Abstract
Real-time PCR is fast, sensitive, specific, and can deliver quantitative data; however, two disadvantages are that this technology is sensitive to inhibition by food and that it does not distinguish between DNA originating from viable, viable nonculturable (VNC), and dead cells. For this reason, real-time PCR has been combined with a novel discontinuous buoyant density gradient method, called flotation, in order to allow detection of only viable and VNC cells of thermotolerant campylobacters in chicken rinse samples. Studying the buoyant densities of different Campylobacter spp. showed that densities changed at different time points during growth; however, all varied between 1.065 and 1.109 g/ml. These data were then used to develop a flotation assay. Results showed that after flotation and real-time PCR, cell concentrations as low as 8.6 × 102 CFU/ml could be detected without culture enrichment and amounts as low as 2.6 × 103 CFU/ml could be quantified. Furthermore, subjecting viable cells and dead cells to flotation showed that viable cells were recovered after flotation treatment but that dead cells and/or their DNA was not detected. Also, when samples containing VNC cells mixed with dead cells were treated with flotation after storage at 4 or 20°C for 21 days, a similar percentage resembling the VNC cell fraction was detected using real-time PCR and 5-cyano-2,3-ditolyl tetrazolium chloride-4′,6′-diamidino-2-phenylindole staining (20% ± 9% and 23% ± 4%, respectively, at 4°C; 11% ± 4% and 10% ± 2%, respectively, at 20°C). This indicated that viable and VNC Campylobacter cells could be positively selected and quantified using the flotation method.
Campylobacter jejuni, Campylobacter coli, and Campylobacter lari have been recognized worldwide to be among the most frequently reported food-borne pathogens in humans (1). The traditional detection of Campylobacter spp. by culture-based methods is time-consuming, laborious, and difficult to adapt for quantitative analysis. Additionally, it has been reported that Campylobacter may go into a viable nonculturable (VNC) state (20), which is not detected by culture-based methods. The significance of this state for Campylobacter diagnostics is still controversial, as some studies have shown that organisms in the VNC state cannot colonize baby chicks, and for that reason the VNC state has been considered a degenerative form (3, 16). Other studies have shown that VNC cells of this organism can in fact colonize baby chicks and suggest that the VNC state is a dormant form of the cell (22, 24). Since no conclusive evidence seems yet available, there is a possibility that VNC cells may become infective and therefore should be detected by diagnostic methods for food-borne pathogens.
Due to the aforementioned problems with Campylobacter detection, PCR and real-time PCR have been increasingly used for detection (12, 17, 21, 25). While this technology has tremendous potential for sensitive and simple identification, further studies addressing the quantification aspect of this technology are required before it can be widely accepted. There are limitations to the application of PCR technology. First of all, detection from food samples such as poultry or poultry rinses may be limited due to the presence of PCR inhibitors (12). Secondly, nucleic acid-based detection methods do not differentiate between nucleic acids originating from dead or viable/VNC cells, introducing a risk for false-positive results (21). This has resulted in a great demand for sample treatment methods that can overcome PCR inhibition and are capable of differentiating between viable and dead cells (17).
Recently, a novel sample treatment method called flotation, which is based on traditional buoyant density centrifugation, was developed (26). Using Yersinia enterocolitica as a model system, it was shown that this method can separate the target organism from environmental sample matrices and background flora (BGF) based on differences in buoyant densities. Further studies showed that this method could be successfully employed to reduce the risk of false-negative results due to detection of DNA originating from dead cells (27). The aim of the present study was to develop a flotation real-time PCR method which can be used to rapidly detect and quantify viable and VNC cells of C. coli, C. jejuni, and C. lari without false-positive results due to detection of DNA originating from dead cells. Also, to ensure correct detection of naturally contaminated samples, the buoyant density behavior of different Campylobacter spp. strains was investigated at different time points during growth. Finally, this method was tested on spiked and naturally contaminated samples.
MATERIALS AND METHODS
Samples.
