved in the degradation of sulfonates and sulfonate esters in C. glutamicum. Therefore, bioinformatic analyses were combined with phenotypic analyses of targeted deletion mutants to infer the involvement of specific genes and to predict functions for their gene products.
Utilization of aliphatic sulfonates in C. glutamicum.
The ssu gene class in C. glutamicum consists of the genes ssuA, ssuB, ssuC, ssuD1, ssuD2, and ssuI. According to their sequences, SsuA, SsuB, and SsuC are similar to the ABC transport system for sulfonates in E. coli or B. subtilis (35, 36), while SsuD1 exhibits similarities to known sulfonatases. As in all other organisms analyzed so far (18), the C. glutamicum ssuD1CBA genes also form a predicted operon.
Deletion of ssuA, ssuB, or ssuC resulted in mutants that were not able to grow on any short-chain aliphatic sulfonate. Therefore, we concluded that the putative ABC transporter consisting of SsuA (extracellular binding protein), SsuB (ATP-binding protein), and SsuC (transmembrane protein) is responsible for the import of this class of compounds. However, utilization of long-chain aliphatic sulfonates like decanesulfonate was found to be not dependent on the ssuCBA genes. Therefore, an additional uptake system in C. glutamicum has to be postulated (for example, uptake via a fatty acid transporter).
It is worthwhile to consider the situation in B. subtilis, in which, as in C. glutamicum, the ssuA and ssuC genes are necessary for utilization of short-chain aliphatic sulfonates (36). In E. coli the situation is different, since in addition to SsuABC a second sulfonate transporter, TauABC, is known (8). While SsuABC is thought to be responsible for the uptake of most sulfonates, including long-chain sulfonates, TauABC is essential only for the uptake of the short-chain sulfonate taurine. As concluded for C. glutamicum, taurine is also imported by the SsuABC transporter in B. subtilis (36).
In this study, C. glutamicum was found to have two genes encoding proteins with high levels of similarity to known SsuD sulfonatases, the ssuD1 and ssuD2 genes encoding broad-range sulfonatases with overlapping substrate spectra. In E. coli and all other bacteria described previously, only a single broad-spectrum sulfonatase, designated SsuD and encoded by the ssuD gene, was found (18). However, it should be mentioned that additional sulfonatases which are responsible for the degradation of specific sulfonate subgroups are known, like TauD in E. coli, which is mainly responsible for the degradation of taurine (33), and MsuD in P. aeruginosa, which is capable of degrading methanesulfonate and other small sulfonates (18). This is in contrast to the situation in C. glutamicum, in which no specialized sulfonatases could be detected and SsuD1 as well as SsuD2 are apparently able to degrade a broad range of sulfonates, including taurine and methanesulfonate.
The C. glutamicum ssuI gene was shown to be essential for efficient degradation of all sulfonates or sulfonate esters tested. From sequence similarity analyses it could only be determined that ssuI encodes a putative reductase. Similar analyses predicted that the ssuD1, ssuD2, and seuABC genes encode FMNH2-dependent monooxygenases with a high level of conservation of the residues important for FMNH2 binding. This type of monooxygenase does not possess a prosthetic flavin group but essentially depends on a reductase which produces FMNH2 by using NAD(P)H as a reducing agent (17). We therefore suggest that ssuI encodes a reductase which is essential and specific for the enzymatic action of all monooxygenases involved in sulfonate and sulfonate ester degradation in C. glutamicum. It is of interest that a BLAST search did not reveal any protein in other organisms with a high level of similarity to SsuI. It can be assumed that ssuI is replaced in other organisms by ssuE encoding the reductase for the sulfonate utilization system (17). The isolated enzymes SsuE and SsuD from E. coli were characterized biochemically, which showed that the action of the sulfonatase SsuD essentially depends on a reductase restoring FMNH2 and that SsuE has this function (7). Interestingly, in all organisms analyzed so far, the ssuE gene was found to be not essential for growth on aliphatic sulfonates (17). It is supposed that SsuE can be replaced by other nonspecific reductases. Possible explanations for the finding that SsuI is essential for sulfonate and sulfonate ester degradation in C. glutamicum are (i) that the SsuD and SeuAB monooxygenases must form a complex with SsuI to be fully functional and (ii) that the monooxygenases cannot efficiently interact with the FMN reductases in C. glutamicum other than SsuI. This is in contrast to the situation found for E. coli, in which the appropriate reductase, SsuE, was shown to efficiently deliver the FMNH2 needed for the sulfonate cleavage mediated by SsuD in vitro, although SsuE is not essential for this process in vivo (7, 35). It has been postulated that SsuD of E. coli and other organisms, like P. aeruginosa and P. putida, is able to interact with several FMN reductases, including SsuE (17).