Campylobacter jejuni CCUG 10937 (Culture Collection University of Göteborg [CCUG], Göteborg, Sweden), C. jejuni ATCC 33291 (American Type Culture Collection [ATCC], Manassas, VA), C. jejuni ATCC 29428 (ATCC), C.coli CCUG 10939 (CCUG), and C. lari CCUG 12774 (CCUG) were grown in Bolton broth (Oxoid, Unipath, Basingstoke, United Kingdom) supplemented with Bolton broth selective supplement (Oxoid) at 42°C under a microaerophilic atmosphere, created by the use of Campygen (Oxoid). Viable counts were performed on Preston agar plates (according to NMKL no. 119 2A, 2nd ed., 1990) under the same conditions. Undefined background flora was grown by adding 20% (vol/vol) chicken rinse in Tryptone soya broth (Oxoid) under conditions similar to those required for Campylobacter growth. Rinse samples were made by mixing 25 g of chicken neck skin with 225 ml of physiological saline, followed by mixing in a stomacher for 5 min. Afterwards, the chicken was removed from the sample. The absence of detectable Campylobacter spp. in the samples was confirmed by real-time PCR and selective plating. When necessary, dilutions of the chicken samples prior to PCR were made in physiological saline.
Buoyant density measurements.
Throughout the study three different colloidal density gradient media, all formulated from RediGrad (Amersham Biosciences AB, Uppsala, Sweden) were used: BactXtractor-L (BX-L), with a density of 1.058 g/ml, pH of 7.5, and osmolality of 322 mOsm/kg; BactXtractor-M (BX-M), with a density of 1.132 g/ml, pH of 7.5, and osmolality of 300 mOsm/kg; and BactXtractor-H (BX-H), with a density of 1.309 g/ml, pH of 7.5, and osmolality of 340 mOsm/kg (BactXtractor is a registered trademark of QRAB, Bålsta, Sweden). For all three media the pH was adjusted with 1 M HCl and the osmolality was adjusted using a Roebling osmometer (Labex Instruments AB, Helsingborg, Sweden) by the addition of ultrapure (99.99%) NaCl. The final densities of BX-L and BX-H were reached after dilution and concentration of RediGrad. After the pH and the osmolality had been set, BX-M had a density of 1.132 g/ml and needed no further adjustment. Densities were measured using a DMA46 density meter (Instrument AB Lambda, Stockholm, Sweden).
Buoyant densities of microorganisms and food particles were determined as described by Pertoft (19). Briefly, 6 ml of BX-M was mixed with 2 ml sample, 2 ml physiological saline, and 50 μl of a density marker bead (DMB) solution (Amersham Biosciences AB) in a plastic 15-ml conical tube. Centrifugation was performed at 10,000 × gav for 30 min at room temperature, using an angle rotor. A plot describing the density of different locations in the self-generating gradient was made by measuring the distance from the bottom of the tube to the different layers of DMB and plotting the known densities of the different DMB (in g/ml) versus distance (in cm).
Flotation conditions.
For flotation prior to quantitative PCR, a one-step flotation setup was used, based on a method described by Wolffs et al. (26) and redesigned for Campylobacter (see Fig. 2). In brief, three layers with different densities were carefully layered below each other. The bottom layer consisted of a high-density solution mixed with the sample to a density of approximately 1.200 g/ml. The middle and top layers had densities of 1.109 and 1.065 g/ml, respectively. The resulting discontinuous gradients were centrifuged for 15 min at 4,500 × gmax in a swing-out rotor, and afterwards 1-ml samples were taken, using sterile 2-ml syringes, for further analysis. The samples were added to 2-ml Eppendorf tubes, diluted with physiological saline to 2 ml (to obtain a density of the solution that allowed pelleting of cells), and centrifuged at 13,000 × gmax in a benchtop Eppendorf centrifuge for 5 min. Afterwards, 1.5 ml of the supernatant was removed and the cells were resuspended in the remaining 0.5 ml to obtain a 2× concentrated sample.
FIG. 2.

Overview of the flotation setup. This setup is used to recover viable and VNC cells with buoyant densities of 1.065 and 1.109 g/ml. The Campylobacter recovery location is at the upper interface. The buoyant densities of the different colloidal solutions used in the two setups are shown. *, the density of the lowest layer may vary slightly because it consists of sample mixed with colloidal silica solution. Since the density of the sample may vary, this can impact the final buoyant density of this layer.
Real-time PCR conditions.
A real-time PCR assay was developed based on primers OT1559 and 18-1 (12). Hybridization probes (PWC1, 5′-GGAAAC CCTGACGCAGCA-fluorescein-3′; PWC2, 5′-LCred640-GCCGCGTGGAGG ATGAC-3′) were developed to detect the 287-bp amplicon. The PCR mixture consisted of 1 U Tth DNA polymerase (Roche Diagnostics, Mannheim, Germany), 1× accompanying buffer, 10 mM deoxynucleoside triphosphate, 0.2 μM of each primer, 0.3 μM of each probe (TIB Molbiol, Berlin, Germany), and 2 μl sample in a final volume of 20 μl. Each amplification was run in a LightCycler instrument (Roche Diagnostics) started with a denaturation step of 1 min at 95°C followed by 40 cycles of 10-s denaturation at 95°C and 10-s annealing at 58°C, and then a single fluorescence measurement and elongation for 25 s at 72°C. Amplification was followed by a melt curve analysis between 65°C and 95°C and, finally, a cooling step for 1 min at 40°C. During amplification, the fluorescence was measured in channels F2/F1. The quantification data, in terms of crossing point (Cp) values, were expressed as fractional cycle numbers determined from the intersection of the log-linear fluorescence curve, with a threshold crossing line based on the second derivative method of the LightCycler software, version 5.32 (Roche Diagnostics).
Campylobacter DNA was purified using an Invitrogen EasyDNA kit (Invitrogen, Groningen, The Netherlands). Tenfold dilutions of Campylobacter between 0.01 and 1.36 × 107 genomic DNA copies were used to obtain standard curves. The number of genomic copies was determined according to previous studies, in one of which Josefsen et al. calculated that 1 C. jejuni genome weighs 1.7 fg (5,15). Standard curves with cells were created using 10-fold dilutions of cell cultures. Cells were lysed by incubation at 95°C for 5 min. All measurements were run at least in independent duplicate runs. After amplification, Cp results were plotted against the log of the initial number of genomes. From this graph, the linear range was determined. After determination of the linear range of amplification, linear regression was used to calculate the slope of the Cp-versus-log initial DNA or cell concentration plot using the points in the linear range. From this slope the amplification efficiency (AE) was calculated using the following equation: AE = (10-1/slope) − 1 (6).
Quantification of viable cells and mixtures of VNC and dead cells.
For the spiked and naturally contaminated samples, chicken samples were spiked or analyzed directly by two different methods: direct plating using Preston agar and real-time PCR. To obtain quantitative real-time PCR measurements, a standard curve created for whole cells was used. For the second experiment, i.e., the VNC/dead cell experiment, cells of C. jejuni ATCC 33291, C. jejuni ATCC 29428, and C. jejuni CCUG 10937 were kept in Bolton broth for a period of 21 days at three different storage temperatures, i.e., 4°C, 20°C, or 42°C, under a microaerobic atmosphere and then stained with CTC (5-cyano-2,3-ditolyl tetrazolium chloride) and DAPI (4′-6-diamino-2-phenylindole, dihydrochloride) as described by Cappelier et al. (2). Briefly, 0.5 ml brain heart infusion medium (Oxoid) was mixed with 0.1 ml of a 0.05 g/liter solution of pyruvic acid (Sigma) and 0.5 ml cell suspension. CTC was added to a final concentration of 5 mM and incubated for 4 h at 37°C. Cells were then harvested by filtration on black isopore polycarbonate membrane filters (0.2-μm pore size and 25-mm diameter; Millipore, Watford, England) and covered with 5 mg/liter DAPI (Sigma) solution for 5 min for counterstaining. Counts were obtained randomly by counting 2 times 20 fields per filter and, for each sample, two filters were counted.
The number of metabolically active cells (VNC) was divided by the total number of cells as determined by DAPI staining in order to acquire a relative amount of VNC cells in the mixture. To ensure the absence of culturable cells, plate counts were performed. After this, 1 ml of each mixture was subjected to flotation. After flotation, 1-ml samples were withdrawn from the Campylobacter recovery location and, as a control, from the top of the lower layer and the top of the upper layer. Afterwards the cells were added to 2-ml Eppendorf tubes, diluted with physiological saline to 2 ml (to obtain a density of the solution that allowed pelleting of cells), and centrifuged at 13,000 × gmax in a benchtop Eppendorf centrifuge for 5 min. Afterwards, 1.75 ml of the supernatant was removed and the cells were resuspended in the remaining 0.25 ml, after which they were heated for 5 min at 95°C to obtain cell lysis. These final samples were analyzed by quantitative PCR. Standard curves for absolute quantification were made with overnight cultures of known CFU/ml diluted in physiological saline. This implied that data acquired from both layers, even when originating from dead and VNC cells, were expressed in terms of CFU/ml.