Utilization of sulfonate esters in C. glutamicum.
The C. glutamicum gene cluster seuABC encodes proteins resembling the FMNH2-dependent Dsz monooxygenases from Rhodococcus sp. strain IGTS8, which are responsible for the utilization of dibenzothiophene as a sulfur source (5). Growth tests with deletion mutants showed that the seuA and seuB genes in C. glutamicum are involved in sulfonate ester utilization. The finding that sulfonate ester utilization in C. glutamicum depends on the ssuI gene encoding a putative reductase supports the assumption that seuA and seuB encode FMNH2-dependent monooxygenases.
The ability of a C. glutamicum ssuD1 ssuD2 double-deletion mutant to grow on most sulfonate esters showed that these substances can be degraded by a pathway different from that used for degradation of sulfonates. Not only the degradation but also the import of sulfonate esters differs from that of sulfonates. The ssuA, ssuB, and ssuC deletion mutants were still able to grow on most sulfonate esters, demonstrating the presence of an as-yet-unidentified transporter for this class of compounds in C. glutamicum. Therefore, sulfonate esters can be considered compounds that represent a distinct group of compounds, which in C. glutamicum is handled differently than the sulfonate group, but the possibility that the seu genes are also responsible for the utilization of another, as-yet-unidentified class of substances cannot be excluded. The results obtained for the utilization of sulfonate esters in C. glutamicum cannot be compared to previously described data since so far the metabolism of these compounds has not been studied in any other organism.
Transcriptional regulation of the ssu and seu genes.
In this study, it was shown that the expression rates for all ssu and seu genes were very high during growth on sulfonates or a sulfonate ester compound compared to growth on sulfate or sulfite. Evidently, the ssu and seu genes exhibit a tight connection not only at the functional level but also at the regulatory level. This situation has also been described for other organisms, like B. subtilis (36) or E. coli (35), in which the ssu genes are highly expressed during growth on different sulfonates but not during growth on sulfate (17). In E. coli it was shown with a transcriptional lacZ reporter gene fusion that sulfate actually repressed ssu gene expression. However, the exact mechanism by which sulfate acts on the transcription of the ssu genes is still not known (34).
No CDS similar to one of the known E. coli regulators of sulfonate utilization, like cysB or cbl (35), was found in the C. glutamicum genome. On the contrary, ssuD1 was shown to be regulated by the McbR repressor (28). This repressor, however, seems to represent a global regulator of C. glutamicum sulfur metabolism and is therefore not a likely candidate for selection between different sulfur sources. Therefore, the question of how gene regulation in C. glutamicum distinguishes between different sulfur sources remains. To identify the regulator(s) involved, additional experiments are necessary.
Supplementary Material
Acknowledgments
D.J.K. and C.R. acknowledge grants from the International NRW Graduate School in Bioinformatics and Genome Research. This work was supported in part by Degussa AG (Düsseldorf, Germany).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autry, A. R., and J. W. Fitzgerald. 1990. Sulfonate S: a major form of forest soil organic sulfur. Biol. Fertil. Soils 10:50-56. [Google Scholar]
- 3.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook, A. M., H. Laue, and F. Junker. 1999. Microbial desulfonation. FEMS Microbiol. Rev. 22:399-419. [DOI] [PubMed] [Google Scholar]
- 5.Denome, S. A., C. Oldfield, L. J. Nash, and K. D. Young. 1994. Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. J. Bacteriol. 176:6707-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichhorn, E., C. A. Davey, D. F. Sargent, T. Leisinger, and T. J. Richmond. 2002. Crystal structure of Escherichia coli alkanesulfonate monooxygenase SsuD. J. Mol. Biol. 324:457-468. [DOI] [PubMed] [Google Scholar]
- 7.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 2000. Deletion analysis of the Escherichia coli taurine and alkanesulfonate transport systems. J. Bacteriol. 182:2687-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 10.Falbe, J., and M. Regitz. 1995. Roempp Chemie Lexikon CD. 9th ed. Thieme, Stuttgart, Germany.