RESULTS
Buoyant densities.
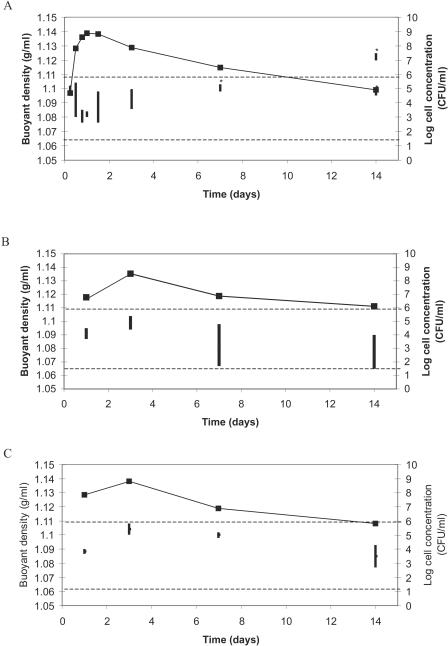
To be able to design a flotation method, information on the buoyant densities of the target organism and the other components in the sample (food particles and background flora) is needed. The buoyant densities of different Campylobacter spp. strains at different time points over a period of 2 weeks were measured (Fig. 1A to C). The results showed that changes in buoyant density were observed over a range of 1.065 to 1.130 g/ml but that different changing patterns were observed between the different strains. All strains showed an initial increase in buoyant density, with a peak after 2 to 4 days, after which a slow drop in buoyant density was observed. In one case, with the increase of time, a split of the population into different densities was observed (Fig. 1A). Further studies showed that the population with the highest density did not result in any viable counts, whereas the population with the lower density did contain viable cells (data not shown). Buoyant density experiments were also performed with C. coli and C. lari (data not shown) and resulted in patterns comparable to Fig. 1B and C, with variations between 1.109 to 1.078 g/ml and 1.102 to 1.076 g/ml, respectively. Therefore, viable Campylobacter cells were observed to have variations in buoyant density between 1.065 and 1.109 g/ml. Analysis of the undefined BGF resulted in three main populations with buoyant densities of 1.048 to 1.049 g/ml, 1.060 to 1.063 g/ml, and 1.082 to 1.083 g/ml. Finally, chicken rinse particles concentrated mainly around a buoyant density of 1.024 to 1.030 g/ml, with two smaller fractions at densities of 1.096 and above 1.108 g/ml.
FIG. 1.
Changes in buoyant densities of three C. jejuni strains during a period of 14 days. (A) C. jejuni CCUG 10937; (B) C. jejuni ATCC 33291; (C) C. jejuni ATCC 29428. ▪, cell counts observed over time (y axis). The vertical lines represent the buoyant density window (z axis). *, faint bands due to low cell densities. The horizontal dotted lines indicate the limits between the two upper flotation solutions. To concentrate the target at the Campylobacter recovery location, the density has to fall between these two marked densities.
Flotation method.
Based on the obtained buoyant densities for BGF, chicken rinse, and Campylobacter, a flotation setup was designed (Fig. 2). The intention of the setup was to include all viable campylobacter (Fig. 1) and to exclude as much of the chicken rinse sample components and the BGF as possible from the Campylobacter recovery location. Running samples containing all three components in the setup showed that fractions of the BGF and the chicken rinse with the lowest buoyant densities were found on the top of the density gradient. The fraction of the chicken rinse with the highest buoyant densities was mainly found at the lower interface, whereas the middle fractions of both BGF and chicken rinse could not be separated from the Campylobacter recovery location. Sampling followed by viable counts and CTC staining in the top layer and the interface of the two lowest layers revealed no viable (including VNC) Campylobacter cells. Finally, the presence of PCR inhibition caused by the sample was evaluated by comparing PCR efficiencies. Results showed that DNA in chicken rinse samples after flotation resulted in an amplification efficiency (0.89 ± 0.04) similar to purified DNA in water (0.87 ± 0.01), whereas DNA in untreated chicken samples showed a significantly lower efficiency (0.67 ± 0.12).
Quantification of viable and VNC Campylobacter cells.
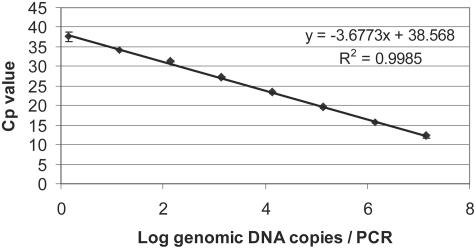
In order to have a highly specific quantitative PCR assay compatible for use with the LightCycler real-time PCR machines, a set of hybridization probes was developed for an already published primer pair. The linear range of amplification for the developed method was between 1.39 and 1.39 × 107 genomic DNA copies per PCR (Fig. 3). The amplification efficiency was calculated from the slope over the linear range of amplification and was 0.87 (R2 = 0.9985). To test the developed method on chicken samples, two experiments were set up. In the first experiment the detection of artificially and naturally contaminated samples with different populations and amounts of viable and dead cells was tested (Table 1). The first group of samples consisted of artificially spiked chicken skin rinses. The samples were tested both with plate counts and with real-time PCR after flotation. Results showed that cell concentrations from 2.6 × 107 CFU/ml to as low as 2.6 × 103 CFU/ml or 10 CFU/PCR mixture could be quantified after performing flotation and quantitative PCR. The cell counts obtained by plate counts and flotation followed by real-time PCR were of the same order of magnitude but in most cases were lower for the flotation. A second group of samples consisted of uninoculated samples that were purchased at local supermarkets and tested for the presence of Campylobacter. After analysis by selective plate counts and flotation combined with quantitative PCR, Campylobacter spp. were detected in only 3 out of 30 samples. Of those samples, two samples had cell counts that fell within the linear range of amplification and could be correctly amplified, whereas one sample had a lower cell number (<2.6 × 103 CFU/ml). Concentrations below the linear range of amplification can still be identified as positive (as was confirmed by melting curve analysis). These results are considered to be semiquantitative, and in this case cell numbers were estimated by extrapolation of the standard curve.
FIG. 3.
Standard curve for hybridization probe Campylobacter real-time PCR assay. Data are the results from duplicate analyses.
TABLE 1.
Quantification of Campylobacter spp. in artificially and naturally contaminated chicken samples
| Sample type | Cell concentration ± SDe (CFU/ml), as determined by:
|
|
|---|---|---|
| Selective plate countsa | Flotation and real-time PCRb | |
| Spiked | 2.6 × 107 ± 5.6 × 106 | 1.3 × 107 ± 1.5 × 106 |
| 2.6 × 106 ± 5.6 × 105 | 1.4 × 106 ± 1.3 × 105 | |
| 2.6 × 105 ± 5.6 × 104 | 1.9 × 105 ± 3.3 × 104 | |
| 2.6 × 104 ± 5.6 × 103 | 1.9 × 104 ± 0.8 × 103 | |
| 2.6 × 103 ± 5.6 × 102 | 3.5 × 103 ± 1.8 × 102 | |
| Naturally contaminated | 0.0c | 0.0c |
| 1.0 × 104 ± 7.1 × 102 | 1.3 × 104 ± 1.2 × 103 | |
| 2.8 × 106 ± 4.2 × 105 | 1.1 × 106 ± 4.1 × 105 | |
| 8.6 × 102 ± 8.5 × 101 | 2.8 × 102 ± 0.8 × 102d | |
Chicken rinse samples were plated on Preston agar and counted to acquire initial cell counts.
Real-time PCR measurements were calculated from a standard curve made for cells instead of DNA, since no DNA was purified after flotation. The standard curve had a linear range of amplification from 2.6 × 107 until 2.6 × 103 and was described by the equation y = −3.673x + 46.285 (R2 = 0.9944).
A total of 27 unspiked samples did not contain any Campylobacter spp. as determined by both methods.