- 11.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Großmann, K., K. Herbster, and M. Mack. 2000. Rapid cloning of metK encoding methionine adenosyltransferase from Corynebacterium glutamicum by screening a genomic library on a high density colony-array. FEMS Microbiol. Lett. 193:99-103. [DOI] [PubMed] [Google Scholar]
- 13.Horton, R. M., H. D. Hunt, Ho, S. N., J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 14.Hüser, A. T., A. Becker, I. Brune, M. Dondrup, J. Kalinowski, J. Plassmeier, A. Pühler, I. Wiegräbe, and A. Tauch. 2003. Development of a Corynebacterium glutamicum DNA microarray and validation by genomewide expression profiling during growth with propionate as carbon source. J. Biotechnol. 106:269-286. [DOI] [PubMed] [Google Scholar]
- 15.Kahnert, A., P. Vermeij, C. Wietek, P. James, T. Leisinger, and M. A. Kertesz. 2000. The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol. 182:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 17.Kertesz, M. A. 1999. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 18.Kertesz, M. A., M. Rist, K. Schmidt-Larbig, and T. Wüest. 1999. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J. Bacteriol. 181:11464-11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Amino acid fermentation. I. Production of l-glutamic acid by various microorganisms. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 20.Liebman, J. F. 1991. Thermochemistry of sulphonic acids and their derivates, p. 283-321. In S. Patai and Z. Rappoport (ed.), The chemistry of sulphonic acids, esters and their derivates. Wiley, New York, N.Y.
- 21.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsubara, T., T. Ohshiro, and I. Y. Nishina Yoshihiro. 2001. Purification, characterization, and overexpression of flavin reductase involved in dibenzothiophene desulfurization by Rhodococcus erythropolis D-1. Appl. Environ. Microbiol. 67:1179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison, R. T., and R. N. Boyd. 1983. Organic chemistry, 4th ed. Allyn and Bacon Inc., Boston, Mass.
- 24.Oldfield, C., O. Pogrebinsky, J. Simmonds, E. S. Olson, and C. F. Kulpa. 1997. Elucidation of the metabolic pathway for dibenzothiophene desulphurization by Rhodococcus sp. strain IGTS8 (ATCC 53968). Microbiology 143:2961-2973. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quadroni, M., P. James, P. Dainese-Hatt, and M. A. Kertesz. 1999. Proteome mapping, mass spectrometric sequencing and reverse transcription-PCR for characterization of the sulfate starvation-induced response in Pseudomonas aeruginosa PAO1. Eur. J. Biochem. 266:986-996. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen, R. 2001. Quantification on the Lightcycler, p. 21-34. In S. Meuer, C. Wittwer, and K. Nakagawara (ed.), Rapid cycle real-time PCR, methods and applications. Springer Verlag, Heidelberg, Germany.
- 28.Rey, D. A., A. Pühler, and J. Kalinowski. 2003. The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103:51-65. [DOI] [PubMed] [Google Scholar]
- 29.Rückert, C., A. Pühler, and J. Kalinowski. 2003. Genome-wide analysis of the l-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. J. Biotechnol. 104:213-228. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 32.Tauch, A., O. Kirchner, B. Löffler, S. Götker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 33.van der Ploeg, J., M. A. Weiss, E. Saller, N. Hiroko, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Ploeg, J. R., E. Eichhorn, and T. Leisinger. 2001. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch. Microbiol. 176:1-8. [DOI] [PubMed] [Google Scholar]
- 35.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 36.van der Ploeg, J. R., T. Leisinger, N. J. Cummings, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 37.Vermeij, P., C. Wietek, A. Kahnert, T. Wüest, and M. A. Kertesz. 1999. Genetic organization of sulphur-controlled aryl desulphonation in Pseudomonas putida S-313. Mol. Microbiol. 32:913-916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.