This sample fell outside the linear range of amplification but was quantified by extrapolation of the standard curve.
Data are from independent triplicate measurements.
In the second experiment, samples containing different amounts of VNC and dead cells were analyzed. After a period of 3 weeks, the presence of VNC cells (as determined by negative plate counts and presence of metabolically active cells as shown by CTC staining) was detected for only one C. jejuni strain tested (CCUG 10937). By incubating the strain at different temperatures, different proportions of VNC and dead cells were obtained. Quantifying the different mixes after flotation by real-time PCR showed that the percentages of cells recovered at the standard Campylobacter recovery location after flotation resembled the amounts of VNC cells in the sample (Table 2).
TABLE 2.
Quantification of VNC C. jejuni after flotation, by real-time PCRe
| Samplea | VNC cell amount (% of total mix of dead and VNC cells) as determined by CTC stainingb | No. of viable cells as determined by flotation and real-time PCRc |
|---|---|---|
| 4°C | 23 ± 4 | 20 ± 9 |
| 20°C | 10 ± 2 | 11 ± 4 |
| 42°C | <1 | —d |
Campylobacter cultures were kept at 4°C, 20°C, or 42°C for 21 days. Viable counts proved that culturable cells were not present in the three cultures.
Percentage of metabolically active cells (VNC cells) compared to dead cells as determined by CTC-DAPI staining.
After flotation, cells were recovered from the two interfaces (Fig. 3) and quantified using real-time PCR. Assuming that the cells recovered from both layers form the total amount of dead plus VNC cells (100%), relative amounts recovered per layer were calculated.
No PCR signal was obtained.
Data are from independent duplicate measurements.
DISCUSSION
Recent studies have shown the potential of flotation combined with real-time PCR detection, using Yersinia enterocolitica in meat juice from pork as a model system (26, 27). To design a flotation method for the detection of Campylobacter spp. in chicken rinse, it was necessary to study the buoyant densities of the target organisms, the chicken sample, and BGF present on the chicken. Most studies, which use buoyant density as a separating factor in sample treatment, have used exponentially growing cells or overnight cultures for density measurements (9, 10, 23, 26). Also, one study observed that no obvious changes in buoyant density of Escherichia coli cells could be noted during different growth phases (7). However, a recent study presented contrasting data showing that E. coli cells shifted to a higher buoyant density and a higher number of populations in the transition from exponential to stationary phase (14). To ensure capture of Campylobacter spp. in all growth phases, different Campylobacter strains were studied over a period of 14 days, and results showed that, in all cases, the buoyant densities of viable cells of the different strains under different conditions varied between 1.065 and 1.109 g/ml. Also, in most cases a pattern was seen where after several days a lowering in the density was observed. Although these data are in contrast with the increase in buoyant density of E.coli reported by Makinoshima et al. (14), they may be explained by earlier studies in which Tholozan et al. described that C. jejuni cells after periods of 15 days and 30 days entered the VNC state with a concomitant increase in cell volume due to the uptake of water (24). This possible uptake of water can explain a decrease in the buoyant cell density. Furthermore, in Fig. 1A a split into two populations with different densities was observed. Results indicated that no viable cells were found in the population with the highest density. These results agree with a previous study showing an increase in buoyant density after cell death (10). Finally, because the observed densities for viable cells of all strains varied between 1.065 and 1.109 g/ml, these limits were chosen for the densities in the flotation setup (Fig. 1). Therefore, viable cells of all strains at all time points should be collected at the Campylobacter recovery location.
Because of the relatively large density window chosen for the flotation setup, it was not possible to avoid concentration of some BGF and chicken particles at the Campylobacter recovery location. Still, no PCR inhibition was observed, which can be explained by the use of the alternative DNA polymerase Tth. Previous studies have already shown that when using this enzyme in real-time PCR, a lower sensitivity towards PCR inhibitors was observed (11, 13).
To optimize possibilities for quantification, fluorescent hybridization probes were designed with an existing primer pair. The specificity of the assay was not extensively tested again, as it is based on specific primers, previously validated against 150 related and nonrelated species (12). The use of fluorescent probes was selected due to an increased sensitivity compared to the use of SYBR Green and to allow possible combination with simultaneous detection of an internal control. With a linear range of amplification stretching from 1.36 genomic copies to 1.36 × 107, this assay was shown to be as sensitive or up to 1 log unit more sensitive than previously published real-time probe-based PCR assays, based on the same (5, 18) and other (4, 8) primer sets.
Finally, applying flotation and the developed real-time PCR assay to different types of samples showed that cell concentrations as low as 860 CFU/ml or 3.4 CFU/PCR mixture could be positively identified without culture enrichment. Also, it confirmed that flotation and real-time PCR combined methods can be applied to a wide range of cell concentrations. Currently, flotation is run without concentration, i.e., 1 ml of sample is subjected to flotation and 1 ml is recovered afterwards. For future applications where lower cell concentrations are expected, it is possible that larger sample volumes can be used in flotation and in determining target concentrations. Although the results between plate counts and flotation followed by real-time PCR were on the same order of magnitude, the samples subjected to flotation showed lower concentrations (on average, 75% of the plate counts). Previous flotation studies have shown that this is due to inefficient sampling from the target recovery location. However, the recovered fractions in this study are higher than previously reported for Y. enterocolitica (26).
Results on dead and VNC cells showed that false-positive results due to detection of DNA originating from dead cells were minimized, since no signals were recovered from the Campylobacter recovery location containing 99% dead cells (Table 2). This is in agreement with previous flotation studies, which have shown that due to the low flotation speed of DNA it does not reach the target recovery location (26, 27). Furthermore, results indicated that VNC cells are detected together in the same density window with viable cells after flotation and real-time PCR. This was the original aim of the procedure, since previous studies indicated that VNC cells are still infectious (22). In the present study it was impossible to obtain fractions of VNC cells large enough to allow visual buoyant density determinations. Nonetheless, it seems possible that, keeping in mind results by Tholozan et al. concerning uptake of water by VNC cells (24) and the observed lowering in buoyant density in most strains after 14 days (Fig. 1), a lower buoyant density could indicate a transition into the VNC state. This suggests that it might be possible to separate VNC and viable cells in the future.
In conclusion, we have developed a new flotation technique for isolation of C. coli, C. jejuni, and C. lari in chicken rinse to be used prior to quantitative PCR, and the combined methods could detect Campylobacter spp. as low as 8.6 × 102 CFU/ml without culture enrichment. The method reduced PCR inhibition by BGF and chicken rinse samples and was able to minimize the risk of false-positive results due to detection of DNA from dead cells. Also, results indicated that VNC cells were recovered with viable cells and, therefore, false-negative results were avoided. Finally, the results emphasized the importance of studies concerning changes in buoyant density in different growth phases, especially when buoyant density is used for separation of target cells in sample treatment.
Acknowledgments
We thank Stefano Rodolfi for excellent technical assistance.
This work was financially supported by the Commission of the European Communities within the program “Quality of Life and Management of Living Resources,” QLK1-1999-00226, the Nordic Innovation Centre, and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, 2001-4068.
REFERENCES
- 1.Allos, B. M., and M. J. Blaser. 1995. Campylobacter jejuni and the expanding spectrum of related infections. Clin. Infect. Dis. 20:1092-1099. [DOI] [PubMed] [Google Scholar]
- 2.Cappelier, J. M., B. Lazaro, A. Rossero, A. Fernandez-Astorga, and M. Federighi. 1997. Double staining (CTC-DAPI) for detection and enumeration of viable but non-culturable Campylobacter jejuni cells. Vet. Res. 28:547-555. [PubMed] [Google Scholar]
- 3.Hald, B., K. Knudsen, P. Lind, and M. Madsen. 2001. Study of the infectivity of saline-stored Campylobacter jejuni for day-old chicks. Appl. Environ. Microbiol. 67:2388-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inglis, G. D., and L. D. Kalischuk. 2004. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time quantitative PCR. Appl. Environ. Microbiol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Josefsen, M. H., N. R. Jacobsen, and J. Hoorfar. 2004. Enrichment followed by quantitative PCR both for rapid detection and as a tool for quantitative risk assessment of food-borne thermotolerant campylobacters. Appl. Environ. Microbiol. 70:3588-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 7.Kubitschek, H. E., W. W. Baldwin, and R. Graetzer. 1983. Buoyant density constancy during the cell cycle of Escherichia coli. J. Bacteriol. 155: 1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaGier, M. J., L. A. Joseph, T. V. Passaretti, K. A. Musser, and N. M. Cirino. 2004. A real-time multiplexed PCR assay for rapid detection and differentiation of Campylobacter jejuni and Campylobacter coli. Mol. Cell. Probes 18:275-282. [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist, R. 1999. Detection of Shigella spp. in food with a nested PCR method-sensitivity and performance compared with a conventional culture method. J. Appl. Microbiol. 86:971-978. [DOI] [PubMed] [Google Scholar]
- 10.Lindqvist, R., B. Norling, and S. T. Lambertz. 1997. A rapid sample preparation method for PCR detection of food pathogens based on buoyant density centrifugation. Lett. Appl. Microbiol. 24:306-310. [DOI] [PubMed] [Google Scholar]
- 11.Löfström, C., R. Knutsson, C. E. Axelsson, and P. Rådström. 2004. Rapid and specific detection of Salmonella spp. in animal feed samples by PCR after culture enrichment. Appl. Environ. Microbiol. 70:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lübeck, P. S., N. Cook, M. Wagner, P. Fach, and J. Hoorfar. 2003. Toward an international standard for PCR-based detection of food-borne thermotolerant campylobacters: validation in a multicenter collaborative trial. Appl. Environ. Microbiol. 69:5670-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lübeck, P. S., P. Wolffs, S. L. W. On, P. Ahrens, P. Rådström, and J. Hoorfar. 2003. Towards an international standard for PCR-based detection of food-borne thermotolerant campylobacters: assay development and analytical validation. Appl. Environ. Microbiol. 69:5664-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makinoshima, H., A. Nishimura, and A. Ishihama. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269-279. [DOI] [PubMed] [Google Scholar]
- 15.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medema, G. J., F. M. Schets, A. W. van de Giessen, and A. H. Havelaar. 1992. Lack of colonization of 1 day old chicks by viable, non-culturable Campylobacter jejuni. J. Appl. Bacteriol. 72:512-516. [DOI] [PubMed] [Google Scholar]
- 17.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perelle, S., M. Josefsen, J. Hoorfar, F. Dilasser, J. Grout, and P. Fach. 2004. A LightCycler real-time PCR hybridization probe assay for detecting food-borne thermophilic Campylobacter. Mol. Cell. Probes 18:321-327. [DOI] [PubMed] [Google Scholar]
- 19.Pertoft, H. 2000. Fractionation of cells and subcellular particles with Percoll. J. Biochem. Biophys. Methods 44:1-30. [DOI] [PubMed] [Google Scholar]
- 20.Rollins, D. M., and R. R. Colwell. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheu, P., K. Berghof, and U. Stahl. 1998. Detection of pathogenic and spoilage micro-organisms in food with the polymerase chain reaction. Food Microbiol. 15:13-31. [Google Scholar]
- 22.Stern, N. J. 1994. Mucosal competitive exclusion to diminish colonization of chickens by Campylobacter jejuni. Poultry Sci. 73:402-407. [DOI] [PubMed] [Google Scholar]
- 23.Thisted-Lambertz, S., R. Lindqvist, A. Ballagi-Pordany, and M. L. Danielsson-Tham. 2000. A combined culture and PCR method for detection of pathogenic Yersinia enterocolitica in food. Int. J. Food Microbiol. 57:63-73. [Google Scholar]
- 24.Tholozan, J. L., J. M. Cappelier, J. P. Tissier, G. Delattre, and M. Federighi. 1999. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 65:1110-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winters, D. K., A. E. O'Leary, and M. F. Slavik. 1997. Rapid PCR with nested primers for direct detection of Campylobacter jejuni in chicken washes. Mol. Cell. Probes 11:267-271. [DOI] [PubMed] [Google Scholar]
- 26.Wolffs, P., B. Norling, and P. Rådström. 2004. Rapid quantification of Yersinia enterocolitica in pork samples by a novel sample preparation method, flotation, prior to real-time PCR. J. Clin. Microbiol. 42:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolffs, P., B. Norling, and P. Rådström. 2005. Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Methods 60:315-323. [DOI] [PubMed] [Google Scholar]